ABSTRACT
Inclusion body disease (IBD) is an infectious fatal disease of snakes typified by behavioral abnormalities, wasting, and secondary infections. At a histopathological level, the disease is identified by the presence of large eosinophilic cytoplasmic inclusions in multiple tissues. To date, no virus or other pathogen has been definitively characterized or associated with the disease. Using a metagenomic approach to search for candidate etiologic agents in snakes with confirmed IBD, we identified and de novo assembled the complete genomic sequences of two viruses related to arenaviruses, and a third arenavirus-like sequence was discovered by screening an additional set of samples. A continuous boa constrictor cell line was established and used to propagate and isolate one of the viruses in culture. Viral nucleoprotein was localized and concentrated within large cytoplasmic inclusions in infected cells in culture and tissues from diseased snakes. In total, viral RNA was detected in 6/8 confirmed IBD cases and 0/18 controls. These viruses have a typical arenavirus genome organization but are highly divergent, belonging to a lineage separate from that of the Old and New World arenaviruses. Furthermore, these viruses encode envelope glycoproteins that are more similar to those of filoviruses than to those of other arenaviruses. These findings implicate these viruses as candidate etiologic agents of IBD. The presence of arenaviruses outside mammals reveals that these viruses infect an unexpectedly broad range of species and represent a new reservoir of potential human pathogens.
IMPORTANCE
Inclusion body disease (IBD) is a common infectious disease of captive snakes. IBD is fatal and can cause the loss of entire animal collections. The cause of the disease has remained elusive, and no treatment exists. In addition to being important to pet owners, veterinarians, breeders, zoological parks, and aquariums, the study of animal disease is significant since animals are the source of virtually every emerging infectious human disease. We searched for candidate causative agents in snakes diagnosed with IBD and found a group of novel viruses distantly related mainly to arenaviruses but also to filoviruses, both of which can cause fatal hemorrhagic fevers when transmitted from animals to humans. In addition to providing evidence that strongly suggests that these viruses cause snake IBD, this discovery reveals a new and unanticipated domain of virus biology and evolution.
Introduction
Inclusion body disease (IBD) is a transmissible, progressive, and ultimately fatal disease of snakes first described several decades ago (1–3). In captive boid snakes, IBD is the most commonly diagnosed disease of suspected viral origin, and there are no treatments or vaccines available. Animals diagnosed with IBD are recommended for euthanasia if there is a risk of transmission to other animals or if there is neurological involvement. It has been speculated that a retrovirus may cause IBD, but to date, no virus or other pathogen has been definitively characterized (3, 4) and no molecular diagnostic method exists. The origin of the disease’s name and the basis for its diagnosis are large eosinophilic inclusions in the cytoplasm of cells of infected animals. A specific 68-kDa protein has been purified from infected tissues, but it is not clear whether this is a viral or endogenous protein or whether the inclusions are directly caused by infection (5). IBD has been described in a variety of species, but it has been best characterized in snakes in the families Boidae and Pythonidae, i.e., boas and pythons (3–10). The clinical and histological manifestations and progression of IBD differ in boid snakes and pythons (1, 3). In pythons, inclusion bodies are primarily found in cells of the central nervous system and clinical signs include abnormal body posture and movement. In boas, inclusions are found in tissues throughout the body and regurgitation is an additional characteristic clinical sign. Pythons typically succumb to disease more rapidly than do boas, which may survive for months or years after symptom onset if provided with supportive care. Although there is evidence that IBD is transmissible, the precise mode of transmission is unknown (3). Conclusive identification of an etiologic agent or agents has the potential to enable IBD treatments, vaccines, diagnostics, and prevention policies.
In addition to being worthy of study in their own right, animal viruses are of great relevance to human health. The simple reason for this is that humans are animals, and many viruses that infect other animals also infect humans. Some of the most medically important human viral pathogens originated from animals or have animal reservoirs, including influenza viruses, HIV-1 and -2, severe acute respiratory syndrome (SARS) coronavirus, henipaviruses, West Nile virus, rabies virus, hantaviruses, filoviruses, and arenaviruses. Furthermore, animal viruses and their hosts often provide excellent models for the study of pathogenic mechanisms, immune response to infection, host-pathogen interactions, treatments, and vaccines.
Unbiased, high-throughput methods are transforming the ability to identify candidate etiologic agents in infectious diseases of unknown cause (11–13). Metagenomic pathogen discovery techniques aim to identify pathogen nucleic acid in infected samples without bias. The first generation of such technologies included the Virochip microarray (14, 15). Now, high-throughput sequencing, such as the Illumina technology, is being increasingly used as its price decreases and throughput increases. The massive depth of sequence combined with increasingly capable assembly and search methods offers greater sensitivity to detect divergent pathogens than ever before. Ultimately, however, sequencing can only ever identify candidate etiologic agents, and demonstration of causality requires significant additional experimental effort.
In this study, we used high-throughput sequencing to search for candidate causes of IBD. We began our investigation using samples from snakes from a local aquarium with confirmed IBD diagnoses and ultimately identified and sequenced the complete genomes of two related viruses, both of which have characteristic attributes of arenaviruses but also share some similarity with filoviruses. Our findings strongly suggest that these viruses may account for at least a significant fraction of IBD cases and reveal the arenavirus family and their hosts to be much broader than had previously been appreciated.
RESULTS
Metagenomic investigation of inclusion body disease.
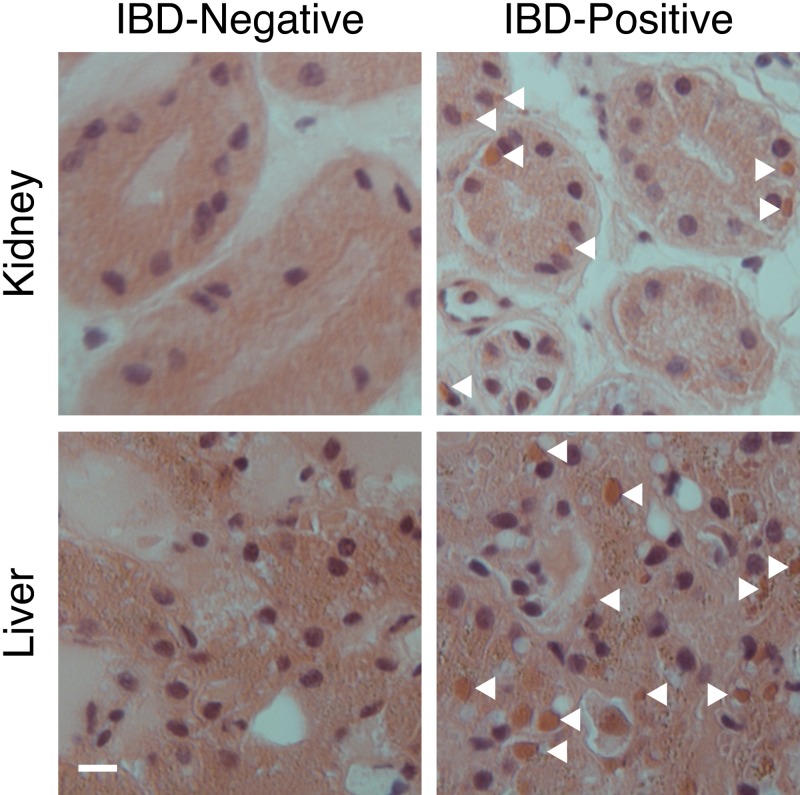
The Steinhart Aquarium at the California Academy of Sciences (CAS) houses a wide range of snake species, including annulated tree boas (Corallus annulatus) and boa constrictors (Boa constrictor). In 2009, several snakes at the aquarium were diagnosed with IBD by histopathology of liver biopsy specimens and/or blood smears (Table 1). Snakes diagnosed as IBD positive as well as one snake (CAS04) that screened negative via blood smear but that had been housed with positive snakes were euthanized and necropsied. Tissue samples were collected and examined by histopathology, revealing inclusions in multiple tissues in positive animals, confirming the antemortem IBD diagnoses (Fig. 1; Table 1).
TABLE 1 .
