ABSTRACT
Subminimal inhibitory concentrations of antibiotics have been shown to induce bacterial biofilm formation. Few studies have investigated antibiotic-induced biofilm formation in Staphylococcus aureus, an important human pathogen. Our goal was to measure S. aureus biofilm formation in the presence of low levels of β-lactam antibiotics. Fifteen phylogenetically diverse methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-sensitive S. aureus (MSSA) strains were employed. Methicillin, ampicillin, amoxicillin, and cloxacillin were added to cultures at concentrations ranging from 0× to 1× MIC. Biofilm formation was measured in 96-well microtiter plates using a crystal violet binding assay. Autoaggregation was measured using a visual test tube settling assay. Extracellular DNA was quantitated using agarose gel electrophoresis. All four antibiotics induced biofilm formation in some strains. The amount of biofilm induction was as high as 10-fold and was inversely proportional to the amount of biofilm produced by the strain in the absence of antibiotics. MRSA strains of lineages USA300, USA400, and USA500 exhibited the highest levels of methicillin-induced biofilm induction. Biofilm formation induced by low-level methicillin was inhibited by DNase. Low-level methicillin also induced DNase-sensitive autoaggregation and extracellular DNA release. The biofilm induction phenotype was absent in a strain deficient in autolysin (atl). Our findings demonstrate that subminimal inhibitory concentrations of β-lactam antibiotics significantly induce autolysin-dependent extracellular DNA release and biofilm formation in some strains of S. aureus.
IMPORTANCE
The widespread use of antibiotics as growth promoters in agriculture may expose bacteria to low levels of the drugs. The aim of this study was to investigate the effects of low levels of antibiotics on bacterial autoaggregation and biofilm formation, two processes that have been shown to foster genetic exchange and antibiotic resistance. We found that low levels of β-lactam antibiotics, a class commonly used in both clinical and agricultural settings, caused significant autoaggregation and biofilm formation by the important human pathogen Staphylococcus aureus. Both processes were dependent on cell lysis and release of DNA into the environment. The effect was most pronounced among multidrug-resistant strains known as methicillin-resistant S. aureus (MRSA). These results may shed light on the recalcitrance of some bacterial infections to antibiotic treatment in clinical settings and the evolution of antibiotic-resistant bacteria in agricultural settings.
Introduction
Staphylococcus aureus continues to be a major cause of health care-related and community-associated infections. The emergence of multidrug-resistant strains such as methicillin-resistant Staphylococcus aureus (MRSA) has contributed to the spread of this bacterium (1). In addition, S. aureus often forms matrix-encased biofilms on tissues and medical devices, which confer additional drug resistance and further complicate treatment (2).
Although S. aureus biofilms have been shown to be resistant to killing by antibiotics at concentrations sufficient to kill planktonic cells, several studies have shown that S. aureus biofilm formation can be stimulated by subminimal inhibitory concentrations (sub-MICs) of some antibiotics. The cell wall-active antibiotics oxacillin (3), cephalothin (4), cephalexin (5), and vancomycin (3) and the protein synthesis inhibitor linezolid (6) have been shown to stimulate S. aureus biofilm formation in vitro by as much as 4-fold when present at sub-MICs. Enhanced biofilm formation in the presence of sub-MICs of antibiotics is a common phenomenon among bacteria and is thought to result from a global response to cell stress (7). Other studies found an inhibitory effect or no effect on S. aureus biofilm formation by sub-MICs of other cell wall-active antibiotics (cefamandole, cefuroxime, nafcillin, teicoplanin, and vancomycin) and protein synthesis inhibitors (tetracycline and roxithromycin) (5, 6, 8, 9). Most of these studies, however, utilized a single strain of S. aureus and tested antibiotics at a single or limited number of antibiotic concentrations, usually ≤1/2× MIC.
Antibiotic-induced biofilm formation may be a clinically relevant process because bacteria are exposed to sub-MIC antibiotics during the normal course of antibiotic therapy (10). The widespread use of antibiotics as growth promoters in agriculture may also expose bacteria to low levels of the drugs (11). The goal of the present study was to measure biofilm formation by a variety of S. aureus strains in vitro in response to sub-MICs of β-lactam antibiotics, a class of antibiotic commonly used in both clinical and agricultural settings.
RESULTS
β-Lactam antibiotics induce S. aureus biofilm formation.
