ABSTRACT
From September to December 2011, 162 New England harbor seals died in an outbreak of pneumonia. Sequence analysis of postmortem samples revealed the presence of an avian H3N8 influenza A virus, similar to a virus circulating in North American waterfowl since at least 2002 but with mutations that indicate recent adaption to mammalian hosts. These include a D701N mutation in the viral PB2 protein, previously reported in highly pathogenic H5N1 avian influenza viruses infecting people. Lectin staining and agglutination assays indicated the presence of the avian-preferred SAα-2,3 and mammalian SAα-2,6 receptors in seal respiratory tract, and the ability of the virus to agglutinate erythrocytes bearing either the SAα-2,3 or the SAα-2,6 receptor. The emergence of this A/harbor seal/Massachusetts/1/2011 virus may herald the appearance of an H3N8 influenza clade with potential for persistence and cross-species transmission.
IMPORTANCE
The emergence of new strains of influenza virus is always of great public concern, especially when the infection of a new mammalian host has the potential to result in a widespread outbreak of disease. Here we report the emergence of an avian influenza virus (H3N8) in New England harbor seals which caused an outbreak of pneumonia and contributed to a U.S. federally recognized unusual mortality event (UME). This outbreak is particularly significant, not only because of the disease it caused in seals but also because the virus has naturally acquired mutations that are known to increase transmissibility and virulence in mammals. Monitoring the spillover and adaptation of avian viruses in mammalian species is critically important if we are to understand the factors that lead to both epizootic and zoonotic emergence.
Introduction
Fatal pulmonary epizootics of influenza have been observed previously in seal populations, including outbreaks of H7N7 in 1979 to 1980 (1, 2), H4N5 in 1983 (3) and H4N5 and H3N3 in 1991 to 1992 (4). Such outbreaks are significant not just because of the detriment they pose to animal health but because influenza in mammals can act as a source for human pandemics (5).
In a <4-month period beginning in September 2011, 162 harbor seals (Phoca vitulina) were found dead or moribund along the New England coast. This number is approximately four times the expected mortality for this period. Most of the affected individuals were less than 6 months old, and common causes of death (including malnourishment) were ruled out. Five of the affected animals were investigated to identify a causative agent, and here we demonstrate that avian influenza virus subtype H3N8 was responsible for the observed clinical and pathological signs in these animals. Unlike any previous outbreak in seals, this H3N8 virus has naturally acquired mutations that reflect adaptation to mammalian hosts and that are known to increase virulence and transmissibility in avian H5N1 viruses infecting mammals. The virus has further acquired the ability to use the SAα-2,6 receptor commonly found in the respiratory tracts of mammals, including humans. The existence of a transmissible and pathogenic influenza is of obvious public concern.
RESULTS AND DISCUSSION
Five animals were submitted for anatomical and microbiological analysis. All were collected from the peak of the outbreak (late September to October) and had pneumonia and ulcerations of the skin and oral mucosa (Fig. 1). Nucleic acids extracted from lung, trachea, liver, kidney, thoracic lymph node, mesenteric lymph node, spleen, skin lesion, and oral mucosa were tested by PCR for the presence of a wide range of pathogens, including herpesviruses, poxviruses, adenoviruses, polyomaviruses, caliciviruses, paramyxoviruses, astroviruses, enteroviruses, flaviviruses, rhabdoviruses, orbiviruses, and influenza viruses. Influenza A virus was detected in several tissues from all five animals, and PCR cloning and sequencing of genes for hemagglutinin (HA) and neuraminidase (NA) revealed the subtype to be H3N8, a subtype typically associated with infection of avian, equine, and canine hosts (6-8). Influenza virus was isolated from the allantoic fluid of specific-pathogen-free (SPF) eggs inoculated with homogenates of PCR-positive tissues, including lung, lymph nodes, tonsil, and kidney, and all isolates were reconfirmed to be H3N8. In accord with conventional nomenclature, the virus is provisionally named A/harbor seal/Massachusetts/1/2011.
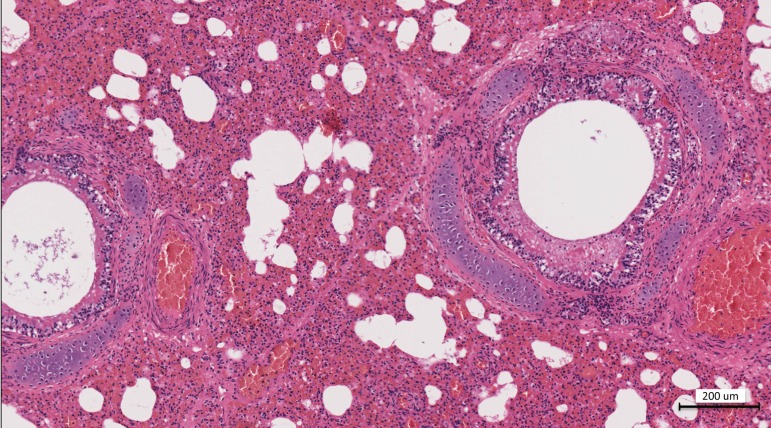
FIG 1 .
Hematoxylin and eosin staining of the lung at a ×10 magnification. There is diffuse acute interstitial pneumonia and a mix of acute hemorrhagic alveolitis with necrotizing bronchitis. Multifocally, alveoli are either filled with hemorrhage and scant inflammatory cells or expanded with emphysema. There is irregularity to the bronchial mucosa due to necrosis, a mild to moderate edema, and mucous partially filling the bronchial lumen. There is mild to moderate expansion of the interlobular septa, with edema and hemorrhage.
In situ hybridization (ISH) using oligonucleotide probes for influenza virus H3N8 segments 4 (HA) and 7 (matrix) and immunohistochemistry using polyclonal antibodies against H3N8 HA antigen confirmed the presence of influenza virus in lung, where signal was concentrated in the bronchiole epithelium and mucosa of the pulmonary parenchyma (Fig. 2 and see Fig. S1 in the supplemental material). The average load of HA and NA RNAs in lung was 300 copies/100 ng of extracted RNA. ISH staining in nonrespiratory tissues was limited to sporadic infection of single cells in intestine, kidney, and lymph node, and the average HA/NA RNA load was five copies/100 ng. These results are consistent with the histological observation that the main site of viral replication is the respiratory tract. Cellular morphology and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) consistent with apoptosis was observed in virus-infected pulmonary epithelial cells (Fig. 3). TUNEL staining also revealed the widespread presence of apoptotic cells in areas where no virus was observed but not in negative-control tissue, suggesting an additional host-mediated response to the infection.