Summary of snakes analyzed
| Sample | Source | Snake | Species | IBD diagnosis | Virus identificationa |
|---|---|---|---|---|---|
| 1 | California Academy of Sciences, San Francisco, CA | CAS01b | Emerald tree boa | − | − |
| 2 | California Academy of Sciences, San Francisco, CA | CAS02 | Annulated tree boa | + | + (CASV) |
| 3 | California Academy of Sciences, San Francisco, CA | CAS03 | Annulated tree boa | + | + (CASV) |
| 4 | California Academy of Sciences, San Francisco, CA | CAS04 | Annulated tree boa | − | − |
| 5 | California Academy of Sciences, San Francisco, CA | CAS05 | Annulated tree boa | + | + (CASV) |
| 6 | California Academy of Sciences, San Francisco, CA | CAS06 | Boa constrictor | + | + (GGV) |
| 7 | California Academy of Sciences, San Francisco, CA | CAS07 | Boa constrictor | + | + (GGV) |
| 8 | Wildwood Veterinary Hospital, Portola Valley, CA | Larry | Dumeril’s boa | − | − |
| 9 | Wildwood Veterinary Hospital, Portola Valley, CA | Romeo | Boa constrictor | − | − |
| 10 | Wildwood Veterinary Hospital, Portola Valley, CA | Juliet | Boa constrictor | − | − |
| 11 | Wildwood Veterinary Hospital, Portola Valley, CA | Bo | Boa constrictor | − | − |
| 12 | Wildwood Veterinary Hospital, Portola Valley, CA | Dumi | Dumeril’s boa | ?c | − |
| 13 | Zoo/Exotic Pathology Services, West Sacramento, CA | v0110067-3d | Boa constrictor | + | − |
| 14 | Zoo/Exotic Pathology Services, West Sacramento, CA | v0103776-7d | Boa constrictor | − | − |
| 15 | Zoo/Exotic Pathology Services, West Sacramento, CA | v0103836-1d | Ball python | − | − |
| 16 | Zoo/Exotic Pathology Services, West Sacramento, CA | v0103824-5d | Boa constrictor | + | − |
| 17 | Zoo/Exotic Pathology Services, West Sacramento, CA | v0717777 | Boa constrictor | − | − |
| 18 | Zoo/Exotic Pathology Services, West Sacramento, CA | v0727595 | Boa constrictor | − | − |
| 19 | Zoo/Exotic Pathology Services, West Sacramento, CA | v0738356 | Ball python | − | − |
| 20 | Zoo/Exotic Pathology Services, West Sacramento, CA | v0861921 | Boa constrictor | − | − |
| 21 | Zoo/Exotic Pathology Services, West Sacramento, CA | v0864039 | Boa constrictor | + | + (CVV) |
| 22 | Zoo/Exotic Pathology Services, West Sacramento, CA | v1063991 | Python species | ?e | − |
| 23 | Zoo/Exotic Pathology Services, West Sacramento, CA | v1069081 | Python species | − | − |
| 24 | Zoo/Exotic Pathology Services, West Sacramento, CA | v1073638 | Ball python | − | − |
| 25 | Zoo/Exotic Pathology Services, West Sacramento, CA | v1034003 | California king snake | − | − |
| 26 | Zoo/Exotic Pathology Services, West Sacramento, CA | v1033991 | Blacktail rattlesnake | − | − |
| 27 | Zoo/Exotic Pathology Services, West Sacramento, CA | v1032134 | California king snake | − | − |
Arenavirus identification by RT-PCR and, for samples 1 to 16, deep sequencing.
Only blood (no frozen tissue) was available for animal CAS01.
No histopathological IBD diagnosis was performed for this sample.
These samples were formalin fixed; the rest were frozen.
Histopathology was unclear for this sample due to tissue autolysis.
FIG 1 .
Histology showing eosinophilic inclusions in IBD-positive tissue samples. Liver and kidney frozen tissue sections from boa constrictors subsequently diagnosed with or without IBD were H&E stained and imaged. Nuclei appear blue/purple, and eosinophilic structures, including cytoplasmic inclusions (some of which are marked with arrowheads), appear orange/brown. Bars, 10 µm.
To search for candidate IBD etiologic agents, we performed an unbiased high-throughput metagenomic analysis. RNA was extracted from frozen brain, lung, liver, kidney, heart, and gastrointestinal (GI) tissue from the animals for which multiple tissues were available (CAS02 to CAS07; Table 1), and libraries were prepared for deep sequencing (see Materials and Methods). Of these six animals, five had been diagnosed as IBD positive, and one (CAS04) was negative. Sequencing on the Illumina HiSeq platform generated approximately 1 × 108 pairs of 100-nucleotide (nt) sequences, on average ~6 million sequences for each of the 35 samples (no sample was available from the heart of snake CAS07). The complete data set for all samples is available from the NCBI Short Read Archive (accession no. SRA053624). To facilitate the search for viral sequences, we first removed low-quality, low-complexity, and host-derived sequences. To remove host sequences, including those deriving from possibly confounding endogenous retroviruses, we removed reads matching the recently sequenced boa constrictor genome (assembly no. 1 [16]). These operations reduced the size of the data sets by an average of 93%.
Identification of two distinct arenavirus-like genomes.
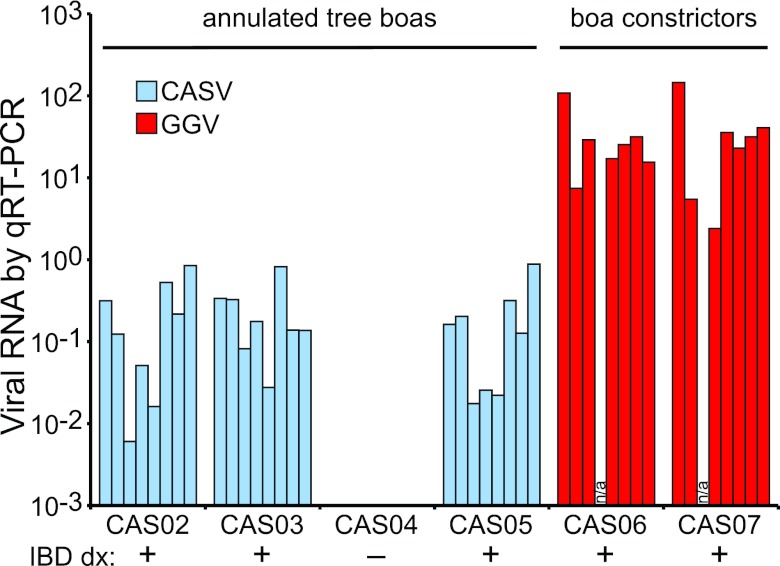
With the remaining sequences, we performed BLASTX searches against a database of viral protein sequences (17). This search revealed the presence of substantial numbers of sequences with similarity to arenavirus protein sequences in all of the IBD-diagnosed samples. We used these BLAST hits to nucleate complete genome assemblies using the PRICE de novo genome assembler (G. Ruby, freely available at http://derisilab.ucsf.edu/software/price/index.html). This analysis revealed that there were actually two distinct (59% pairwise nucleotide identity) but related viruses in the snakes from the aquarium: one from the IBD-positive annulated tree boas and one from the IBD-positive boa constrictors (Table 1; Fig. 2 and 3). We then used PCR, rapid amplification of cDNA ends (RACE), and Sanger sequencing to validate the assemblies. Retrospective mapping of the sequences revealed that the individual IBD-positive tissue data sets contained between 8,422 and 227,134 viral sequences (between 0.13% and 3.8% of the total reads). Overall genome coverage ranged from 825-fold to 3,335-fold (the average number of sequences covering each base). The complete genomes for both viral species are available from GenBank (accession numbers JQ717261 to JQ717264). The data sets from IBD-negative CAS04 contained very low frequencies of sequences mapping to the two viruses (<5 × 10−5), a phenomenon that was also observed for the boa constrictor virus in the annulated tree boa samples and vice versa. These low-copy-number sequences likely resulted from intersample cross-contamination or bar code misregistration during sequencing (18). This explanation was corroborated by quantitative reverse transcription-PCR (qRT-PCR), which demonstrated that all tissues of snake CAS04 were negative for viral RNA (there was no amplification) (Fig. 2). This analysis also confirmed that the two viruses segregated perfectly with the two snake species. By sequencing and by quantitative PCR (qPCR), viral RNA was detected in every tissue examined, a pattern reminiscent of the histopathological detection of inclusions in all of these tissues (Fig. 2).
FIG 2 .
Viral RNA is detected in all tissues of IBD-diagnosed snakes. The relative amount of L segment viral RNA detected by qRT-PCR for each tissue in each snake is shown. Results are normalized to the detected levels of two snake mRNAs (glyceraldehyde-3-phosphate dehydrogenase and RPS2). Virtually identical results were obtained for S segment RNA. Each bar represents one tissue: from left to right, brain, GI tissue, heart, kidney, liver, lung, serum, and blood cells. The snakes’ histopathology-based IBD diagnoses are indicated. Snake designations are given in Table 1. A sample was unavailable for CAS07 heart tissue, and data were unavailable for the CAS06 kidney.