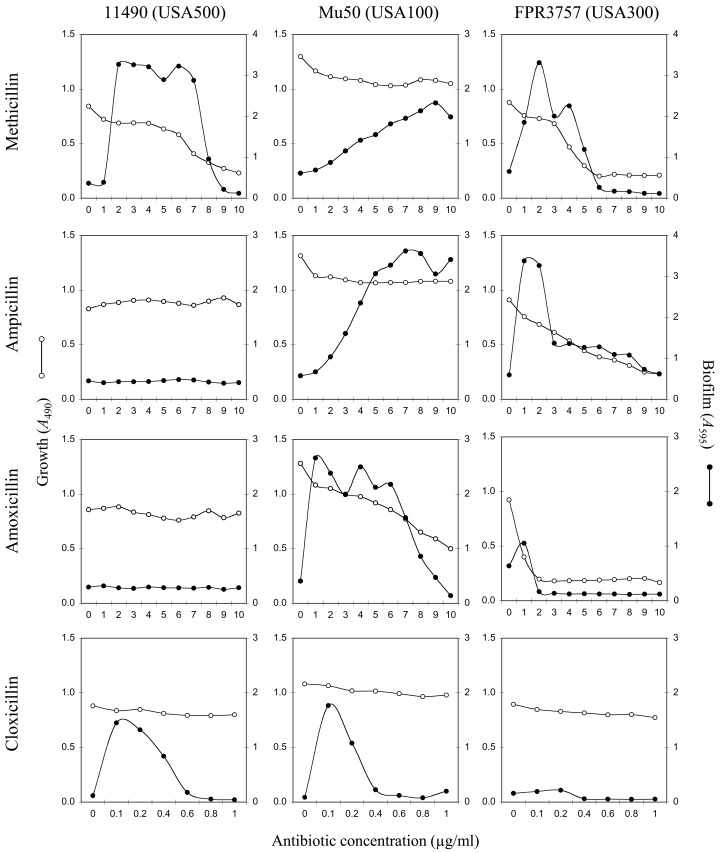
Figure 1 shows growth (A490) and biofilm formation (A595) by methicillin-resistant S. aureus (MRSA) strains 11490 (USA500), Mu50 (USA100), and FPR3757 (USA300) in the presence of methicillin, ampicillin, amoxicillin, and cloxacillin at concentrations ranging from 0 to 10 µg/ml. The MIC values for these four antibiotics against these three strains ranged from 2 to >10 µg/ml. All four antibiotics significantly induced biofilm in at least one strain, although the pattern of biofilm induction was strain and antibiotic dependent. Only methicillin induced biofilm formation in all three strains. Most antibiotics that induced biofilm formation exhibited a biphasic dose-response curve characterized by low-dose stimulation of biofilm formation and high-dose inhibition.
FIG 1 .
Bacterial growth and biofilm formation by three MRSA strains (strains 11490, Mu50, and FPR3757) in the presence of sub-MICs of four β-lactam antibiotics. Bacterial growth (A490) is indicated on the left-hand y axes, biofilm formation (A595) is indicated on the right-hand y axes, and antibiotic concentration is indicated on the x axes. Values show average absorbance values for duplicate wells. Error bars were omitted for clarity.
Methicillin induces biofilm formation in strains that exhibit a weak biofilm phenotype.
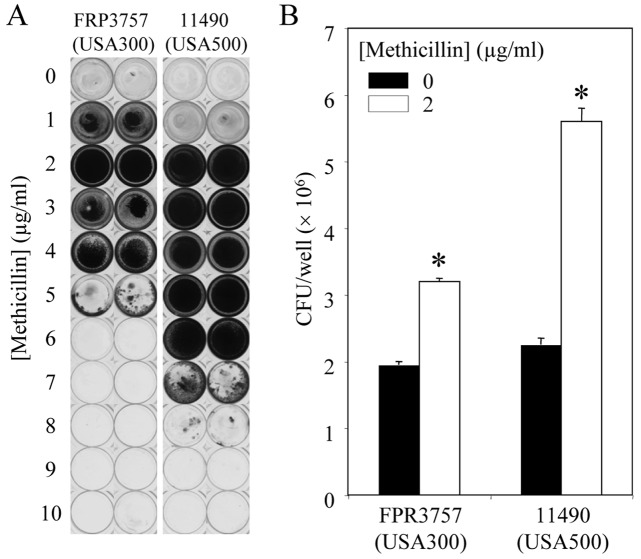
Since sub-MICs of methicillin induced biofilm in all three MRSA strains, we tested the ability of sub-MICs of methicillin to induce biofilm formation in a total of 15 MRSA and methicillin-sensitive S. aureus (MSSA) strains of diverse origin (Table 1). Biofilm formation was significantly induced in six strains, five of which were MRSA strains. The methicillin concentrations that induced maximum biofilm formation in these five strains (highest A595 value) ranged from 1 to 7 µg/ml. The maximum fold level of biofilm induction was inversely correlated to the amount of biofilm produced by the test strain in the absence of methicillin (P < 0.05). Figure 2A shows photographs of crystal violet-stained wells that were inoculated with S. aureus 11490 (USA500) and FPR3757 (USA300) and cultured in the presence of increasing concentrations of methicillin. Increased crystal violet binding at 2 µg/ml methicillin correlated with a significant increase in biofilm CFU values (Fig. 2B).
TABLE 1 .