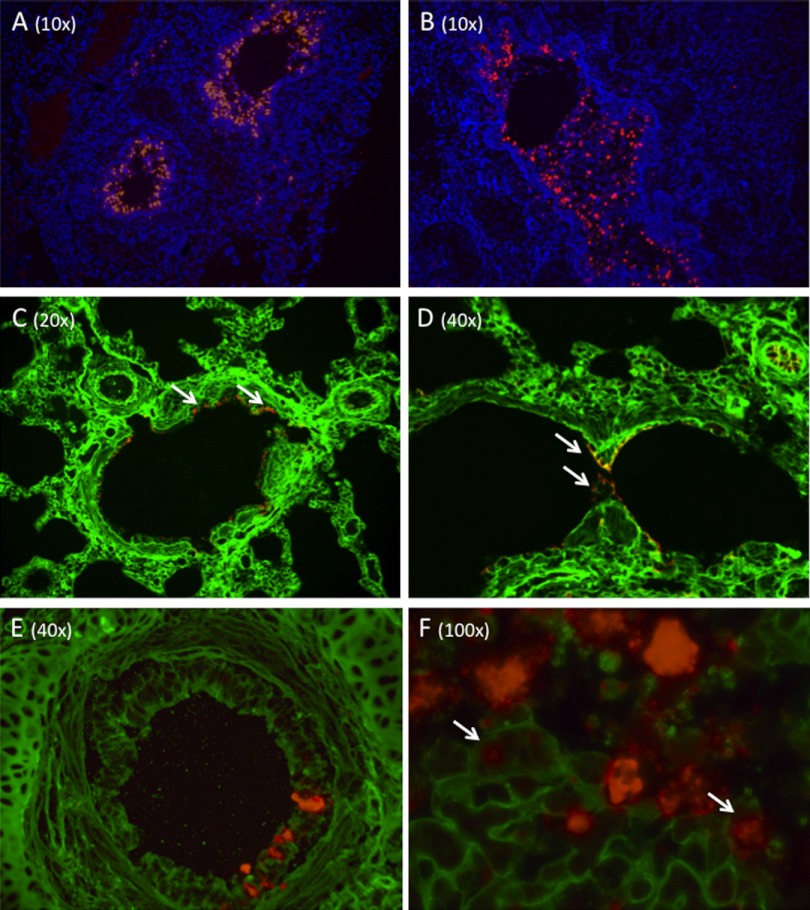
FIG 2 .
(A and B) Fluorescent in situ hybridization (ISH) of H3N8 virus-infected seal cells with DAPI counterstaining. A probe targeting the viral hemagglutinin demonstrates diffuse infection of the bronchial mucosal epithelium. ISH was also performed using probes for the matrix gene. Staining was identical to that shown here for HA. (C and D) Lectin staining to demonstrate the distribution of SAα-2,3 and SAα-2,6 in seal pulmonary parenchyma. The SAα-2,6 (green) was detected using fluorescein-labeled Sambucus nigra agglutinin (SNA) lectin, while SAα-2,3 (red) was detected using Maackia amurensis II (MAL II) lectin. Both infected and uninfected control tissues were stained, and the results were consistent for both. High levels of SAα-2,6 are observed on bronchiole and alveolar epithelial cells and on endothelial cells. The images in panels C and D were selected because they show staining for both sialic acids; however, the expression of SAα-2,3 was rarely observed (arrows) and limited to bronchiole luminal (C) and occasional alveolar (D) epithelia. (E and F) Costaining of SAα-2,6 and H3N8 HA. SAα-2,6 (green) is expressed on the respiratory epithelium of an intrapulmonary bronchus (E). H3N8 virus-infected cells (red) are present. A serial section was also stained for SAα-2,3, and none was detected. A high-magnification image of infected mucosa clearly shows H3N8 virus infection of cells expressing SAα-2,6 (arrows). All composite images are presented separately (single stains) in Fig. S1 to S3.
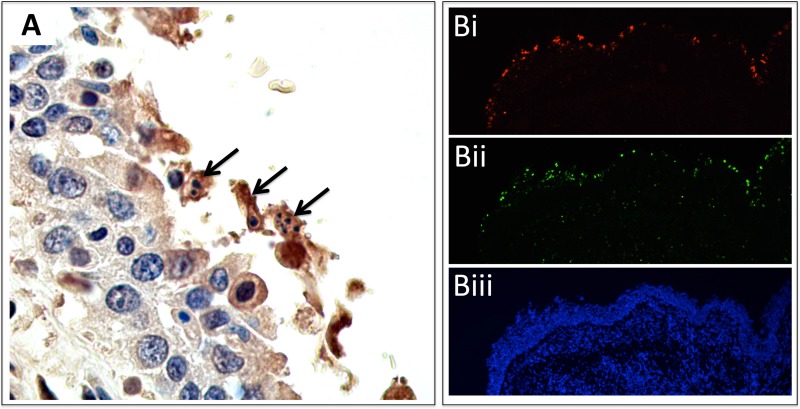
FIG 3 .
(A) Immunohistochemistry (IHC) of seal bronchus. Polyclonal antibodies were raised against H3N8 virus-specific HA antigen. Brown staining (DAB reporter system) indicates the presence of viral antigen. There is irregularity of the mucosal surface, with sloughed epithelium. IHC demonstrates the presence of viral antigen, pyknosis, and apoptosis (arrows). No viral antigen or apoptosis was seen on negative-control tissue. (Bi) ISH staining of H3N8 virus in lung epithelium (HA probe). (Bii) TUNEL staining (green) in the same region (serial section) of the lung, showing the presence of apoptotic cells. Comparison of virus and TUNEL staining shows localization of apoptosis to virally infected cells. (Biii) DAPI (with dihydrochloride) staining for cell nuclei.
Full-genome sequencing was completed following PCR cloning of all eight influenza genome segments directly from infected tissue. Sequences were submitted to GenBank and assigned accession numbers JQ433879 to JQ433882. Phylogenetic analyses of these nucleotide sequences with avian, canine, and equine H3N8 influenza virus genomes demonstrated the closest relationship to a virus identified in North American waterfowl in all 8 segments (Fig. 4). These data are consistent with the recent transmission of the H3N8 virus from wild birds to seals. The closest avian relative, A/blue-winged teal/Ohio/926/2002, had an overall 96.07% nucleotide sequence identity across the genome (Hamming distance), with no individual segment having less than 94% identity. This level of similarity across all segments with an isolate separated by a span of 10 years suggests that this virus has been circulating in the aquatic bird populations since at least 2002.