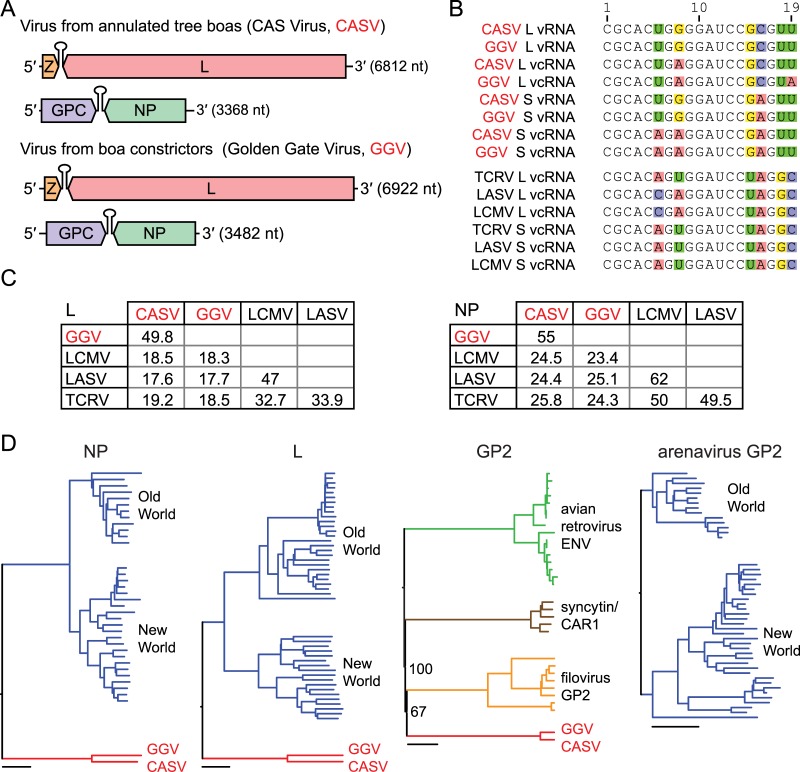
FIG 3 .
New viruses mix arenavirus characteristics and filovirus-like glycoproteins. (A) Genome organization of the viruses recovered from CAS (California Academy of Sciences) snakes. Features are to scale, and segments are in the predicted genomic sense. Predicted coding regions and intergenic hairpins are depicted. (B) An alignment of the terminal 19 5′ nucleotides of the indicated genome (vRNA) or antigenome (vcRNA) segments. Variable residues are highlighted. For comparison, terminal sequences from three reference rodent arenaviruses are shown: TCRV (NC_004292.1), LASV (NC_004296.1), and LCMV (NC_004294.1). (C) Matrices of pairwise percent amino acid identity between snake viruses and reference rodent arenaviruses. Values are based on multiple sequence alignments generated using ClustalW software (see Materials and Methods). Sequence accession numbers are the same as those for panel B. (D) Phylogenies of predicted snake virus NP, L, and GP2 protein sequences and related sequences. Old World and New World designate major clades of previously described arenaviruses. Bars, 0.2 substitutions per site. The bootstrap percentages for select nodes are indicated.
In addition to arenavirus-like sequences, reads with similarity to retroviral sequences were identified in all of the annulated tree boa samples (CAS02 to CAS05). These retrovirus-like sequences were most closely related to previously described endogenous retroviral sequences from pythons (19), were not found in the boa constrictor genome sequence or the boa constrictor samples (CAS06 and CAS07), and thus likely derive from annulated tree boa endogenous retroviruses.
The recovery of high numbers of arenavirus-related sequences from the IBD-diagnosed snakes but not the single IBD-negative snake suggested that the viruses might play a role in disease pathogenesis (Fig. 2). To investigate this possibility, we obtained and screened 21 additional samples from IBD-positive and -negative snakes of various species for viral RNA (Table 1). We performed RT-PCR with multiple sets of primers, including consensus primers designed to amplify sequence from both novel viruses, and by additional deep sequencing in some cases. A third related virus was detected by RT-PCR in a boa constrictor from Collierville, TN, diagnosed with IBD in 2008. Although the complete genome of this third virus has not yet been fully determined, alignments of the recovered sequences reveal that this third virus is more closely related (~80% nucleotide identity) to the virus recovered from the CAS boa constrictors (see Fig. S1 in the supplemental material). Viral RNA was not detected in the other three confirmed IBD cases. Thus, 6/8 IBD-positive samples were positive for one of these divergent but related viruses. We did not detect viral RNA in any of 18 control samples (Table 1). These results show that multiple related arenavirus-like viruses are associated with geographically and temporally widespread IBD cases and also reveal a high degree of diversity in this clade. Until all available samples can be analyzed by deep sequencing, we cannot rule out the presence of additional divergent species in the samples that have tested negative by RT-PCR.
By convention, species in the family Arenaviridae and several other single-stranded RNA (ssRNA) virus families are named after the geographical location from which they were isolated. We propose naming the virus species isolated from the annulated tree boas CAS virus (CASV; for California Academy of Sciences virus) and naming the two species from boa constrictors Golden Gate virus (GGV) and Collierville virus (CVV; Table 1).
Genome analysis.
Analysis of the recovered genome sequences revealed that they possess attributes characteristic of arenavirus genomes, including a bisegmented ambisense organization, with two opposite-sense open reading frames (ORFs) on each of two genome segments (Fig. 3A) (20, 21). Also, as is the case with other members of the family, on both segments the intergenic regions separating the ORFs were predicted to form hairpin structures, with predicted Gibbs free energy changes (ΔG) of between −50 and −91 kcal/mol at 30°C (22). The inter-ORF sequences of the S and L segments of GGV and CASV share 38% and 42% pairwise nucleotide identity, respectively, in global alignments. Also consistent with arenavirus genome structure, the terminal 19 nucleotides (nt) at the 5′ and 3′ ends of the L segment are nearly reverse complements of each other and are similar to the 5′- and 3′-end sequences of the S segment (Fig. 3B). The terminal sequences of these viruses are conserved at 13/19 residues with the terminal sequences of previously described arenaviruses (Fig. 3B). The L and S segments of CASV measure 6,812 nt and 3,368 nt, respectively, while the L and S segments of GGV measure 6,922 nt and 3,482 nt, respectively.
Phylogenetic relationships.
To further investigate the relationship between these viruses and previously described arenaviruses, we conducted comparative and phylogenetic analyses of the four major open reading frames (L, NP, Z, and GPC) present on CASV and GGV. In arenaviruses, L is the large RNA-dependent polymerase involved in viral genome replication and transcription (20, 21, 23). NP, the viral nucleoprotein, forms a circular ribonucleoprotein complex with the genome-length viral RNAs (21, 24). The predicted snake proteins share 23 to 26% and 17 to 19% pairwise amino acid identity with the lymphocytic choriomeningitis virus (LCMV), Lassa virus (LASV), and Tacaribe virus (TCRV) L and NP proteins, respectively (Fig. 3C). These viruses were selected for comparison as representative members of the two major clades of previously described arenaviruses: the Old World (OW) and New World (NW) arenaviruses, also known as the Lassa/LCMV and Tacaribe serocomplexes (20, 21, 25, 26). The CASV and GGV L and NP protein sequences are 55% and 50% identical in amino acids, respectively, or roughly as similar as the L and NP proteins of LCMV and LASV, which are 47% and 62% identical, respectively. In phylogenies, the snake virus proteins form a monophyletic clade separate from those formed by the Old World and New World arenaviruses (Fig. 3D).
Previously described arenaviruses encode a small RING domain-containing zinc binding protein (Z) on the L segment and a glycoprotein precursor (GPC) on the S segment (20, 21, 27, 28). There are ORFs encoding proteins with similar predicted functional domains at the same genomic positions of the snake viruses, but their evolutionary relationship to the arenavirus Z and GPC genes is less clear. The predicted protein sequences did not possess detectable direct homology with previously described arenavirus Z and GPC sequences, as measured by BLASTP against the nr database using an expect value cutoff of 0.1. Instead, they were more similar to other viral and nonviral proteins.
The predicted small “Z” proteins of CASV and GGV (115 and 116 amino acids, respectively) were most similar to nonviral RING domain-containing proteins, with the best BLASTP (NCBI nr database) hit for both being that to various zinc finger and RING domain-containing proteins. These alignments were driven largely by conserved cysteine residues and had poor expect (E) values of greater than 0.1. Using the more sensitive HHPred software (29), which detects remote homologies and makes structural predictions of proteins of unknown structure, the best hits were also to RING domain-containing ubiquitin ligases, with probabilities of homologous relationship of >92%.
Arenavirus Z proteins are essential for virus budding and are myristoylated on the amino (N) terminus (30–32). The snake virus Z proteins do not possess N-terminal glycine residues typically associated with myristoylation but instead have predicted transmembrane domains in their first 50 amino acids, which may serve a similar role. Most arenavirus Z proteins also contain carboxyl (C)-terminal “late” domains with characteristic motifs (commonly PTAP or PPPY) that recruit cellular proteins that help drive virus budding (33). The CASV and GGV Z proteins do not contain recognizable late domain motifs in their C termini. However, the C-terminal sequences of the NPs of these viruses do contain short motifs similar to late motifs (PKPV and PTPA), and it is possible that, as is the case for Marburg virus, NP contains a functional late domain (34).