Induction of S. aureus biofilm formation by sub-MICs of methicillin
|
S. aureus strain (lineage) |
Maximum biofilm inductiona |
Basal biofilm formationb |
Methicillin MIC (µg/ml) |
SCCmec genotypec |
agr genotype |
Reference or source |
|---|---|---|---|---|---|---|
| LAC (USA300) | 11.8* | 15 | >8 | IV | I | 31 |
| 11490 (USA500) | 10.5* | 12 | >8 | IV | I | PHRIe |
| MW2 (USA400) | 8.9* | 13 | >8 | IVa | III | 32 |
| Newman | 6.2* | 19 | 2 | − | I | 33 |
| FPR3757 (USA300) | 5.8* | 16 | 6 | IV | I | 34 |
| Mu50 (USA100) | 5.1* | 12 | >8 | II | II | 35 |
| 476 | 1.8 | 12 | 3 | − | III | 36 |
| COL | 1.4 | 16 | >8 | I | I | 37 |
| RF122 | 1.2 | 15 | 3 | − | II | 38 |
| 252 | 1.2 | 61 | >8 | II | III | 36 |
| 383 | 1.1 | 104 | >8 | − | NDd | 39 |
| SH1000 | 1.0 | 100 | 1 | − | I | 40 |
| MZ100 | 1.0 | 122 | 1 | − | I | 41 |
| N315 | 1.0 | 119 | 2 | II | II | 35 |
| SA113 | 1.0 | 119 | 1 | − | I | 42 |
Maximum fold increase in crystal violet binding in the presence of sub-MICs of methicillin. The method used to calculate the value is described in Materials and Methods. Values are averages from 2 or 3 experiments. Standard errors were ±7% on average. Values that were significantly different (P < 0.05 by Student’s t test) from the values for the no-antibiotic control are indicated by an asterisk.
Relative amount of crystal violet binding in the absence of methicillin compared to that of strain SH1000 in the absence of methicillin. The values were calculated as follows: (absorbance of the test strain/absorbance of strain SH1000) × 100.
The staphylococcal cassette chromosome mec element (SCCmec) genotype is shown. A minus sign indicates the absence of SCCmec and that the strain is an MSSA strain.
ND, not determined.
Barry Kreiswirth at the Public Health Research Institute (PHRI), Newark, NJ, supplied this strain.
FIG 2 .
Biofilm formation by S. aureus strains FPR3757 (USA300) and 11490 (USA500) in the presence of sub-MICs of methicillin. (A) Photographs of crystal violet-stained microtiter plate wells. Duplicate wells are shown. (B) Average biofilm CFU/well values for biofilms cultured in 0 or 2 µg/ml methicillin. Error bars indicate ranges. Values that were significantly different (P < 0.05) from those of the no-methicillin control are indicated by an asterisk.
Methicillin-induced biofilm formation is inhibited by DNase.
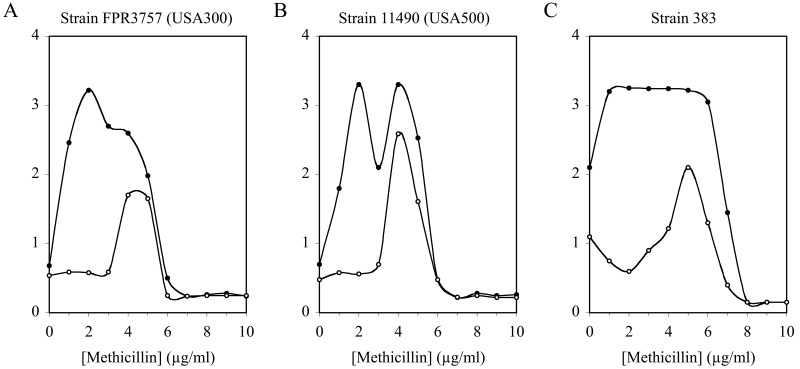
Previous studies showed that extracellular DNA (eDNA) constitutes a major adhesive polymer in S. aureus biofilms (12). To determine whether eDNA plays a role in methicillin-induced biofilms, we cultured strains 11490 (USA500) and FPR3757 (USA300) in sub-MICs of methicillin in the presence or absence of recombinant human DNase I (rhDNase), a potent inhibitor of S. aureus biofilm formation (13). Figure 3A and B show that rhDNase significantly inhibited methicillin-induced biofilm formation by both strains. Inhibition was more efficient at low methicillin concentrations (0 to 3 µg/ml) than at higher methicillin concentrations (4 to 5 µg/ml). For comparison, we measured the effect of rhDNase on biofilm formation by noninducible S. aureus strain 383 cultured in the presence of sub-MICs of methicillin (Fig. 3C). Like strains 11490 (USA500) and FPR3757 (USA300), strain 383 exhibited a biofilm phenotype that was inhibited by rhDNase more efficiently at low methicillin concentrations than at high methicillin concentrations. Dispersin B, an enzyme that degrades S. aureus poly-N-acetylglucosamine (PNAG) biofilm polysaccharide (12), had no effect on methicillin-induced biofilm formation in any of the three strains, whereas proteinase K caused nearly complete inhibition of biofilm formation by all three strains at all methicillin concentrations (data not shown). These findings are consistent with the observation that MRSA strains produce predominantly protein- and DNA-based biofilms (14). None of the three enzymes had a significant effect on the MIC of methicillin against any of the three test strains (data not shown).
FIG 3 .