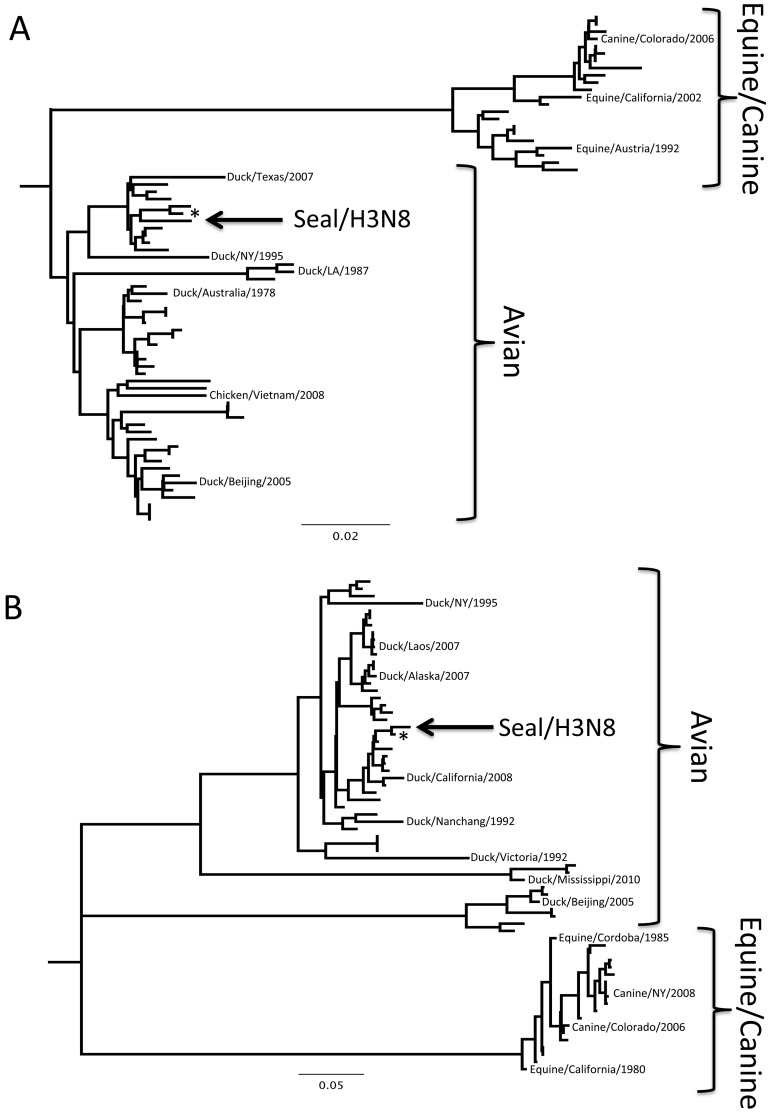
FIG 4 .
Phylogenetic trees of representative influenza H3N8 genome segments. Nucleotide sequence alignments for all genome segments were created using ClustalW, and trees were produced using neighbor-joining, maximum-likelihood, and Bayesian algorithms. Models of evolution were selected using ModelTest, and a tree was selected based on a consensus of the results of the three algorithms. Only the trees for HA and NA are shown; however, all eight segments showed strong association with sequences of avian origin. Trees are constructed with H3N8 viruses only, and published sequences were selected to represent variation in the year, host, and location of isolation. *, A/blue-winged teal/Ohio/926/2002; NY, New York; LA, Louisiana.
A total of 37 amino acid substitutions separate the seal H3N8 and avian H3N8 viruses, which are summarized in Table 1. The corresponding amino acids found in other seal influenza viruses, in the canine and equine H3N8 viruses, and in selected human influenza viruses are included for comparison. Of these, mutations PB2-701N and HA-260M are shared exclusively by the seal and mammalian (canine/equine) H3N8 viruses, while mutations NA-399R, PB2-382V, and PA-184N are shared by the seal H3N8 and human H3N2 viruses, all of which suggests adaptation to mammalian hosts. Mutations PB2-60N, PB2-376R, PB1-174I, PB1-309G, PB1-359G, PB1-376V, PB1-377G, PB1-464E, NP-63V, NP-128G, and NP-296H are all exclusively found in the seal H3N8 virus (Table 1). Future studies will be required to assess the functional significance of many of these mutations, especially in seal H3N8 PB1, where a significant number of exclusive mutations were observed. Given the importance of PB1 in viral replication, it is probable that these mutations represent adaptive selection to accommodate host-specific differences in intracellular replication.
TABLE 1 .
List of amino acid substitutions between seal H3N8 and avian H3N8 virusesa
| Segment (protein) | nt position |
aa position |
Amino acid substitution |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Seal H3N8 (2011) |
Avian H3N8 |
Seal H7N7 (1980) |
Seal H4N5 (1982) |
Seal H3N3 (1992) |
Equine H3N8 |
Canine H3N8 |
Human H3N2 |
Human H1N1 (seasonal) |
Human H1N1 (pandemic) |
|||
| 1 (PB2) | 178 | 60 | N | D | D | D | D | D | D | D | D | D |
| 441 | 147 | M | I | I | I | I | V | V | I | I | T | |
| 1127 | 376 | R | K | K | K | K | K | K | K | K | K | |
| 1144 | 382 | V | I | I | I | I | I | I | V | I | I | |
| 2101 | 701 | N | D | D | D | D | N | N | D | D | D | |
| 2 (PB1) | 522 | 174 | I | M | M | M | M | M | M | M | M | M |
| 925 | 309 | G | W | W | W | W | W | W | W | W | W | |
| 1075 | 359 | G | S | S | S | S | S | S | S | S | S | |
| 1126 | 376 | V | I | I | I | I | I | I | I | I | I | |
| 1130 | 377 | G | D | D | D | D | D | D | D | D | D | |
| 1392 | 464 | E | D | D | D | D | D | D | D | D | D | |
| 3 (PA) | 253 | 85 | A | T | T | T | T | T | T | T | T | I |
| 551 | 184 | N | S | S | S | S | S | S | N | S | S | |
| 794 | 265 | L | P | P | T | P | P | P | P | P | P | |
| 4 (HA) | 242 | 81 (65) | K | T | G | D | T | T | T | T | S | N |
| 323 | 108 (92) | S | N | E | T | N | S | N | K | N | S | |
| 329 | 110 (94) | S | F | S | V | F | F | F | Y | E | D | |
| 452 | 151 (135) | E | G | A | K | G | R | R | T | V | V | |
| 527 | 176 (160) | V | A | A | A | A | S | S | K | L | S | |
| 713 | 238 (222) | L | W | Q | W | W | W | L | R | K | K | |
| 778 | 260 (244) | M | V | T | V | V | M | M | L | I | T | |
| 802 | 268 (252) | V | I | I | I | I | V | V | I | I | V | |
| 859 | 287 (271) | N | D | D | A | D | D | D | D | N | D | |
| 1114 | 372 | K | Q | Q | Q | Q | Q | Q | Q | Q | Q | |
| 1247 | 416 | L | S | T | E | S | S | S | S | N | N | |
| 5 (NP) | 187 | 63 | V | I | I | I | I | I | I | I | I | I |
| 383 | 128 | G | D | D | D | D | D | D | D | D | D | |
| 886 | 296 | H | Y | Y | Y | Y | Y | Y | Y | Y | Y | |
| 1336 | 446 | G | R | R | R | R | R | R | R | K | R | |
| 6 (NA) | 440 | 147 | E | V | I | None | I | V | I | V | V | I |
| 849 | 283 | D | E | N | None | D | E | E | Y | T | S | |
| 937 | 313 | R | G | Q | None | G | G | G | S | G | G | |
| 958 | 320 | S | P | L | None | H | P | P | V | F | F | |
| 1186 | 396 | D | N | N | None | N | N | D | R | I | I | |
| 1195 | 399 | R | W | W | None | W | W | W | R | W | W | |
| 1295 | 432 | A | E | A | None | N | E | E | E | R | K | |
| 8 (NS1/NS2) | 263 | 88 | H | R | R | R | R | R | R | R | R | R |
A total of 40 amino acid substitutions were observed in a comparison of seal H3N8 virus with avian H3N8 virus. Sequences of other seal influenza viruses, canine and equine H3N8 viruses, and selected human influenza viruses were included for comparison. Amino acid positions presented in parentheses represent corresponding H3 numbering. None, no sequence available for comparison; nt, nucleotide; aa, amino acid.