CASV and GGV encode a predicted transmembrane glycoprotein at the same genomic position as arenavirus GPCs. However, like Z, these proteins did not contain detectable homology to arenavirus glycoproteins as measured by BLAST (versus nr) or HHPred. Instead, by BLASTP analysis the snake virus proteins were related to the glycoproteins of filoviruses (e.g., Ebola and Marburg viruses) and avian retroviruses (e.g., avian leukosis virus) and the cellular syncytin, a repurposed endogenous retroviral gene (Fig. 3D) (35, 36). The predicted GPs of CASV and GGV are 393 and 427 amino acids long and, like other class 1 viral fusion proteins, may be proteolytically cleaved into two functional domains: the GP1 receptor binding and the GP2 transmembrane/fusion domains (37, 38). Amino acids 222 to 393 of CASV GP and 256 to 427 of GGV GP are 82% identical. This relatively conserved domain likely corresponds to the GP2 domain and was the region with detectable similarity by BLAST. Using HHPred, the top hit for this domain of both CASV and GGV was to the Sudan Ebola virus GP2 (PDB accession no. 3S88; probability of homologous relationships, 99.98%). Like filovirus GP2s, the CASV and GGV domains are predicted to form C-terminal transmembrane domains that anchor the protein in the viral membrane. In contrast to the relatively conserved GP2 domain, the predicted N-terminal GP1 domains of CASV and GGV glycoproteins are only 31% identical, and neither domain contains detectable homology to known proteins (by BLASTP versus nr with an E value of <1). Analysis of the predicted GP1/GP2 boundaries did not reveal obvious candidate protease cleavage sites present in both the GGV and CASV sequences.
In vitro virus culture.
To enable further characterization of these viruses, we developed in vitro cell culture growth conditions that permitted virus propagation. We first attempted to infect reptile cell lines available from the ATCC (viper heart VH-2 and iguana heart IGH-2). Inoculum was prepared from the kidney and liver of the boa constrictors CAS06 and CAS07, the samples from which sufficient frozen tissue remained. Viral RNA levels in culture supernatant were monitored by qRT-PCR. Viral RNA was initially detectable in the supernatant but rapidly disappeared as culture medium was replaced and was undetectable by 7 days postinoculation and over the course of 18 days. We observed similar negative results with an African green monkey-derived Vero cell line, known to be permissive for the replication of many arenaviruses. This suggested that these lines or the culture conditions were not permissive for replication or that the inoculum that we used lacked replication-competent virus.
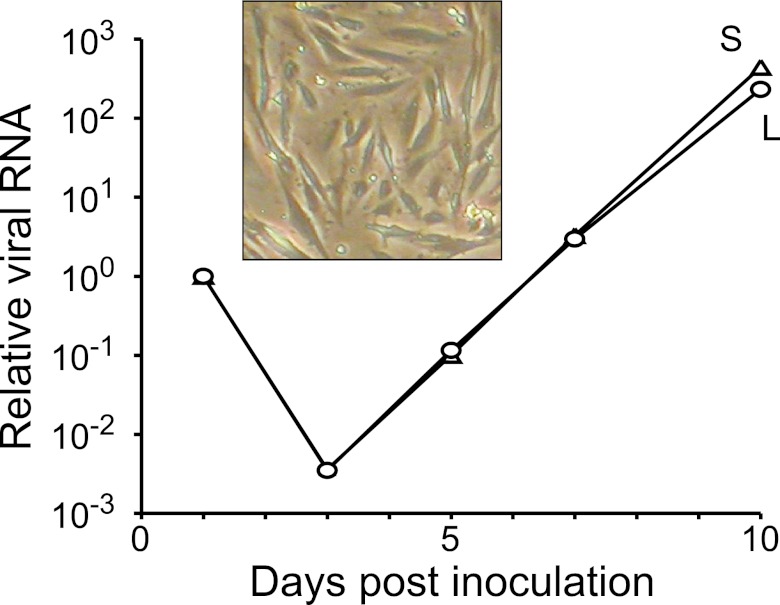
Hypothesizing that boa constrictor cells might be permissive for replication of the boa constrictor-derived virus, we harvested tissue from a 22-year-old female boa constrictor (“Juliet”) who died of lymphoma and attempted to create continuous cell lines (39). Of the multiple tissues and conditions attempted, proliferation was evident only in a culture of adherent cells derived from Juliet’s kidneys (Fig. 4, inset). This cell line (JK cells) continues to proliferate, although it is not yet clear whether it is immortal. We inoculated a culture of JK cells with liver and kidney homogenates from the CAS boa constrictors and observed an exponential increase of viral RNA in culture supernatant after an initial decrease (Fig. 4). Viral RNA was not detectable in mock-inoculated JK cultures. Infected cells did not exhibit gross cytopathic effects over the course of 21 days.
FIG 4 .
Viral replication in a boa constrictor continuous cell line. Relative levels of viral L and S segment RNA measured by qRT-PCR in the supernatant of JK cells inoculated with homogenate from snake CAS06 kidney and liver. Inset, image of JK cells.
We next developed and validated an antibody against the viral nucleoprotein as an additional tool to study the virus and its relationship to disease. This polyclonal antibody was raised in rabbits against a synthetic peptide corresponding to the C-terminal 14 residues of the NP encoded by GGV. We chose this antigen for several reasons. First, for previously described arenaviruses, NP is the most abundant viral protein in infected cells and is the most antigenic viral protein in vivo (21). Second, the predicted molecular mass of this protein (67 kDa) is similar to the reported mass (68 kDa) of an abundant protein specific to IBD-positive tissues (5). Furthermore, the NP of other arenaviruses is reported to localize to intracytoplasmic inclusions in infected cells (40, 41).
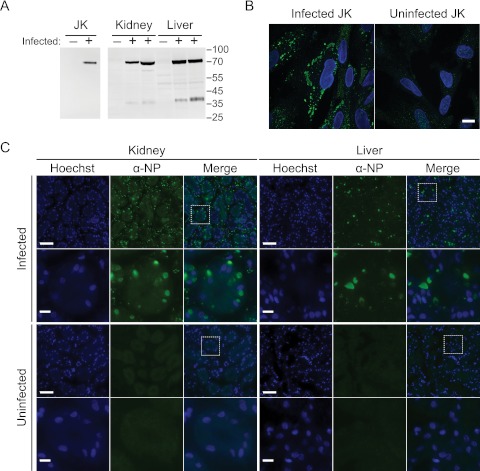
By Western blotting, the anti-NP antiserum specifically detected a protein of the predicted mass in infected JK cells and tissues (Fig. 5A) but not in tissues from a virus-negative/IBD-negative snake. Further, in infected JK cells, this antibody stained large cytoplasmic aggregates (Fig. 5B). In contrast, uninfected cells displayed only diffuse background staining (Fig. 5B), as did preimmune serum staining of infected cells. In liver and kidney sections from infected boa constrictors, staining with this antibody showed that the viral nucleoprotein is localized to large cytoplasmic inclusions in cells throughout the tissues (Fig. 5C). In contrast, only diffuse background from nonspecific staining and autofluorescence was evident in uninfected tissue sections.
FIG 5 .
An antibody to the viral nucleoprotein stains cytoplasmic aggregates in infected cells and tissues. (A) Western blot with a polyclonal anti-NP peptide antibody against infected and mock-infected JK cells and against liver and kidney tissue homogenates from one uninfected and two infected boa constrictors. Numbers represent molecular masses in kDa. (B) Immunofluorescence using the same antibody on infected and mock-infected JK cells. Images show merged Hoechst stain and anti-NP signals. Bar, 10 µm. (C) Immunofluorescence of liver and kidney tissue sections from an infected and an uninfected boa constrictor. Rows 2 and 4 show the boxed areas of the images in rows 1 and 3. Bars, 50 µm (rows 1 and 3) and 10 µm (rows 2 and 4).
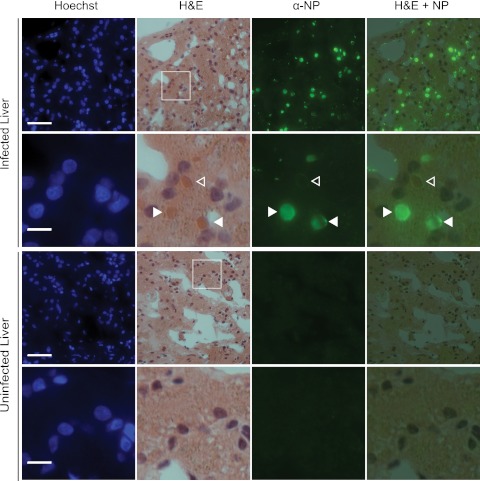
We wondered whether the large NP inclusions that we observed in infected JK cells and infected boa constrictor tissues corresponded to the inclusion bodies that are diagnostic for IBD. We performed hematoxylin and eosin (H&E) staining on sections previously visualized by immunofluorescence and found that this was indeed the case (Fig. 6). Many eosinophilic inclusions stained brightly with NP while others appeared to be ringed by NP fluorescence (closed and open arrowheads in Fig. 6). This could be explained by differential cross-sectioning of the inclusions during tissue sectioning, for instance, if the inclusions were roughly spherical and coated with NP protein. In this scenario, in equatorial cross sections the NP fluorescence would ring the inclusions. And if the section captured the top or bottom surface of the inclusion, then the fluorescence would appear as filled circles. Alternatively, there may be multiple, structurally distinct types of inclusions, or inclusions may be differentially accessible to antibody during staining.