Effect of rhDNase on biofilm formation by S. aureus in sub-MICs of methicillin. Strains FPR3757 (USA300), 11490 (USA500), and 383 were cultured in broth supplemented with 10 µg/ml rhDNase (open circles) or no enzyme (filled circles). Values show average absorbance values for duplicate wells. Error bars were omitted for clarity.
Sub-MICs of methicillin induce autoaggregation and eDNA release.
Autoaggregation plays an important role in maintaining the stability of S. aureus biofilm colonies (15). We used a visual test tube settling assay to investigate the effects of sub-MICs of methicillin on autoaggregation by S. aureus 11490 (USA500) and FPR3757 (USA300) (Fig. 4). Cells cultured in broth supplemented with 2 µg/ml methicillin rapidly settled to the bottom of the tube, whereas control cells cultured without methicillin remained in suspension. Cells cultured in the broth supplemented with 2 µg/ml methicillin and then treated with 10 µg/ml rhDNase remained in suspension, indicating that sub-MICs of methicillin induce eDNA-dependent intercellular adhesin. Supernatants of S. aureus FPR3757 (USA300) biofilms cultured in 1 to 3 µg/ml methicillin, as well as supernatants of S. aureus 11490 (USA500) biofilms cultured in 3 to 4 µg/ml methicillin, contained high-molecular-weight DNA (Fig. 5), which correlated with biofilm formation (Fig. 2) and is consistent with eDNA release (16, 17).
FIG 4 .
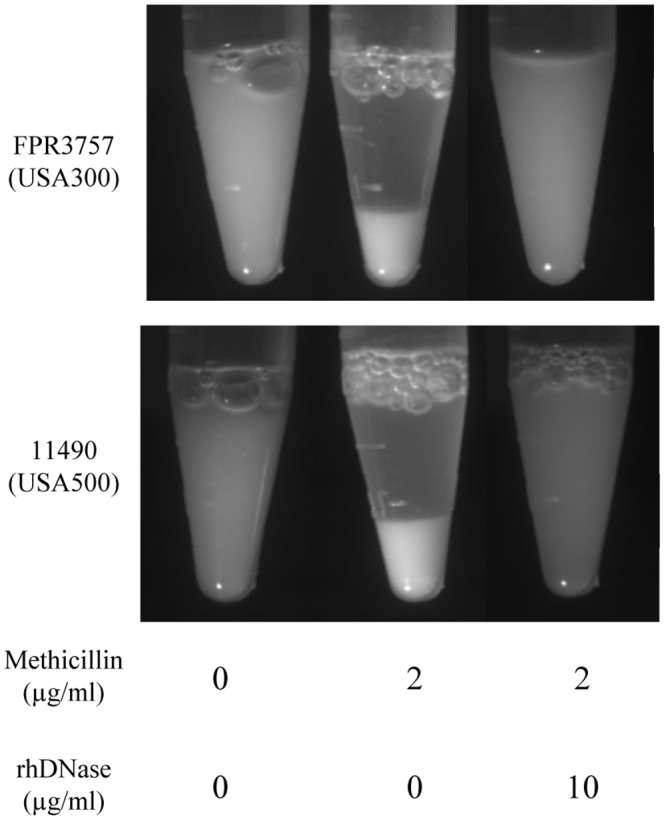
Sub-MICs of methicillin induce autoaggregation in S. aureus. Strains FPR3757 (USA300) and 11490 (USA500) were cultured for 18 h in broth supplemented with 0 or 2 µg/ml methicillin and then treated with 0 or 10 µg/ml rhDNase. The cells were rinsed with saline and transferred to a microcentrifuge tube. The tubes were incubated statically for 10 min and then photographed.
FIG 5 .
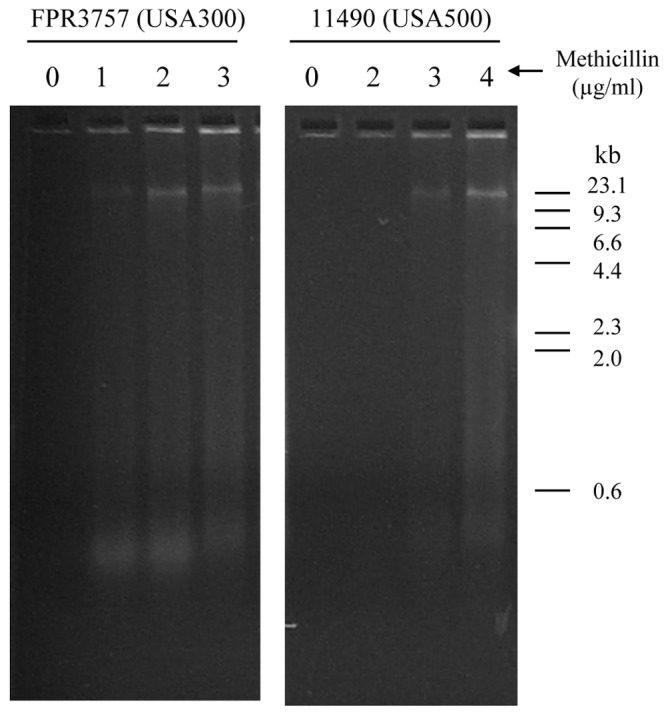
Sub-MICs of methicillin induce eDNA release in S. aureus. Biofilms of strains FPR3757 (USA300) and 11490 (USA500) were cultured for 18 h in broth supplemented with increasing concentrations of methicillin. Biofilm supernatants were analyzed by agarose gel electrophoresis. The sizes (in kilobases) of DNA molecular size markers electrophoresed in an adjacent lane are shown to the right of the gels.