The seal H3N8 genome was interrogated for any genetic features that might contribute to enhanced transmissibility and virulence in seals. Expression of a second peptide (PB1-F2) from segment 2 has been associated with an increase in pathogenicity by inducing apoptosis and increasing both inflammation and secondary bacterial pneumonia (9, 10). The seal H3N8 virus contains an intact open reading frame for the pathogenic version of this accessory protein, which includes a serine at amino acid position 66 (9). All five seals had evidence of apoptosis and secondary bacterial pneumonia.
Glycosylation can also affect pathogenicity in influenza viruses (11-14). Six potential glycosylation sites were detected in the seal H3N8 HA, based on the sequence X−2X−1NX(S/T) X+1, at amino acid positions 24, 38, 54, 181, 301, and 499. None had features suggestive of inactivity or reduced efficiency, as was previously demonstrated for H5N2 (12). Many H5N1 viruses have an additional glycosylation site at positions 158 to 160, and previous work demonstrated that the deletion of this site is critical for H5N1 viruses to bind to human-SAα2,6-like receptors and to transmit between mammals (15). This glycosylation site is missing in the seal H3N8 HA.
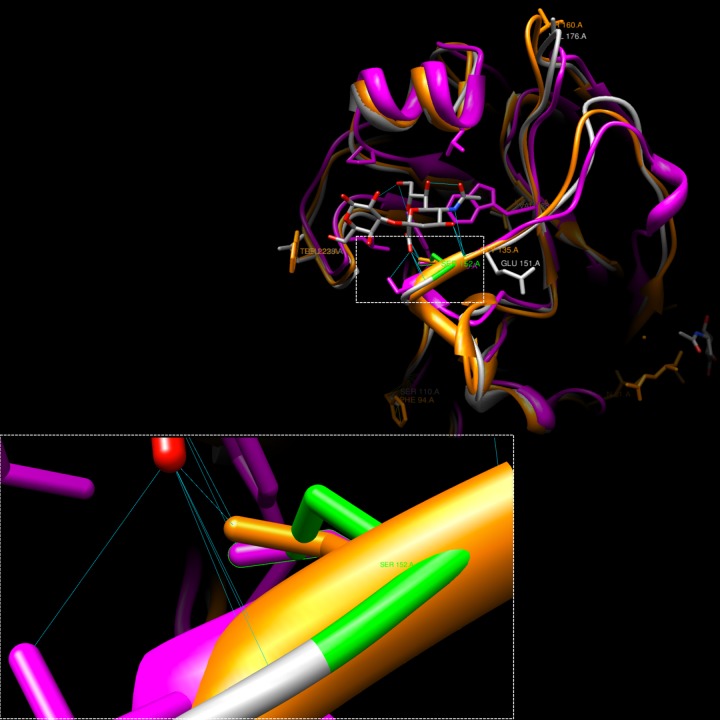
In order to investigate the specificity of sialic acid binding, the seal virus was compared to avian H5 and swine H9, both of which bind to the sialyloligosaccharide SAα2,3 ligand in a structural-homology model (Fig. 5). All structures confirmed the presence of a highly conserved serine at position 152 (corresponding to S136 by H3 numbering [16, 17]), which lies in a binding pocket, where its hydroxyl group contacts the axial carboxylate of sialic acid (17). While the seal virus contains the same conserved S152, it also harbors a neighboring G151E mutation (Table 1), which introduces a large residue capable of both donating and receiving hydrogen bonds with residues in close proximity to the ligand-binding pocket. Rotamer hydrogen bond analysis of the modeled seal structure indicates that HA’s altered conformation results in reduced hydrogen bonding between the conserved serine and SAα2,3 compared to that of H5 and H9 influenza viruses. Such changes in sialic acid binding play important roles in novel host adaptation (18).
FIG 5 .
Structural-homology model showing the interaction of influenza HA with the SAα2,3 ligand. Seal H3 (gray), avian H5 (orange) (Protein Data Bank [PDB] accession number 1JSO), and swine H9 (pink) (PDB accession number 1JSH) were compared. The mutation G151E causes a conformational shift and interrupts H bonding between seal H3 S152 and SAα2,3, which suggests a reduction in SAα2,3 binding efficiency. A lost H bond in seal H3 is depicted in green.
Mutations at positions 226 and 228 (H3 numbering) in the H3 HA can also affect receptor-binding preferences and can either completely abrogate (Q226L) or reduce (G228S) affinity for the avian-preferred SAα-2,3 interaction (18, 19). Seal H3N8 virus maintains the avian phenotype at positions 226 (Q) and 228 (G), which correlates with a continued ability to use SAα-2,3. Together, these findings suggest that the seal virus may still be able to use SAα2,3, but perhaps with less efficiency than in its original avian host. Given this, we investigated whether seal H3N8 may have adapted and acquired an additional or increased affinity for the SAα-2,6 receptors that are more prevalent in mammalian respiratory tissue (20-23).
The pulmonary distribution of SAα-2,3 and SAα-2,6 influenza receptors was investigated using the receptor-specific lectins Sambucus nigra agglutinin (SNA) for SAα-2,6 and Maackia amurensis lectin II (MAL II) for SAα-2,3. SAα-2,6 was widely expressed in both infected and noninfected pulmonary parenchyma, with the highest concentration seen on endothelial cells, followed by alveolar/bronchiole epithelia (Fig. 2 and see Fig. S2 in the supplemental material). SAα-2,3 was also observed, but less frequently, and was generally limited to the luminal surfaces of epithelial cells of the bronchioles. This broadly agrees with the expression of these SA saccharides in humans and pigs (20-23) and demonstrates that seals do express receptors that would allow avian viruses to initiate infection. This observation is supported by the H3N3 seal virus from the 1991 epizootic (4), which was shown to preferentially bind SAα-2,3 in vitro (19). However, the limited prevalence of SAα-2,3 in the lower lung suggests that the process of infection is inefficient and may help to explain why epizootics of avian influenza occur but are infrequent in harbor seals.