FIG 6 .
In infected tissues, viral nucleoprotein localizes to eosinophilic inclusion bodies. The same tissue sections were first imaged by fluorescent microscopy and then H&E stained and reimaged. Rows 2 and 4 show the boxed areas of the images in rows 1 and 3. Closed arrowheads, eosinophilic inclusions fully staining; open arrowheads, inclusions ringed by NP fluorescence (see text). Bars, 50 µm (rows 1 and 3) and 10 µm (rows 2 and 4).
DISCUSSION
Here we described the discovery and characterization of two complete viruses and partial sequence from a related, yet potentially distinct third virus, isolated from cases of snake inclusion body disease. These viruses share a typical arenavirus-like genome organization, but the protein sequences of Z and GPC imply a more complicated evolutionary relationship to previously characterized arenaviruses. These viruses were detected in 6/8 snakes with confirmed IBD diagnoses and 0/18 IBD-negative animals. In infected animals, viral RNA and protein were detected in tissues with cytoplasmic inclusions, and indeed, viral nucleoprotein was found to localize to the same eosinophilic inclusions that give the disease its name. Although formal confirmation that these novel arenavirus-like agents cause disease in snakes will require experimental challenge studies, their detection in reptiles raises an array of intriguing questions about the host range, evolution, basic biology, and mechanisms of pathogenesis associated with this unusual branch of the virus phylogeny.
The arenaviruses are a family of viruses that had been previously believed to infect only mammals (21, 25, 26, 42). Rodents are thought to be the natural hosts of arenaviruses, and individual virus species are associated with specific hosts. Infection is typically chronic and asymptomatic in rodents. However, when arenaviruses zoonotically infect humans or other mammals, severe disease can result. Scientific study has focused on these viruses for two principal reasons: (i) some of these viruses (e.g., LASV) can cause fatal hemorrhagic fever in humans, and (ii) the arenavirus LCMV provides an excellent tool to study the immune response in its natural host, Mus musculus (43). Infection of members of the Reptilia class of animals demonstrates that arenavirus infection is not limited to mammals.
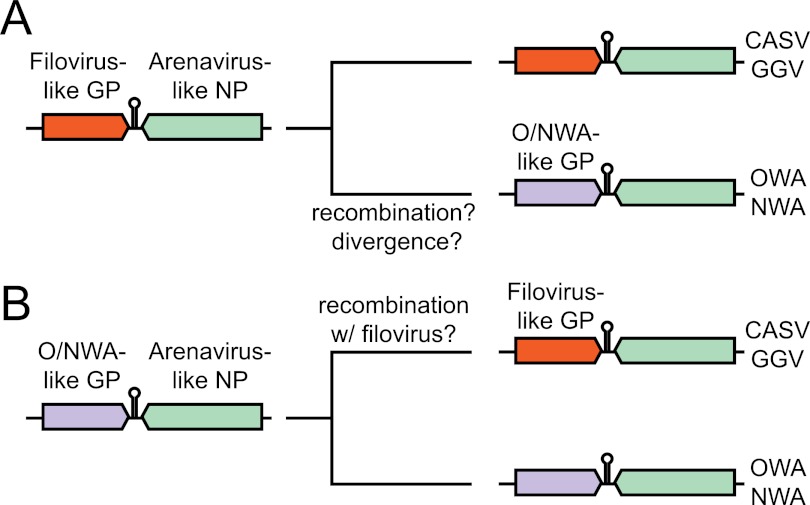
The discovery of viruses that are in most ways arenavirus-like with filovirus-like GP2 domains relates to the previously stated hypothesis that envelope glycoproteins from filoviruses and arenaviruses share an ancient common ancestor (44, 45). There are several possible models to explain the configurations of the extant arenavirus species. One possibility is that the GPC gene of the arenavirus common ancestor was more similar to the GPC gene of CASV and GGV and therefore to those of filoviruses and avian retroviruses (Fig. 7A). In this scenario, recombination or significant divergence occurred on the lineage leading to the Old World (OW) and New World (NW) arenaviruses. An alternate model is that the GP gene of the ancestral arenavirus was similar to that of the present-day rodent arenaviruses and may itself have derived from an ancestor common with the filoviruses (Fig. 7B). Then, along the lineage leading to snake arenaviruses, recombination with a filovirus or retrovirus introduced a new GP gene. Intrasegmental recombination between arenaviruses leading to speciation has been documented, so a precedent for these models exists (46).
FIG 7 .
Models for evolution of arenavirus glycoprotein genes. S segments of extant and presumed ancestral arenaviruses are shown. See text for additional explanation. O/NWA, Old/New World arenavirus.
Whether infection by these viruses causes disease in snakes is perhaps the principal open question following from this study. It is possible that—as is often the case in rodents—arenavirus infection of snakes is chronic and subclinical. Indeed, the CAS snakes diagnosed as IBD and virus positive (Table 1) were not showing any of the typical signs of regurgitation, anorexia, or central nervous system abnormalities at the time of sample collection, though it could have been relatively early in the course of infection. In contrast, the snake from Tennessee that was diagnosed as IBD and arenavirus positive (Table 1) was diagnosed in a postmortem exam. This snake had produced a clutch of infertile eggs (slugs), developed dysecdysis (incomplete shedding), continued to decline, and ultimately died. It is clear that arenavirus infection does result in large viral protein-containing inclusions, a diagnostic finding believed to be prognostic of a fatal outcome, although disease progression has proven variable, with animals surviving for weeks to months after first clinical manifestation, and with the caveat that “IBD” in different snake species may ultimately prove attributable to different causes.
We do not know if snakes are the natural host of these viruses or if snakes are infected adventitiously, in a manner akin to the zoonotic transmission of rodent arenaviruses to humans. Likewise, it is unclear how the virus is transmitted. One possibility is that virus is transmitted from snake to snake by blood-feeding mites, infestations of which have anecdotally been associated with IBD outbreaks (1). Snakes eat rodents; thus, another possibility is that these viruses are transmitted when snakes eat infected mice or rats. This possibility is not unprecedented: “callitrichid hepatitis virus” was originally identified as the agent responsible for outbreaks of fatal hepatitis in captive marmosets and tamarins in zoos (47). This virus was subsequently shown to be identical to LCMV, which was being transmitted to the zoo animals via infected mice that they were fed (48–50). In contrast, the viruses identified here are highly divergent from Old and New World arenaviruses. Additional experiments to test the viruses’ host range in cell lines and animals will help answer these questions.
Two of the eight snakes diagnosed as IBD positive in this study tested negative for arenavirus infection (Table 1). There are a number of other possible causes of the pathology observed in these cases. One obvious alternative is infection by other viruses not detected by our methods, including additional divergent arenaviruses. A precedent for this possibility is the case of avian proventricular dilatation disease, where follow-up studies identified numerous additional genogroups of avian bornavirus, the causative agent (15, 51–54). Deep sequencing of additional IBD-positive samples will help resolve this question. Alternatively, infection by nonviral pathogens may be responsible in some cases, though we did not detect any obvious such organisms in our metagenomic analyses. Alternatively, the cytoplasmic inclusions may be a by-product of some other disease state or cellular stress, although the localization of viral nucleoprotein to these inclusions that we observed would appear to contradict this alternate hypothesis. Expanded association studies will more firmly determine the proportion of IBD cases attributable to arenavirus infection and may identify etiologic agents responsible for nonarenavirus IBD diagnoses.
The findings presented here raise the possibility of improved IBD diagnostics, prevention, and treatment. IBD is currently diagnosed by relatively insensitive blood analysis or invasive biopsy and histopathology. Viral RNA was as abundant in blood cells as in infected tissues (Fig. 2), so RT-PCR using RNA purified from whole blood is the approach offering the best combination of specificity, sensitivity, and ease of sample collection, though proper controls and procedures will have to be used to minimize the possibility of PCR false positives due to contamination. Primers targeting the region of the genomes conserved among the three viruses described here are listed in Table S1 in the supplemental material. Antibodies against viral proteins offer an additional diagnostic approach, but antibodies against conserved viral epitopes would have to be generated and validated.