Methicillin-induced biofilm formation is dependent on autolysin activity.
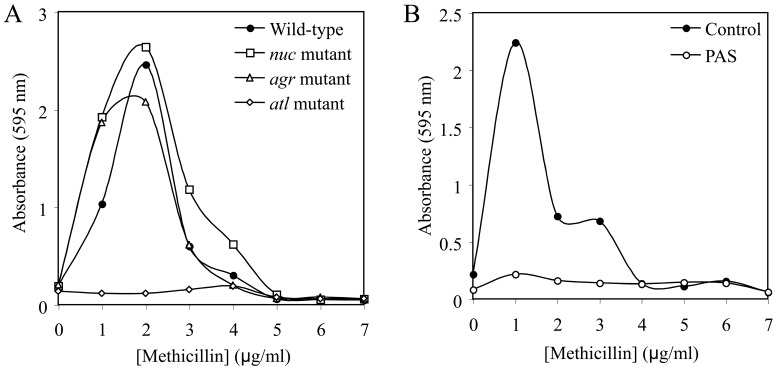
The production of eDNA in S. aureus has been shown to result from cell lysis mediated by the autolytic enzyme AtlA (18). To determine whether AtlA plays a role in methicillin-induced biofilm formation, we cultured S. aureus strain LAC and isogenic atl mutant strain KB4051 in sub-MICs of methicillin and measured growth and biofilm formation (Fig. 6). We also tested strains deficient in the production of thermonuclease (nuc) and the accessory gene regulator (agr) quorum-sensing system, both of which have been shown to play a role in S. aureus biofilm formation and biofilm dispersal in some strains (19, 20). The biofilm induction phenotype was absent in the atl mutant strain, whereas inactivation of the nuc locus or agr gene had little or no effect on methicillin-induced biofilm formation. All three mutants exhibited the same MIC for methicillin as parental strain LAC (data not shown). We also measured biofilm induction by strain LAC cultured in the presence of polyanethole sulfonate (PAS), an inhibitor of S. aureus autolysis (17, 18). PAS completely eliminated the biofilm induction phenotype (Fig. 6B) but did not affect bacterial growth or the methicillin MIC (data not shown).
FIG 6 .
Biofilm formation by S. aureus strain LAC in sub-MICs of methicillin. (A) Biofilm formation by wild-type S. aureus strain LAC and isogenic nuc, agr, and atl mutant strains in sub-MICs of methicillin. (B) Biofilm formation by strain LAC in sub-MICs of methicillin in the presence or absence of 500 µg/ml of the autolysis inhibitor polyanethole sulfonate (PAS). Values in both panels show average absorbance for duplicate wells. Error bars were omitted for clarity.
DISCUSSION
The results of the present study demonstrate that subminimal inhibitory concentrations of four different β-lactam antibiotics significantly induce biofilm formation in some strains of S. aureus. We found that sub-MICs of methicillin induced biofilm formation in S. aureus strains that exhibited a low basal level of biofilm in the absence of antibiotics, but not in strains that exhibited a high basal level of biofilm. These findings suggest that the antibiotic-induced pathway is turned on in strains that exhibit a strong biofilm phenotype such as strain SH1000, but not in strains that exhibit a weak biofilm phenotype such as strain LAC. A similar inverse relation between basal biofilm production and biofilm inducibility was exhibited by Staphylococcus epidermidis in response to sub-MICs of vancomycin, tigecycline, novobiocin, linezolid, and various fluoroquinolones (21, 22). Screening for mutants deficient in the biofilm induction phenotype may be a useful strategy for discovering the effectors of these antibiotic-induced pathways.
The patterns of β-lactam-induced biofilm formation were strain and antibiotic dependent (Fig. 1). When biofilm induction occurred, the fundamental nature of the dose response was biphasic, characterized by low-dose stimulation of biofilm formation and high-dose inhibition. Multiphasic dose-response curves are characteristic of many chemicals, drugs, hormones, biological molecules, and physical stressors (23). In addition, patterns of β-lactam-induced biofilm formation exhibited a bimodal dose response, with each peak exhibiting a different level of sensitivity to inhibition by rhDNase (Fig. 3). These findings suggest that S. aureus exhibits two distinct biofilm phenotypes, each with a different biofilm matrix composition.