Importantly, the rare expression of SAα-2,3 is insufficient to explain the diffuse infection seen throughout the pulmonary parenchyma (Fig. 2). In contrast, the wide distribution of SAα-2,6 is far more consistent with the level of infection observed. Costaining of infected lung with viral HA and SAα-2,3 or SAα-2,6 demonstrated clear infection of SAα-2,6-positive cells, in which no SAα-2,3 was seen (Fig. 2 and S3).
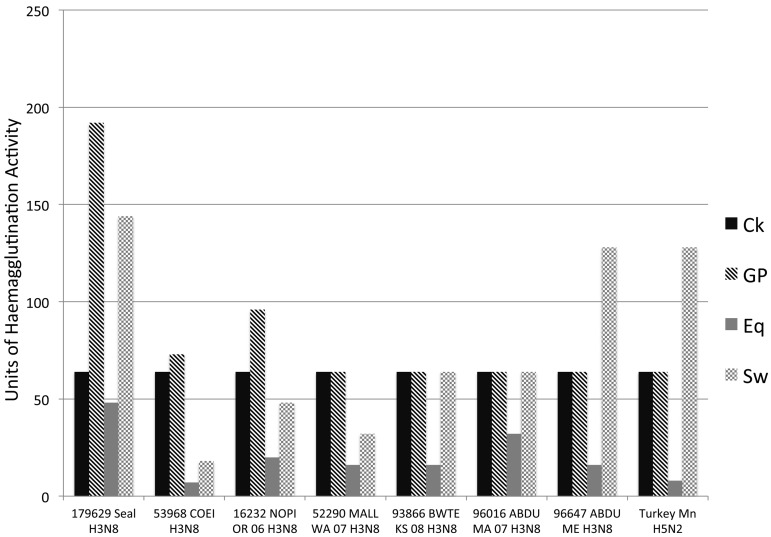
Hemagglutination assays were also performed to confirm sialic acid binding preferences. Seal H3N8 isolates were first sequenced to confirm that passage in eggs had not altered the HA phenotype detected in the infected tissues, and the viruses were then tested for their ability to agglutinate erythrocytes that preferentially express SAα-2,3 (horse) or SAα-2,6 (guinea pig, pig) (24, 25), relative to several avian H3N8 viruses (Fig. 6). Average agglutination titers for seal H3N8 virus with horse erythrocytes (1:48) show that the virus can still bind to SAα-2,3. However, titers were appreciably higher with guinea pig (1:192) and pig (1:144) erythrocytes, demonstrating a preference for SAα-2,6. These findings show that seal H3N8 can use both avian and mammalian receptors and add to previous studies that have demonstrated changes in receptor preferences following a host switch event (26). The patterns of SAα-2,3 and SAα-2,6 binding to seal H3N8 virus also agree with the patterns observed for H3 avian viruses adapting to humans (18).
FIG 6 .
Hemagglutination assays were performed on an isolate of the H3N8 seal virus (A/harbor seal/New Hampshire/179629/2011) to confirm sialic acid binding preferences. Viruses were tested for their ability to agglutinate erythrocytes that preferentially express SAα-2,3 (horse) or SAα-2,6 (guinea pig, pig). Average agglutination titers for seal H3N8 with horse erythrocytes (1:48) show that the virus can still bind to SAα-2,3, though weakly. Titers were appreciably higher with guinea pig (1:192) and pig (1:144) erythrocytes, demonstrating a preference for SAα-2,6. Given that horse erythrocytes express SAα-2,3, it is interesting that the avian H3N8 viruses did not agglutinate with horse RBCs (red blood cells) efficiently, even following repeated attempts. Ito et al. (25) showed that avian H3N8 viruses from Asia in the early 1980s could bind to horse RBCs, while Wiriyarat et al. (43) gave examples of avian viruses (albeit not H3 viruses) that did not bind to horse RBCs. It is not known whether the avian viruses included here simply have a preference for the N-acetyl (NeuAc) sialic acid species, which is not found on horse RBCs, while the seal virus uses N-glycolyl (NeuGc) SAα-2,3. Ck, chicken; GP, guinea pig; Eq, equine; Sw, swine; 179629 Seal H3N8, A/harbor seal/New Hampshire/179629/2011; 53968 COEI H3N8, A/common eider/Massachusetts/20507-001/2007 (H3N8) virus; 16232 NOPI OR 06 H3N8, A/northern pintail/Oregon/44249-547/2006 (H3N8) virus; 52290 MALL WA 07 H3N8, A/mallard/Washington/44338-052/2007 (H3N8) virus; 93866 BWTE KS 08 H3N8, A/blue-winged teal/Kansas/44440-003/2008 (H3N8) virus; 96016 ABDU MA 07 H3N8, A/American black duck/Maine/44411-174/2008 (H3N8) virus; 96647 ABDU ME H3N8, A/American black duck/Maine/44411-532/2008 (H3N8) virus; Turkey Mn H5N2, A/turkey/Minnesota/3689-1551/1981 (H5N2) virus.
A further mutation was observed in HA, this time at position 110 (Table 1). In avian H3 viruses, phenylalanine (Phe/F) is consistently seen, while seal H3N8 uses Ser (F110S). The significance of this (if any) is currently unknown; however, previous work has suggested that this amino acid (position 110) is a critical component of the influenza fusion peptide (27), and given the essential role of fusion in viral replication and the host-specific differences that presumably exist in this process, the F110S substitution may well represent further adaption of this virus to mammalian replication.
The ability of avian influenza viruses to adapt to SAα-2,6-mediated cell entry and replication is regarded as a significant driving force in the emergence of global pandemics (19, 28-30), especially for viruses with phenotypes that confer increased virulence. Such phenotypes are often, though not exclusively, dictated by mutations in segment 1 (PB2), which is an important determinant of host range for influenza viruses. Previous studies have experimentally demonstrated the effect of various PB2 substitutions on virulence and transmissibility in mammalian hosts (15, 31-38), including the modification of the aspartic acid (D) avian phenotype to an asparagine (N) mammalian phenotype at amino acid 701 (15, 31, 32, 36, 39, 40). This D701N mutation has been experimentally introduced into an adapted version of the H7N7 seal influenza virus isolated in 1982 (1, 2) and was shown to increase the pathogenicity of the virus to mice (32).
The seal H3N8 virus from the 2011 outbreak has naturally acquired this D701N substitution (Table 1), which was confirmed by clonal sequencing directly from infected tissue (50 clones/animal) to be the only phenotype present in all five animals. None of the previous outbreaks of influenza in seals showed this 701N phenotype, but it is consistently found in H3N8 viruses from horses and dogs, demonstrating further adaptation to replication in mammalian hosts. These observations raise significant concern about the virulence and transmission of this virus between mammals. Interestingly, analysis of HA sequences over the course of the outbreak show the introduction and maintenance of two nucleotide polymorphisms (Table 2), and while this is insufficient to convincingly demonstrate seal-to-seal transmission, it leads us to postulate that mammalian spread might already have occurred.