IBD is an important disease of captive snakes. While the specific mode of transmission is unknown, it is likely that the elimination of diseased snakes will greatly reduce the likelihood of transmission, even if there exists an intermediate vector. The availability of a diagnostic test, whether by RT-PCR or by virus-specific antibodies, will prevent the introduction of infected, but possibly asymptomatic, snakes into healthy collections. Ultimately, diagnostically driven surveillance by veterinarians will likely identify outbreaks or hot spots of the disease and perhaps one day lead to adequate control of this previously vexing condition. Furthermore, vaccines exist or are in development for some arenaviruses (21, 55–57), and ribavirin and other drugs have been shown to be effective in decreasing the severity of disease caused by hemorrhagic fever-causing arenaviruses (21, 58, 59). This offers hope that arenavirus-targeting vaccines or treatments could be effective against snake IBD.
MATERIALS AND METHODS
Sample collection and processing.
At necropsy, portions of tissues were fixed in formalin or frozen at −80°C until further processing. For histopathological analysis, tissue samples were preserved in 10% neutral buffered formalin solution, embedded in paraffin, sectioned at 5 µm, mounted on glass slides, and stained with hematoxylin and eosin.
For RNA extraction, 100-mg tissue pieces were added to 1 ml of Trizol reagent (Invitrogen) and a ball bearing in a centrifuge tube. Tissue was disrupted using the TissueLyzer (Qiagen) for 2 min at 30 Hz. Samples were clarified by centrifugation at 10,000 × g for 2 min, and then 200 µl chloroform was added. Samples were mixed, incubated for 2 min at room temperature, and then centrifuged for 15 min at 12,000 × g at 4°C. The RNA in the aqueous phase was further purified using an RNA Clean and Concentrator column (Zymo Research) according to the manufacturer’s protocol, including the optional on-column DNase digestion to remove residual DNA. RNA quantity and quality were determined by spectroscopy.
Library preparation and sequencing.
Sequencing libraries were prepared essentially as previously described (60). Two hundred fifty nanograms of RNA was added to 10 µl reverse transcription (RT) reaction mixtures containing 50 pmol oligonucleotide MDS-187, 1× reaction buffer, 5 mM dithiothreitol, 1.25 mM (each) deoxynucleoside triphosphates (dNTPs), and 100 U Superscript III (Invitrogen). The sequences of all oligonucleotides are listed in Table S1 in the supplemental material. Reaction mixtures were incubated for 60 min at 42°C and then 15 min at 70°C. Then, 10 U APE1 and 1 U UDG (NEB) diluted in 5 µl 1× Sequenase buffer (Affymetrix) were added to reaction mixtures to remove the oligo(dU/dT) RT primer. Reaction mixtures were incubated for 30 min at 37°C and then 94°C for 2 min. To generate end-tagged molecules, primer MDS-4 and 2 U of Sequenase DNA polymerase (Affymetrix) in 5 µl of 1× buffer were added to samples, which were ramped from 10°C to 37°C over 8 min and then incubated at 37°C for 8 min. These primer extension reactions were performed twice to generate doubly end-tagged molecules, which were subsequently amplified by PCR. PCR mixtures contained 1× reaction buffer, 2 µM primer MDS-189, 0.25 mM dNTPs, 2 U KlenTaq DNA polymerase (Sigma), and 2 µl library template. Thermocycling conditions were 95°C for 2 min; 2 cycles of 95°C for 30 s, 40°C for 30 s, and 72°C for 1 min; and then 15 cycles with a 55°C annealing temperature. PCR mixtures were cleaned using the Ampure XP reagent (Agencourt) according to the manufacturer’s protocol but using a 1.4:1 ratio of beads to sample. Ten nanograms of library template was then added to PCR mixtures containing 1× reaction buffer, 0.25 mM dNTPs, 0.01 µM (each) primers MDS-9 and MDS-10 (bar code), and 12.5 U KlenTaq DNA polymerase. Thermocycling conditions were 95°C for 2 min and 2 cycles of 95°C for 30 s, 40°C for 30 s, and 72°C for 1 min. Then, primers MDS-200 and -201 were added to reaction mixtures to a final concentration of 0.2 µM, and 6 more cycles with a 58°C annealing temperature were performed. Reaction mixtures were cleaned again with Ampure XP reagent, and their relative concentrations were quantified in qPCR mixtures containing 1× LC480 Sybr Green master mix (Roche) and 0.1 µM (each) of primers MDS-200 and -201. Equivalent amounts of each sample were then pooled, and the pool was cleaned with a DNA Clean and Concentrator column (Zymo Research) according to the manufacturer’s protocol. The pooled libraries were then size selected (400 to 650 nt) using a LabChip XT device (Caliper) and subjected to a final round of amplification in PCR mixtures containing 1× reaction buffer, 0.2 mM dNTPs, 0.2 µM (each) MDS-142 and -199, and 12.5 U KlenTaq polymerase. Reaction conditions were 95°C for 2 min and 18 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min. The pooled libraries were finally purified using a DNA Clean and Concentrator column and quantified spectroscopically and by using the Illumina library quantification kit (Kapa Biosystems). Sequencing was performed on an Illumina HiSeq 2000 instrument. The sequencing data have been deposited in the NCBI SRA under accession SRA053624.
Sequence analysis.
In addition to the default Illumina quality filtering, low-quality sequences containing any N’s were removed from further analysis. Low-complexity sequences with an LZW ratio less than 0.45 (the ratio of the length of the Lempel-Ziv-Welch compressed sequence to the uncompressed length) were additionally removed (60, 61). The first 6 bases of each sequence, corresponding to the random hexamer used to tag library molecules, were trimmed from sequences. Snake sequences were then filtered using the BLASTN alignment tool (version 2.2.21 [17]) to query a database composed of a draft (assembly 1) of the boa constrictor genome (16). Sequences aligning with an expect value of less than 10−8 were filtered. Similarly, sequences that aligned with the Illumina adapter sequences (see Table S1 in the supplemental material) or to φX174 control sequence were removed. This filtering removed between 90 and 97% of sequences. The remaining sequences were searched against databases of viral protein sequences using the BLASTX algorithm, and sequences matching any viral protein sequence with an expect value of less than 2 were further examined. False positives were eliminated by using BLAST to align putative viral sequences to the NCBI nonredundant nucleotide (nt) and protein (nr) databases. Only sequences whose best hit and whose pair’s best hit were to viral sequences were further considered. The PRICE de novo targeted genome assembler was used to generate initial contiguous virus sequences (G. Ruby; freely available at: http://derisilab.ucsf.edu/software/price/index.html). For coverage information, reads were aligned to Sanger validated assemblies using the Bowtie2 alignment software, version 2.0.0 (62).
Sanger sequencing and RACE.
Virus genome sequences assembled from deep sequencing reads were validated using Sanger sequencing and RACE. Primer sequences are listed in Table S1 in the supplemental material. PCR mixtures contained 1× reaction buffer, 0.25 µM primer, 0.2 mM dNTPs, 2 U Taq DNA polymerase, and 0.25 µl cDNA. Thermocycling consisted of 95°C for 2 min and then 30 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 2 min. Amplicons were purified from agarose gels using the PureLink gel extraction kit (Invitrogen) and cloned into the pCR2.1 TOPO vector (Invitrogen) according to the manufacturer’s protocols. Cloned amplicons were Sanger sequenced (Quintara Biosciences). Because the viral genome segments are predicted to exist in genomic and antigenomic forms, 5′ RACE was used to obtain both end sequences, essentially as described elsewhere (63), with primers listed in Table S1. Multiple RACE amplicons were cloned and sequenced as described above. In cases where RACE amplicons did not extend to the end of the deep sequencing assemblies, the deep sequence coverage was sufficient to determine the terminal sequences. The complete genome sequences of CASV and GGV have been deposited with the NCBI under accession numbers JQ717261 to JQ717264.
Antibody production.
A peptide corresponding to the C-terminal 14 amino acids (SGGKKTKDPTPATI) of the nucleoprotein of GGV was synthesized and injected into rabbits for polyclonal antibody production (Pacific Immunology).
Quantitative PCR.
Quantitative PCRs to monitor viral RNA levels in tissues and in culture replication experiments were performed on an LC480 instrument (Roche). RNA was extracted from tissue as described above or from tissue culture supernatant using the ZR viral RNA kit (Zymo Research). Reverse transcription reactions were performed as described for library preparation but used random hexamer priming. qPCR mixtures contained 1× LC480 Sybr Green master mix (Roche), 0.1 µM (each) primer (listed in Table S1 in the supplemental material), and 5 µl of 1:20-diluted cDNA.
Phylogenetic analysis.
The BLASTP alignment tool was used to query the NCBI nr database for sequences similar to predicted viral protein sequences, and sequences producing alignments with an expect value of less than 10−6 were downloaded. The CD-HIT software version 4.0 (64) was used to collapse sequences with greater than 90% pairwise amino acid identity. Multiple-sequence alignments were created using Clustal (version 2.1) using default parameters (65). Maximum likelihood phylogenies were created using PhyML (PhyML plugin for Geneious version 2.0.12) using the LG model of amino acid substitutions, 100 bootstrap replicates, and otherwise default parameters (66).
Western blotting.