Our findings demonstrate that β-lactam-induced S. aureus biofilm formation is dependent on the production of eDNA. Previous studies showed that eDNA constitutes a major biofilm matrix adhesin in S. aureus biofilms cultured in the absence of antibiotics (12, 17–20). The production of eDNA in S. aureus has been shown to result from cell lysis mediated by the autolysin AtlA (18). Consistent with these results, we found that an atl mutant strain of S. aureus exhibited reduced biofilm induction (Fig. 6A) and that polyanethole sulfonate, an inhibitor of autolysis, inhibited the biofilm induction response (Fig. 6B). It is possible that low-level β-lactams induce S. aureus biofilm formation by upregulating AtlA production either directly or through modulation of the expression of other regulatory proteins. Proteins that regulate S. aureus atl expression include the alternative sigma factor σB (18), the DNA-binding protein SarA (24), and the glycopeptide resistance-associated two-component system GraRS (25).
S. aureus thermonuclease encoded by the nuc gene has been shown to be a negative regulator of biofilm formation (16, 20, 24). Kiedrowski et al. (20) found that an S. aureus USA300 strain carrying a mutation in nuc exhibited increased biofilm formation and increased high-molecular-weight eDNA, phenotypes identical to those exhibited by strain FPR3757 (USA300) cultured in low-level methicillin (Fig. 2 and 4). These findings suggest that low-level methicillin may suppress nuc expression. However, we found that a nuc mutation in USA300 strain LAC did not alter the amount of biofilm formed in the absence of antibiotics or the amount of biofilm induction caused by low-level methicillin (Fig. 6). One possible explanation for these findings is that thermonuclease mediates biofilm dispersal, but only under conditions of strong biofilm induction (16).
The agr quorum-sensing system has been shown to regulate S. aureus biofilm formation through modulation of biofilm dispersal (19). Subrt et al. (4) showed that the β-lactam antibiotic cephalothin, when present at 1/4× MIC, induced S. aureus biofilm formation but did not affect agr expression. Consistent with these results, we found that an agr mutation in USA300 strain LAC did not affect the amount of biofilm induction caused by low-level methicillin (Fig. 6). Thus, agr quorum sensing evidently does not play a role in β-lactam-induced biofilm formation in S. aureus.
Biofilm formation by MRSA strains is often dependent on the expression of fibronectin-binding proteins (FnBPs), whereas biofilm formation by MSSA strains usually depends on the production of poly-N-acetylglucosamine (PNAG) exopolysaccharide (14) and eDNA (17). Consistent with these observations, we found that the PNAG-degrading enzyme dispersin B had no effect on methicillin-induced biofilm formation by MRSA strains. Bisognano et al. (26) showed that expression of S. aureus FnBPs was increased in some highly quinolone-resistant strains when cultured in medium supplemented with 1/4× MIC ciprofloxacin. It is possible that sub-MICs of methicillin also induce FnBP expression. Another potential target for sub-MIC β-lactam induction may be spa (encoding protein A), which may play a role in biofilm formation (27) and was shown to be induced by low concentrations of cephalothin (4).
Our findings may have clinical relevance because β-lactam antibiotics are commonly used in both clinical and agricultural settings. In the clinic, bacteria may be exposed to sub-MICs of antibiotics at the beginning and end of a dosing regimen, between doses, or continuously during low-dose therapy (10). In addition, cells buried deep within a biofilm colony may be exposed to sub-MICs of antibiotics because of diffusion gradients (28). Given the high level of biofilm induction and the observed variability among antibiotics and strains, it may be useful to test clinical isolates for biofilm inducibility in order to optimize antibiotic chemotherapy in clinical settings. The widespread use of β-lactams as growth promoters in agriculture may also expose bacteria to low levels of these drugs, and there is concern that subtherapeutic antibiotics may promote antibiotic resistance in agricultural settings (29). It is possible that enhanced biofilm formation by S. aureus in response to low-dose β-lactams may foster genetic exchange and contribute to the spread of antibiotic resistance genes, which may represent an additional drawback of administering β-lactams to farm animals.
MATERIALS AND METHODS
Antibiotics, enzymes and chemicals.
Methicillin was purchased from Sigma-Aldrich (St. Louis, MO), ampicillin was from IBI Scientific (Peosta, IA), and amoxicillin and cloxacillin were from MP Biomedicals (Solon, OH). Stock solutions of antibiotics were prepared in sterile H2O at a 1-mg/ml concentration. Recombinant human DNase I (rhDNase) was obtained from Genentech (South San Francisco, CA). Dispersin B (DspB), a poly-N-acetylglucosamine (PNAG)-specific glycoside hydrolase (12), was obtained from Kane Biotech (Winnipeg, Manitoba, Canada). Proteinase K was purchased from Sigma-Aldrich. Sodium polyanethole sulfonate was purchased from Polysciences, Inc. (Warrington, PA).
Bacterial strains and growth conditions.
The S. aureus strains used in this study are listed in Table 1. In some experiments, strains KB4051 (LAC Δatl), AH1680 (LAC nuc::LtrB) and AH1292 (LAC Δagr::TetM) (20, 30) were also employed. Strains were passaged weekly on blood agar and stored at 4°C. Bacteria were cultured in filter-sterilized tryptic soy broth (Becton, Dickinson, Heidelberg, Germany) supplemented with 6 g/liter yeast extract and 8 g/liter glucose. All cultures were incubated statically at 37°C.