TABLE 2 .
Sequence analysis of the HA genes isolated from various tissuesa
| Animal | Sample | Date | Nucleotide at position: |
|
|---|---|---|---|---|
| 1347 | 1499 | |||
| 278-Pv | Kidney | 28 Sept 2011 | C | C |
| 286-Pv | Trachea | 29 Sept 2011 | C | C |
| Mes LN | C | C | ||
| Kidney | C | C | ||
| 295-Pv | Mes LN | 3 Oct 2011 | C | C |
| Lung | C | C | ||
| Tonsil | T | C | ||
| Kidney | T | A | ||
| Tonsil | 3 Oct 2011 | T | A | |
| Trachea | T | A | ||
| Cerv LN | T | A | ||
Two polymorphisms were observed in HA at positions 1347 and 1499, relative to avian H3N8 sequence CY041887. Isolates from animals earlier in the outbreak showed C at position 1347 and C at position 1499. The variations C1347T and C1499A were observed in animal 295-Pv, in addition to the wild-type sequence. Animal 294-Pv showed only the variant genotype.
Together, the adaptations observed in A/harbor seal/Massachusetts/1/11 suggest that it may be able to persist within the seal population and evolve into a new clade within the H3N8 group, as happened with the canine and equine viruses. An additional concern is the potential zoonotic threat that this virus poses, as it has already acquired mutations in both PB2 and HA that are often, though perhaps not exclusively, regarded as prerequisites for pandemic spread (19-23, 28, 30, 37) and it is uncertain how any persistence of the virus in mammals may continue to alter its phenotype. A comparison of A/harbor seal/Massachusetts/1/11 with human H3N2 viruses revealed three substitutions that are already common to both seal H3N8 and human H3N2 viruses. These are NA-W399R, PB2-I382V, and PA-S184N (Table 1). In all cases, these substitutions are shared by the seal H3N8 and human H3N2 viruses but are not found in influenza viruses isolated previously from seals, in avian, equine, or canine H3N8 viruses, or in either seasonal or pandemic H1N1 viruses. Further studies will be required to establish the functional significance of these substitutions; however, the natural epizootic emergence at this time of a pathogenic virus that can transmit between mammals, found in a species that can become infected with multiple influenza virus subtypes, must be considered a significant threat to both wildlife and public health.
MATERIALS AND METHODS
Extractions, PCR, and sequencing.
RNA was extracted from all tissues using Trizol reagent, and cDNA was synthesized using Superscript III (Invitrogen) according to the manufacturer’s instructions. PCR for the detection of influenza A virus was performed using primers FLUAV-M-U44 (GTCTTCTAACCGAGGTCGAAACG) and FLUAV-M-L287 (GCATTTTGGACAAAGCGTCTACG), to produce a 243-bp product of segment 7 (coding for matrix protein). For full-genome sequencing, full-length cDNAs were amplified for all eight influenza segments. Primers were designed to target terminal sequences for each segment, based on alignments of avian, canine, and equine H3N8 sequences from the Influenza Research Database (http://www.fludb.org). All PCRs were performed using fast-cycling chemistry (Qiagen), according to the manufacturer’s instructions. Amplified products were cloned into the pGEM T-easy vector (Promega) and sent for commercial sequencing.
Virus isolation and intravenous pathogenicity index test.
Homogenates from PCR-positive tissues were inoculated into SPF embryonated chicken eggs, and virus growth was determined by PCR. Tissue homogenates were also used to infect the Vero and MDCK cell lines in the presence of trypsin. Virus isolates were sent to the National Veterinary Services Laboratory (Ames, IA), where the chicken intravenous pathogenicity index test was performed according to the OIE manual (41).
Molecular pathology.
Fluorescent in situ hybridization (FISH) was performed using the Quantigene ViewRNA ISH tissue assay (Affymetrix), according to the manufacturer’s instructions. FISH conditions were optimized to include a 10-min boiling and 20-min protease treatment. Oligonucleotide probes were designed commercially by Affymetrix using sequences of HA and M (accession numbers JQ433879 and JQ433882, respectively). Immunohistochemistry (IHC) was performed by pretreating deparaffinized tissue sections with a 1:10 dilution of antigen retrieval solution (DAKO) for 20 min in a steamer. Samples were then washed three times in distilled water (dH2O), incubated in 3% hydrogen peroxide (in phosphate-buffered saline [PBS]) for 10 min, washed again twice in dH2O and once in PBS, and then blocked (10% normal goat serum, 0.1% bovine serum albumin [BSA]) for 20 min. Sections were treated with HA polyclonal H3N8 antibody (Novus Biologicals; catalogue number NBP1-46796) at a 1:250 dilution for 2 h at room temperature. Following three washes in PBS, sections were incubated in Signal Stain Boost IHC reagent (Cell Signaling; catalogue number 8112) for 30 min at room temperature. Sections were again washed three times in PBS, stained with 3,3-diaminobenzidine (DAB; Dako), and counterstained with hematoxylin. TUNEL staining was performed using the in situ cell death detection kit and fluorescein (Roche) with deparaffinization and protease treatment as described for the FISH protocol.
Simultaneous detection of SAα-2,3 and SAα-2,6 glycans.
Deparaffinized tissue sections (5 µM) were blocked with 1× Carbo-Free solution (Vector Laboratories; catalogue number SP-5040) for 1 h at room temperature. Sections were then stained for SAα-2,6 using fluorescein SNA (Vector Laboratories; catalogue number FL-1301) at 10 µg/ml for 30 min at room temperature, and rinsed twice for 3 min each time in PBS. Sections were then reblocked in 1× Carbo-Free solution for 30 min, before being stained for SAα-2,3 with 10 µg/ml of biotinylated MAL II (Vector Laboratories; catalogue number B-1265) for 30 min at room temperature. The MAL II was poured off, and Texas Red streptavidin (Vector Laboratories; catalogue number SA-5006) was laid over the sections at 10 µg/ml for a further 30 min. Sections were rinsed twice for 3 min each time in PBS and mounted with Vectashield hard-set mounting solution with DAPI (4′,6-diamidine-2-phenylindole) counterstaining.
Hemagglutination assays.