Tissues and cells were homogenized in ice-cold buffer containing 40 mM Tris (pH 7.4), 120 mM NaCl, 0.5% Triton X-100, 0.3% SDS, and Complete protease inhibitors (Roche). Samples were rotated for 30 min and then clarified by centrifugation for 10 min at 10,000 × g at 4°C. Protein concentration was determined using the bicinchoninic acid (BCA) protein assay reagent (Pierce). Five micrograms (cells) or 25 µg (tissues) of total protein was fractionated by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was probed with anti-NP polyclonal antiserum, which was detected with a fluorescently labeled secondary antibody. The blot was scanned on an Odyssey scanner (LiCor).
Tissue culture.
VH-2 and IGH-2 cells were obtained from the ATCC and were grown at 30°C in Eagle’s minimum essential medium with Hanks’ balanced salt solution (University of California San Francisco [UCSF] cell culture facility) supplemented with 10% fetal bovine serum, 50 units/ml penicillin, 50 µg/ml streptomycin, and 25 mM HEPES (pH 7.4) (cMEM). Vero cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin, 50 µg/ml streptomycin at 37°C and 5% CO2.
To generate a continuous boa constrictor cell line (JK), tissues from a boa constrictor that had died of lymphoma were collected at necropsy and stored on ice until processing. Tissue was minced with scalpels, incubated overnight in 0.25% trypsin and 1 mM EDTA in saline A at 4°C, and then plated into culture dishes in cMEM and incubated at 30°C. Culture medium was replaced every 5 days. After 5 weeks, proliferation of adherent cells in a kidney-derived culture was evident and these cells were subsequently passaged weekly.
For inoculation experiments, 500-mg portions of frozen virus-positive and -negative kidneys and livers were finely minced with scalpels, resuspended in serum-free MEM containing 25 mM HEPES (pH 7.4), and homogenized with a Dounce homogenizer. Homogenates were clarified by centrifugation at 10,000 × g for 2 min and filtered through an 0.45-µm filter. Filtrates were diluted between 1:10 and 1:40 and used to inoculate cell cultures.
Immunofluorescence.
Infected or uninfected cells were plated in collagen-coated glass-bottom culture dishes (MatTek) in cMEM. After 1 day of culture, cells were rinsed twice in phosphate-buffered saline (PBS), then fixed in 4% formaldehyde in PBS for 30 min, and then washed 3 times for 3 min in PBS. Cell membranes were permeabilized in 0.1% Triton X-100 in PBS for 5 min, washed, and then blocked in 1% bovine serum albumin (BSA) for 1 h. Cells were incubated with anti-NP antiserum diluted in blocking buffer for 1 h and then washed 6 times for 4 min in PBS. Cells were then incubated in Alexa Fluor 488-conjugated donkey anti-rabbit antibody (Life Technologies), diluted in blocking buffer for 30 min, and then washed again. The penultimate wash contained 0.5 µg/ml Hoechst 33342 (Life Technologies) to stain DNA. Cells were mounted in Vectashield medium (Vector Labs) and imaged. Images were analyzed using ImageJ version 1.43 software (National Institutes of Health). In cases where brightness and contrast were adjusted, this was done so linearly and equally to all images in a set.
Immunohistochemistry.
Tissues were fixed in 4% formaldehyde in PBS and then embedded in paraffin, sectioned, and mounted on slides. Sections were deparaffinized in xylene, rehydrated in graded alcohol, incubated in 1 mM EDTA at 99°C for 20 min, and then rinsed in water. Sections were washed 2 times in TBST (50 mM Tris, pH 7.5, 150 mM NaCl, 0.025% Tween 20), then permeabilized in 0.1% Triton X-100 in PBS for 10 min and washed 3 times in TBST, and then blocked in Tris-buffered saline (TBS) plus 1% BSA plus 5% donkey serum (Sigma). Sections were then incubated for 30 min in anti-NP antiserum diluted in TBS plus 1% BSA, washed in TBST, and then incubated for 30 min in Alexa Fluor 488-conjugated donkey anti-rabbit secondary antibody diluted in TBS plus 1% BSA and washed again. The penultimate wash contained 2 µg/ml Hoechst 33342. The sections were mounted in Vectashield (Vector Labs), coverslipped, and imaged as described above. Sections were subsequently stained with hematoxylin and eosin and reimaged.
Nucleotide sequence accession numbers.
The complete data set for all samples is available from the NCBI Short Read Archive (accession no. SRA053624). The complete genome sequences of CASV and GGV have been deposited with the NCBI under accession numbers JQ717261 to JQ717264.
SUPPLEMENTAL MATERIAL
Oligonucleotides used in this study.
Alignments and tables of pairwise percent nucleotide identity between regions of the S and L segments of CAS virus (California Academy of Sciences virus, from annulated tree boas), GGV (Golden Gate virus, from boa constrictors), and CVV (Collierville virus, from a boa constrictor). Download Figure S1, PDF file, 0.3 MB.
ACKNOWLEDGMENTS
J.L.D. is supported by the Howard Hughes Medical Institute. M.D.S. is supported by NIH grant 5T32HL007185-34 and the Pacific Southwest Regional Center of Excellence (PSWRCE, NIH grant U54 AI065359). J.Y.F. was supported by the HHMI Extraordinary Research Opportunities Program (EXROP).
We thank Tara Christiansen, Elisabeth Krow-Lucal, and Peter Fellowes for logistical and technical assistance; Taryn Hook for helping bring IBD to our attention; the snakes Juliet, Larry, and Balthazar; Kurt Thorn and the Nikon Imaging Center at UCSF; Clement Chu, Charles Runckel, Jessica Lund, and the Center for Advanced Technology at UCSF for assistance with sequencing; Illumina and the organizers (Ian Korf, David Haussler, and Keith Bradnam) and participants in Assemblathon 2 (16) for sequencing and assembly of the boa constrictor genome; the UCSF Helen Diller Family Comprehensive Cancer Center Mouse Pathology Core; and the members of the Bay Area Amphibian and Reptile Society and Michael Buchmeier and Jonathon Abraham for helpful discussions.
Footnotes
Citation Stenglein MD, et al. 2012. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: candidate etiological agents for snake inclusion body disease. mBio 3(4):e00180-12. doi:10.1128/mBio.00180-12.