Biofilm assay.
Inocula were prepared in fresh broth from 18-h-old agar colonies as previously described (12). Aliquots of inocula (200 µl each, ca. 104 to 105 CFU/ml) were transferred to the wells of a 96-well microtiter plate (catalog no. 353936; Falcon) and incubated for 18 to 24 h. For crystal violet staining, the wells were rinsed with water to remove loosely adherent cells and then stained for 1 min with 200 µl of Gram’s crystal violet. The wells were then rinsed with water and dried. The amount of biofilm biomass was quantitated by destaining the wells with 200 µl of 33% acetic acid and then measuring the absorbance of the crystal violet solution in a microplate spectrophotometer set at 595 nm. In cases where the absorbance values were above the dynamic range of the instrument (4.0 optical density units [OD]), the samples were diluted with an equal volume of 33% acetic acid prior to the measurements. To quantitate the amount of biofilm induction, strains were cultured in broth containing 0 to 10 µg/ml of methicillin in 1-µg increments. The maximum biofilm induction value for each strain (Table 1) was equal to the highest observed absorbance value divided by the absorbance value at 0 µg/ml methicillin. For biofilm CFU enumeration, the wells were rinsed three times with sterile phosphate-buffered saline (PBS) and then treated with 10 µg/ml rhDNase to detach the biofilm cells. Biofilm CFU values were enumerated by dilution plating on agar.
Autoaggregation assay.
Bacteria were cultured in 100-mm-diameter tissue culture-treated petri dishes (catalog no. 353003; Falcon) in 20 ml of broth supplemented with 0 or 2 µg/ml methicillin. After 18 to 24 h of growth, the broth was carefully removed, and 500 µl of PBS was added to the dish. The cells were scraped from the surface using a cell scraper and transferred to a 1.5-ml microcentrifuge tube. The cells were resuspended by gentle pipetting, and the tubes were then incubated statically for 10 min and photographed. Some samples were treated with 10 µg/ml of rhDNase for 10 min prior to resuspension.
Extracellular DNA assay.
Bacteria were cultured in 6-well tissue culture-treated polystyrene microtiter plate wells (catalog no. 353046; Falcon) in 4 ml of broth supplemented with increasing concentrations of methicillin. After 18 to 24 h of growth, the broth was carefully removed and 1 ml of TE buffer (10 mM Tris, 1 mM EDTA [pH 8]) was added to the wells. The cells were scraped from the surface using a cell scraper, and the cells were transferred to a 1.5-ml microcentrifuge tube. The tubes were centrifuged at 13,000 rpm for 25 s, and the supernatant was removed. The cell pellets were vigorously resuspended in 200 µl of TE buffer, and the tubes were recentrifuged. A total of 30 µl of the supernatant was electrophoresed through a 1% agarose gel. DNA was visualized by staining with ethidium bromide.
Reproducibility of results and statistics.
All biofilm assays were carried out in duplicate or triplicate wells, which exhibited an average variation in absorbance values of 7%. All assays were repeated 2 to 5 times, and in all cases, the observed patterns of biofilm induction and inhibition were reproducible. The significance of differences between absorbance values was calculated using a Student’s t test. Correlation was measured using a Pearson correlation coefficient.
ACKNOWLEDGMENTS
We thank Barry Kreiswirth (Public Health Research Institute, Newark, NJ) for providing bacterial strains.
This work was supported by NIH grants AI82392 (to J.B.K.), AI83211 (to A.R.H.), and AI83211 and AI038901 (to K.W.B.) and a grant from Genentech (to J.B.K.).
Footnotes
Citation Kaplan JB, et al. 2012. Low levels of β-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. mBio 3(4):e00198-12 doi:10.1128/mBio.00198-12.