Hemagglutination assays were performed according to the WHO diagnostic manual (42). Briefly, red blood cells (RBCs) from rooster chicken, guinea pig, horse, and pig were obtained from Lampire Biological Laboratories (Ottsville, PA). The RBCs were washed in PBS and resuspended to 0.5% (chicken) or 0.75% (guinea pig and pig). Equine RBCs were resuspended to 1% in PBS with 0.5% BSA (43). Viruses were diluted to 64 HA units using chicken red blood cells. Serial dilutions were then made, added to equal volumes of washed RBCs of each species, and incubated in U-bottom plates, with the exception of chicken RBCs, which were incubated in V-bottom plates. Reaction mixtures were incubated at room temperature for 1 h, with the exception of those with chicken RBCs, which were incubated for 30 min. The HA titer endpoint is the reciprocal of the highest dilution which causes complete hemagglutination. The seal H3N8 virus was compared with several avian H3N8 isolates, including A/common eider/Massachusetts/20507-001/2007 (H3N8), A/northern pintail/Oregon/44249-547/2006 (H3N8), A/mallard/Washington/44338-052/2007 (H3N8), A/blue-winged teal/Kansas/44440-003/2008 (H3N8), A/American black duck/Maine/44411-174/2008 (H3N8), and A/American black duck/Maine/44411-532/2008 (H3N8). An H5N2 virus was also included: A/turkey/Minnesota/3689-1551/1981 (H5N2) virus.
Sequence analysis.
Nucleotide sequences were aligned using ClustalW. Phylogenetic trees were constructed using neighbor-joining, maximum-likelihood, and Bayesian algorithms. Models of evolution were selected using ModelTest, and a representative tree was selected based on a consensus of the results of the three algorithms. Published H3N8 sequences included in the analyses were selected to represent the diversity of year, host, and location of isolation.
Structural modeling.
To create a homology model of the seal 2012 outbreak HA sequence, 10 template models were selected based on their super-secondary structures, with use of the LOMETS meta-threading approach (44, 45). Continuous fragments excised from these templates were then reassembled into full-length models by replica exchange Monte Carlo simulations (44). Ab initio modeling of threaded unaligned regions was then used to complete the structure. Low free-energy states were subsequently identified through clustering of simulation decoys by the SPICKER near-native model selection algorithm (46). Chimera was utilized for structural analysis and visualization (47). Hydrogen bonding analysis was based on geometric criteria established through survey of small-molecule crystal systems and Dunbrack rotamer libraries (48, 49).
Nucleotide sequence accession numbers.
The sequences of all eight influenza genome segments were submitted to GenBank and assigned accession numbers JQ433879 to JQ433882.
SUPPLEMENTAL MATERIAL
Fluorescent ISH of seal H3N8 with DAPI counterstaining. Images are at a ×10 magnification. (A and Bi) A hemagglutinin (HA) probe demonstrates diffuse hemagglutination of bronchial mucosal epithelium. (A and Bii) DAPI binding demonstrates blue nuclear counterstaining of all cells. (A and Biii) Localization of the HA probe reaction to bronchial epithelial cells. ISH was also performed using probes for the matrix gene. Staining was identical to that shown here for HA. Download Figure S1, JPG file, 0.1 MB.
Distribution of SAα-2,3 and SAα-2,6 in seal pulmonary parenchyma. SAα-2,6 (green) was detected using fluorescein-labeled SNA lectin, while SAα-2,3 (red) was detected using Maackia amurensis II (MAL II) lectin. Both infected and uninfected control tissues were stained, and the results were consistent for both. High levels of SAα-2,6 were observed on bronchiole (Ai to -iii) and alveolar (Biiii) epithelial cells and on endothelial cells. The images in panels A and B were selected because they show staining for both sialic acids; however, the expression of SAα-2,3 was rarely observed and limited to bronchiole luminal (A) and occasional alveolar (B) epithelia. Download Figure S2, JPG file, 0.2 MB.
Costaining of SAα-2,6 and H3N8 HA. (Ai to -iii) SAα-2,6 is expressed on the respiratory epithelium of an intrapulmonary bronchus (green), and some cells are infected with H3N8 (red). (Bi to -iii) H3N8 virus infection of SAα-2,6-positive cells is clear. When the level of virus infection is low, SAα-2,6 is still present on the cell surface (arrow 1). As infection increases, SAα-2,6 is reduced, presumably cleaved by the viral NA (arrow 2). In heavily infected cells, SAα-2,6 is no longer detectable (arrow 3). These results demonstrate that the virus can infect SAα-2,6-positive cells. Staining for SAα-2,3 was also performed, but no expression was observed in these regions. Download Figure S3, JPG file, 0.2 MB.
ACKNOWLEDGMENTS
We acknowledge funding from the NIH: AI57158 (NBC-Lipkin), LM010140, and CA121852; NIH/NSF TW005769; USAID PREDICT; and DTRA.
We thank Nicole Arrigo for editing and Wendy Barclay and Jennifer Howard for comments on the manuscript. We thank Johnice Miller for technical assistance.
Footnotes
Citation Anthony SJ, et al. 2012. Emergence of fatal avian influenza in New England harbor seals. mBio 3(4):e00166-12. doi:10.1128/mBio.00166-12.