REFERENCES
- 1. Chang L-W, Jacobson ER. 2010. Inclusion body disease, a worldwide infectious disease of boid snakes: a review. J. Exot. Pet Med. 19:216–225 [Google Scholar]
- 2. Axthelm M. 1985. Clinicopathologic and virologic observations of a probable viral disease affecting boid snakes. Proc. Annu. Meet. Am. Assoc. Zoo Vet. 1985:108 [Google Scholar]
- 3. Schumacher J, Jacobson E, Homer B, Gaskin J. 1994. Inclusion body disease in boid snakes. J. Zoo Wildl. Med. 25:511–524 [Google Scholar]
- 4. Jacobson ER, et al. 2001. Partial characterization of retroviruses from boid snakes with inclusion body disease. Am. J. Vet. Res. 62:217–224 [DOI] [PubMed] [Google Scholar]
- 5. Wozniak E, et al. 2000. Isolation and characterization of an antigenically distinct 68-kd protein from nonviral intracytoplasmic inclusions in boa constrictors chronically infected with the inclusion body disease virus (IBDV: Retroviridae). Vet. Pathol. 37:449–459 [DOI] [PubMed] [Google Scholar]
- 6. Pees M, Schmidt V, Marschang RE, Heckers KO, Krautwald-Junghanns M-E. 2010. Prevalence of viral infections in captive collections of boid snakes in Germany. Vet. Rec. 166:422–425 [DOI] [PubMed] [Google Scholar]
- 7. Vancraeynest D, et al. 2006. Inclusion body disease in snakes: a review and description of three cases in boa constrictors in Belgium. Vet. Rec. 158:757–760 [DOI] [PubMed] [Google Scholar]
- 8. Raymond JT, Garner MM, Nordhausen RW, Jacobson ER. 2001. A disease resembling inclusion body disease of boid snakes in captive palm vipers (Bothriechis marchi). J. Vet. Diagn. Invest. 13:82–86 [DOI] [PubMed] [Google Scholar]
- 9. Orós J, Tucker S, Jacobson ER. 1998. Inclusion body disease in two captive boas in the Canary Islands. Vet. Rec. 143:283–285 [DOI] [PubMed] [Google Scholar]
- 10. Carlisle-Nowak MS, et al. 1998. Inclusion body disease in two captive Australian pythons (Morelia spilota variegata and Morelia spilota spilota). Aust. Vet. J. 76:98–100 [DOI] [PubMed] [Google Scholar]
- 11. Tang P, Chiu C. 2010. Metagenomics for the discovery of novel human viruses. Future Microbiol. 5:177–189 [DOI] [PubMed] [Google Scholar]
- 12. Relman DA. 2011. Microbial genomics and infectious diseases. N. Engl. J. Med. 365:347–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mokili JL, Rohwer F, Dutilh BE. 2012. Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2:63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang D, et al. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. U. S. A. 99:15687–15692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kistler AL, et al. 2008. Recovery of divergent avian bornaviruses from cases of proventricular dilatation disease: identification of a candidate etiologic agent. Virol. J. 5:88 http://dx.doi.org/10.1186/1743-422X-5-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Genome Center. University of California, Davis 2011-2012. Assemblathon 2. Genome Center, University of California, Davis, CA. http://assemblathon.org/pages/download-data [Google Scholar]
- 17. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kircher M, Sawyer S, Meyer M. 2012. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40:e3 http://dx.doi.org/10.1093/nar/gkr771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huder JB, et al. 2002. Identification and characterization of two closely related unclassifiable endogenous retroviruses in pythons (Python molurus and Python curtus). J. Virol. 76:7607–7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salvato MS, et al. 2011. Arenaviridae, p 715–723 In King AMQ, Adams MJ, Carstens EB, Lefkowitz E, Virus taxonomy: classification and nomenclature of viruses: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, San Diego, CA. [Google Scholar]
- 21. Buchmeier MJ, de la Torre JC, Peters CJ. 2007. Arenaviridae: the viruses and their replication, p 1791–1828 In Knipe DM, Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 22. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Salvato M, Shimomaye E, Oldstone MB. 1989. The primary structure of the lymphocytic choriomeningitis virus L gene encodes a putative RNA polymerase. Virology 169:377–384 [DOI] [PubMed] [Google Scholar]
- 24. Young PR, Howard CR. 1983. Fine structure analysis of Pichinde virus nucleocapsids. J. Gen. Virol. 64:833–842 [DOI] [PubMed] [Google Scholar]
- 25. Emonet SF, de la Torre JC, Domingo E, Sevilla N. 2009. Arenavirus genetic diversity and its biological implications. Infect. Genet. Evol. 9:417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gonzalez JP, Emonet S, de Lamballerie X, Charrel R. 2007. Arenaviruses. Curr. Top. Microbiol. Immunol. 315:253–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Salvato MS, Shimomaye EM. 1989. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology 173:1–10 [DOI] [PubMed] [Google Scholar]
- 28. Buchmeier MJ, Parekh BS. 1987. Protein structure and expression among arenaviruses. Curr. Top. Microbiol. Immunol. 133:41–57 [DOI] [PubMed] [Google Scholar]
- 29. Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Urata S, de la Torre JC. 2011. Arenavirus budding. Adv. Virol. 2011:180326 http://dx.doi.org/10.1155/2011/180326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Strecker T, et al. 2003. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles. J. Virol. 77:10700–10705 (Erratum, 77:12927.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perez M, Greenwald DL, de la Torre JC. 2004. Myristoylation of the RING finger Z protein is essential for arenavirus budding. J. Virol. 78:11443–11448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bieniasz PD. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55–63 [DOI] [PubMed] [Google Scholar]
- 34. Dolnik O, Kolesnikova L, Stevermann L, Becker S. 2010. Tsg101 is recruited by a late domain of the nucleocapsid protein to support budding of Marburg virus-like particles. J. Virol. 84:7847–7856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mi S, et al. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789 [DOI] [PubMed] [Google Scholar]
- 36. Gallaher WR. 1996. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell 85:477–478 [DOI] [PubMed] [Google Scholar]
- 37. Kielian M, Rey FA. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4:67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. 1998. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. U. S. A. 95:5762–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Freshney RI. 2010. Culture of animal cells: a manual of basic technique and specialized applications, 6th ed. Wiley-Blackwell, Hoboken, NJ [Google Scholar]
- 40. Ortiz-Riaño E, Cheng BYH, de la Torre JC, Martínez-Sobrido L. 2011. The C-terminal region of lymphocytic choriomeningitis virus nucleoprotein contains distinct and segregable functional domains involved in NP-Z interaction and counteraction of the type I interferon response. J. Virol. 85:13038–13048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Young PR, Chanas AC, Lee SR, Gould EA, Howard CR. 1987. Localization of an arenavirus protein in the nuclei of infected cells. J. Gen. Virol. 68:2465–2470 [DOI] [PubMed] [Google Scholar]
- 42. Charrel RN, et al. 2011. Arenaviruses and hantaviruses: from epidemiology and genomics to antivirals. Antiviral Res. 90:102–114 [DOI] [PubMed] [Google Scholar]
- 43. Oldstone MBA. 2006. Viral persistence: parameters, mechanisms and future predictions. Virology 344:111–118 [DOI] [PubMed] [Google Scholar]
- 44. Gallaher WR, DiSimone C, Buchmeier MJ. 2001. The viral transmembrane superfamily: possible divergence of Arenavirus and Filovirus glycoproteins from a common RNA virus ancestor. BMC Microbiol. 1:1 http://dx.doi.org/10.1186/1471-2180-1-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Igonet S, et al. 2011. X-ray structure of the arenavirus glycoprotein GP2 in its postfusion hairpin conformation. Proc. Natl. Acad. Sci. U. S. A. 108:19967–19972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Charrel RN, et al. 2002. Phylogeny of new world arenaviruses based on the complete coding sequences of the small genomic segment identified an evolutionary lineage produced by intrasegmental recombination. Biochem. Biophys. Res. Commun. 296:1118–1124 [DOI] [PubMed] [Google Scholar]
- 47. Montali RJ, et al. 1989. A new transmissible viral hepatitis of marmosets and tamarins. J. Infect. Dis. 160:759–765 [DOI] [PubMed] [Google Scholar]
- 48. Stephensen CB, et al. 1991. Isolation of an arenavirus from a marmoset with callitrichid hepatitis and its serologic association with disease. J. Virol. 65:3995–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Montali RJ, et al. 1993. A common-source outbreak of callitrichid hepatitis in captive tamarins and marmosets. J. Infect. Dis. 167:946–950 [DOI] [PubMed] [Google Scholar]
- 50. Stephensen CB, Park JY, Blount SR. 1995. cDNA sequence analysis confirms that the etiologic agent of callitrichid hepatitis is lymphocytic choriomeningitis virus. J. Virol. 69:1349–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kistler AL, Smith JM, Greninger AL, Derisi JL, Ganem D. 2010. Analysis of naturally occurring avian bornavirus infection and transmission during an outbreak of proventricular dilatation disease among captive psittacine birds. J. Virol. 84:2176–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mirhosseini N, Gray PL, Tizard I, Payne S. 2012. Complete genome sequence of avian bornavirus genotype 1 from a macaw with proventricular dilatation disease. J. Virol. 86:7023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rinder M, et al. 2009. Broad tissue and cell tropism of avian bornavirus in parrots with proventricular dilatation disease. J. Virol. 83:5401–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weissenböck H, et al. 2009. Avian bornaviruses in psittacine birds from Europe and Australia with proventricular dilatation disease. Emerg. Infect. Dis. 15:1453–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ambrosio A, Saavedra M, Mariani M, Gamboa G, Maiza A. 2011. Argentine hemorrhagic fever vaccines. Hum. Vaccin. 7:694–700 [DOI] [PubMed] [Google Scholar]
- 56. Maiztegui JI, et al. 1998. Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. AHF Study Group. J. Infect. Dis. 177:277–283 [DOI] [PubMed] [Google Scholar]
- 57. Fisher-Hoch SP, McCormick JB. 2004. Lassa fever vaccine. Expert Rev. Vaccines 3:189–197 [DOI] [PubMed] [Google Scholar]
- 58. McCormick JB, et al. 1986. Lassa fever. Effective therapy with ribavirin. N. Engl. J. Med. 314:20–26 [DOI] [PubMed] [Google Scholar]
- 59. Emonet SE, Urata S, de la Torre JC. 2011. Arenavirus reverse genetics: new approaches for the investigation of arenavirus biology and development of antiviral strategies. Virology 411:416–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yozwiak NL, et al. 2012. Virus identification in unknown tropical febrile illness cases using deep sequencing. PLoS Negl. Trop. Dis. 6:e1485 http://dx.doi.org/10.1371/journal.pntd.0001485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bozdech Z, et al. 2003. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 4:R9 http://dx.doi.org/10.1186/gb-2003-4-2-r9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Scotto-Lavino E, Du G, Frohman MA. 2006. 5′ end cDNA amplification using classic RACE. Nat. Protoc. 1:2555–2562 [DOI] [PubMed] [Google Scholar]
- 64. Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659 [DOI] [PubMed] [Google Scholar]
- 65. Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 66. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligonucleotides used in this study.
Alignments and tables of pairwise percent nucleotide identity between regions of the S and L segments of CAS virus (California Academy of Sciences virus, from annulated tree boas), GGV (Golden Gate virus, from boa constrictors), and CVV (Collierville virus, from a boa constrictor). Download Figure S1, PDF file, 0.3 MB.