REFERENCES
- 1. Harbarth S. 2006. Control of endemic methicillin-resistant Staphylococcus aureus–recent advances and future challenges. Clin. Microbiol. Infect. 12:1154–1162 [DOI] [PubMed] [Google Scholar]
- 2. Archer NK, et al. 2011. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2:445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mirani ZA, Jamil N. 2011. Effect of sub-lethal doses of vancomycin and oxacillin on biofilm formation by vancomycin intermediate resistant Staphylococcus aureus. J. Basic Microbiol. 51:191–195 [DOI] [PubMed] [Google Scholar]
- 4. Subrt N, Mesak LR, Davies J. 2011. Modulation of virulence gene expression by cell wall active antibiotics in Staphylococcus aureus. J. Antimicrob. Chemother. 66:979–984 [DOI] [PubMed] [Google Scholar]
- 5. Haddadin RN, Saleh S, Al-Adham IS, Buultjens TE, Collier PJ. 2010. The effect of subminimal inhibitory concentrations of antibiotics on virulence factors expressed by Staphylococcus aureus biofilms. J. Appl. Microbiol. 108:1281–1291 [DOI] [PubMed] [Google Scholar]
- 6. Frank KL, Reichert EJ, Piper KE, Patel R. 2007. In vitro effects of antimicrobial agents on planktonic and biofilm forms of Staphylococcus lugdunensis clinical isolates. Antimicrob. Agents Chemother. 51:888–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaplan JB. 2011. Antibiotic-induced biofilm formation. Int. J. Artif. Organs 34:737–751 [DOI] [PubMed] [Google Scholar]
- 8. Carsenti-Etesse H, et al. 1992. Effect of subinhibitory concentrations of cefamandole and cefuroxime on adherence of Staphylococcus aureus and Staphylococcus epidermidis to polystyrene culture plates. Eur. J. Clin. Microbiol. Infect. Dis. 11:732–737 [DOI] [PubMed] [Google Scholar]
- 9. Carsenti-Etesse H, et al. 1993. Effects of subinhibitory concentrations of vancomycin and teicoplanin on adherence of staphylococci to tissue culture plates. Antimicrob. Agents Chemother. 37:921–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Odenholt I. 2001. Pharmacodynamic effects of subinhibitory antibiotic concentrations. Int. J. Antimicrob. Agents 17:1–8 [DOI] [PubMed] [Google Scholar]
- 11. Smith DL, Harris AD, Johnson JA, Silbergeld EK, Morris JG., Jr. 2002. Animal antibiotic use has an early but important impact on the emergence of antibiotic resistance in human commensal bacteria. Proc. Natl. Acad. Sci. U. S. A. 99:6434–6439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Izano EA, Amarante MA, Kher WB, Kaplan JB. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaplan JB, et al. 2012. Recombinant human DNase I decreases biofilm and increases antimicrobial susceptibility in staphylococci. J. Antibiot. (Tokyo) 65:73–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pozzi C, et al. 2012. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 8:e1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heilmann C. 2011. Adhesion mechanisms of staphylococci. Adv. Exp. Med. Biol. 715:105–123 [DOI] [PubMed] [Google Scholar]
- 16. Mann EE, et al. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rice KC, et al. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 104:8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Houston P, Rowe SE, Pozzi C, Waters EM, O’Gara JP. 2011. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immun. 79:1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boles BR, Horswill AR. 2008. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kiedrowski MR, et al. 2011. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One 6:e26714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaplan JB, Jabbouri S, Sadovskaya I. 2011. Extracellular DNA-dependent biofilm formation by Staphylococcus epidermidis RP62A in response to subminimal inhibitory concentrations of antibiotics. Res. Microbiol. 162:535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pérez-Giraldo C, et al. 1994. In-vitro slime production by Staphylococcus epidermidis in presence of subinhibitory concentrations of ciprofloxacin, ofloxacin and sparfloxacin. J. Antimicrob. Chemother. 33:845–848 [DOI] [PubMed] [Google Scholar]
- 23. Calabrese EJ, Baldwin LA. 2001. Hormesis: U-shaped dose responses and their centrality in toxicology. Trends Pharmacol. Sci. 22:285–291 [DOI] [PubMed] [Google Scholar]
- 24. Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. 2008. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS One 3:e3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herbert S, et al. 2007. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog. 3:e102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bisognano C, Vaudaux PE, Lew DP, Ng EY, Hooper DC. 1997. Increased expression of fibronectin-binding proteins by fluoroquinolone-resistant Staphylococcus aureus exposed to subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 41:906–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Merino N, et al. 2009. Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 191:832–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Singh R, Ray P, Das A, Sharma M. 2010. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J. Antimicrob. Chemother. 65:1955–1958 [DOI] [PubMed] [Google Scholar]
- 29. Marshall BM, Levy SB. 2011. Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24:718–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bose JL, Lehman MK, Fey PD, Bayles KW. Contribution of the Staphylococcus aureus AtlA AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS One, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baba T, et al. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827 [DOI] [PubMed] [Google Scholar]
- 33. Duthie ES, Lorenz LL. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95–107 [DOI] [PubMed] [Google Scholar]
- 34. Diep BA, et al. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739 [DOI] [PubMed] [Google Scholar]
- 35. Kuroda M, et al. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225–1240 [DOI] [PubMed] [Google Scholar]
- 36. Holden MT, et al. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 101:9786–9791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gill SR, et al. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Herron LL, et al. 2002. Genome sequence survey identifies unique sequences and key virulence genes with unusual rates of amino acid substitution in bovine Staphylococcus aureus. Infect. Immun. 70:3978–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eleaume H, Jabbouri S. 2004. Comparison of two standardisation methods in real-time quantitative RT-PCR to follow Staphylococcus aureus genes expression during in vitro growth. J. Microbiol. Methods 59:363–370 [DOI] [PubMed] [Google Scholar]
- 40. Horsburgh MJ, et al. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shanks RM, et al. 2005. Heparin stimulates Staphylococcus aureus biofilm formation. Infect. Immun. 73:4596–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jonsson P, Lindberg M, Haraldsson I, Wadström T. 1985. Virulence of Staphylococcus aureus in a mouse mastitis model: studies of alpha hemolysin, coagulase, and protein A as possible virulence determinants with protoplast fusion and gene cloning. Infect. Immun. 49:765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]