REFERENCES
- 1. Geraci JR, et al. 1982. Mass mortality of harbor seals: pneumonia associated with influenza A virus. Science 215:1129–1131 [DOI] [PubMed] [Google Scholar]
- 2. Webster RG, et al. 1981. Characterization of an influenza A virus from seals. Virology 113:712–724 [DOI] [PubMed] [Google Scholar]
- 3. Hinshaw VS, et al. 1984. Are seals frequently infected with avian influenza viruses? J. Virol. 51:863–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Callan RJ, Early G, Kida H, Hinshaw VS. 1995. The appearance of H3 influenza viruses in seals. J. Gen. Virol. 76(Part 1):199–203 [DOI] [PubMed] [Google Scholar]
- 5. Vijaykrishna D, et al. 2010. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328:1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crawford PC, et al. 2005. Transmission of equine influenza virus to dogs. Science 310:482–485 [DOI] [PubMed] [Google Scholar]
- 7. Guo Y, et al. 1992. Characterization of a new avian-like influenza A virus from horses in China. Virology 188:245–255 [DOI] [PubMed] [Google Scholar]
- 8. Wilcox BR, et al. 2011. Influenza-A viruses in ducks in Northwestern Minnesota: fine scale spatial and temporal variation in prevalence and subtype diversity. PLoS One 6:e24010 http://dx.doi.org/10.1371/journal.pone.0024010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. 2007. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 3(10):e141 http://dx.doi.org/10.1371/journal.ppat.0030141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McAuley JL, et al. 2007. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2:240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goto H, Kawaoka Y. 1998. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 95:10224–10228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawaoka Y, Naeve CW, Webster RG. 1984. Is virulence of H5N2 influenza viruses in chickens associated with loss of carbohydrate from the hemagglutinin? Virology 139:303–316 [DOI] [PubMed] [Google Scholar]
- 13. Kawaoka Y, Webster RG. 1988. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 85:324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li S, Schulman J, Itamura S, Palese P. 1993. Glycosylation of neuraminidase determines the neurovirulence of influenza A/WSN/33 virus. J. Virol. 67:6667–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao Y, et al. 2009. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 5:e1000709 http://dx.doi.org/10.1371/journal.ppat.1000709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kasson PM, Ensign DL, Pande VS. 2009. Combining molecular dynamics with Bayesian analysis to predict and evaluate ligand-binding mutations in influenza hemagglutinin. J. Am. Chem. Soc. 131:11338–11340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569 [DOI] [PubMed] [Google Scholar]
- 18. Stevens J, et al. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312:404–410 [DOI] [PubMed] [Google Scholar]
- 19. Matrosovich M, et al. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J. Virol. 74:8502–8512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nelli RK, et al. 2010. Comparative distribution of human and avian type sialic acid influenza receptors in the pig. BMC Vet. Res. 6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shinya K, et al. 2006. Avian flu: influenza virus receptors in the human airway. Nature 440:435–436 [DOI] [PubMed] [Google Scholar]
- 22. Trebbien R, Larsen LE, Viuff BM. 2011. Distribution of sialic acid receptors and influenza A virus of avian and swine origin in experimentally infected pigs. Virol. J. 8:434 http://dx.doi.org/10.1186/1743-422X-8-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Poucke SG, Nicholls JM, Nauwynck HJ, Reeth K. 2010. Replication of avian, human and swine influenza viruses in porcine respiratory explants and association with sialic acid distribution. Virol. J. 7:38 http://dx.doi.org/10.1186/1743-422X-7-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harvey R, Martin AC, Zambon M, Barclay WS. 2004. Restrictions to the adaptation of influenza A virus H5 hemagglutinin to the human host. J. Virol. 78:502–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ito T, et al. 1997. Receptor specificity of influenza A viruses correlates with the agglutination of erythrocytes from different animal species. Virology 227:493–499 [DOI] [PubMed] [Google Scholar]
- 26. Chutinimitkul S, et al. 2010. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J. Virol. 84:11802–11813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu S, et al. 2011. CL-385319 inhibits H5N1 avian influenza A virus infection by blocking viral entry. Eur. J. Pharmacol. 660:460–467 [DOI] [PubMed] [Google Scholar]
- 28. Connor RJ, Kawaoka Y, Webster RG, Paulson JC. 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205:17–23 [DOI] [PubMed] [Google Scholar]
- 29. Imai M, et al. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tumpey TM, et al. 2007. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 315:655–659 [DOI] [PubMed] [Google Scholar]
- 31. de Jong MD., et al. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12:1203–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gabriel G, et al. 2005. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. U. S. A. 102:18590–18595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hatta M, Gao P, Halfmann P, Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840–1842 [DOI] [PubMed] [Google Scholar]
- 34. Hatta M, et al. 2007. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 3(10):e133 http://dx.doi.org/10.1371/journal.ppat.0030133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salomon R, et al. 2006. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 203:689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Steel J, Lowen AC, Mubareka S, Palese P. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 5:e1000252 http://dx.doi.org/10.1371/journal.ppat.1000252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Van Hoeven N, et al. 2009. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc. Natl. Acad. Sci. U. S. A. 106:3366–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamada S, et al. 2010. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 6:e1001034 http://dx.doi.org/10.1371/journal.ppat.1001034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Le QM, Sakai-Tagawa Y, Ozawa M, Ito M, Kawaoka Y. 2009. Selection of H5N1 influenza virus PB2 during replication in humans. J. Virol. 83:5278–5281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Z, et al. 2005. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 79:12058–12064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. OIE 2012. Manual of diagnostic tests and vaccines for terrestrial animals, 6th ed. World Organisation for Animal Health, Paris, France [Google Scholar]
- 42. WHO 2002. WHO Manual on Animal Influenza Diagnosis and Surveillance. WHO, Geneva, Switzerland [Google Scholar]
- 43. Wiriyarat W, et al. 2010. Erythrocyte binding preference of 16 subtypes of low pathogenic avian influenza and 2009 pandemic influenza A (H1N1) viruses. Vet. Microbiol. 146:346–349 [DOI] [PubMed] [Google Scholar]
- 44. Roy A, Xu D, Poisson J, Zhang Y. 2011. A protocol for computer-based protein structure and function prediction. J. Vis. Exp. 57:e3259 http://www.jove.com/details.php?id=3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu S, Zhang Y. 2007. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 35:3375–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y, Skolnick J. 2004. SPICKER: approach to clustering protein structures for near-native model selection. J. Comput. Chem. 25:865–871 [DOI] [PubMed] [Google Scholar]
- 47. Pettersen EF, et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 48. Dunbrack RL. 2002. Rotamer libraries in the 21st century. Curr. Opin. Struct. Biol. 12:431–440 [DOI] [PubMed] [Google Scholar]
- 49. Mills JE, Dean PM. 1996. Three-dimensional hydrogen-bond geometry and probability information from a crystal survey. J. Comput. Aided Mol. Des. 10:607–622 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fluorescent ISH of seal H3N8 with DAPI counterstaining. Images are at a ×10 magnification. (A and Bi) A hemagglutinin (HA) probe demonstrates diffuse hemagglutination of bronchial mucosal epithelium. (A and Bii) DAPI binding demonstrates blue nuclear counterstaining of all cells. (A and Biii) Localization of the HA probe reaction to bronchial epithelial cells. ISH was also performed using probes for the matrix gene. Staining was identical to that shown here for HA. Download Figure S1, JPG file, 0.1 MB.
Distribution of SAα-2,3 and SAα-2,6 in seal pulmonary parenchyma. SAα-2,6 (green) was detected using fluorescein-labeled SNA lectin, while SAα-2,3 (red) was detected using Maackia amurensis II (MAL II) lectin. Both infected and uninfected control tissues were stained, and the results were consistent for both. High levels of SAα-2,6 were observed on bronchiole (Ai to -iii) and alveolar (Biiii) epithelial cells and on endothelial cells. The images in panels A and B were selected because they show staining for both sialic acids; however, the expression of SAα-2,3 was rarely observed and limited to bronchiole luminal (A) and occasional alveolar (B) epithelia. Download Figure S2, JPG file, 0.2 MB.
Costaining of SAα-2,6 and H3N8 HA. (Ai to -iii) SAα-2,6 is expressed on the respiratory epithelium of an intrapulmonary bronchus (green), and some cells are infected with H3N8 (red). (Bi to -iii) H3N8 virus infection of SAα-2,6-positive cells is clear. When the level of virus infection is low, SAα-2,6 is still present on the cell surface (arrow 1). As infection increases, SAα-2,6 is reduced, presumably cleaved by the viral NA (arrow 2). In heavily infected cells, SAα-2,6 is no longer detectable (arrow 3). These results demonstrate that the virus can infect SAα-2,6-positive cells. Staining for SAα-2,3 was also performed, but no expression was observed in these regions. Download Figure S3, JPG file, 0.2 MB.