Abstract
Transcription of the human adenovirus 5 genome in transformed rat embryo cells (DFK3) was investigated using two different approaches. Preferential digestion of transcribed viral sequences by DNase I was analysed using kinetics of renaturation of 32P-labeled Ad5 HpaI restriction fragments in the presence of material which was stable after nuclease treatment. The second approach was the hybridization of 32P-labeled nuclear RNA from transformed cells with Ad5 restriction fragments which were attached to a nitrocellulose filter. These two methods gave similar results. It was found that not all integrated regions of the Ad5 genome are active in transformed cells. 2,5 copies of the HpaI-E fragment of Ad5 DNA were found in transformed DFK3 cell line. Nuclear RNA from these cells hybridized to HpaI-E fragment of Ad5 DNA, but only about half of sequences of the integrated HpaI-E fragment was sensitive to DNase I digestion.
Full text
PDF



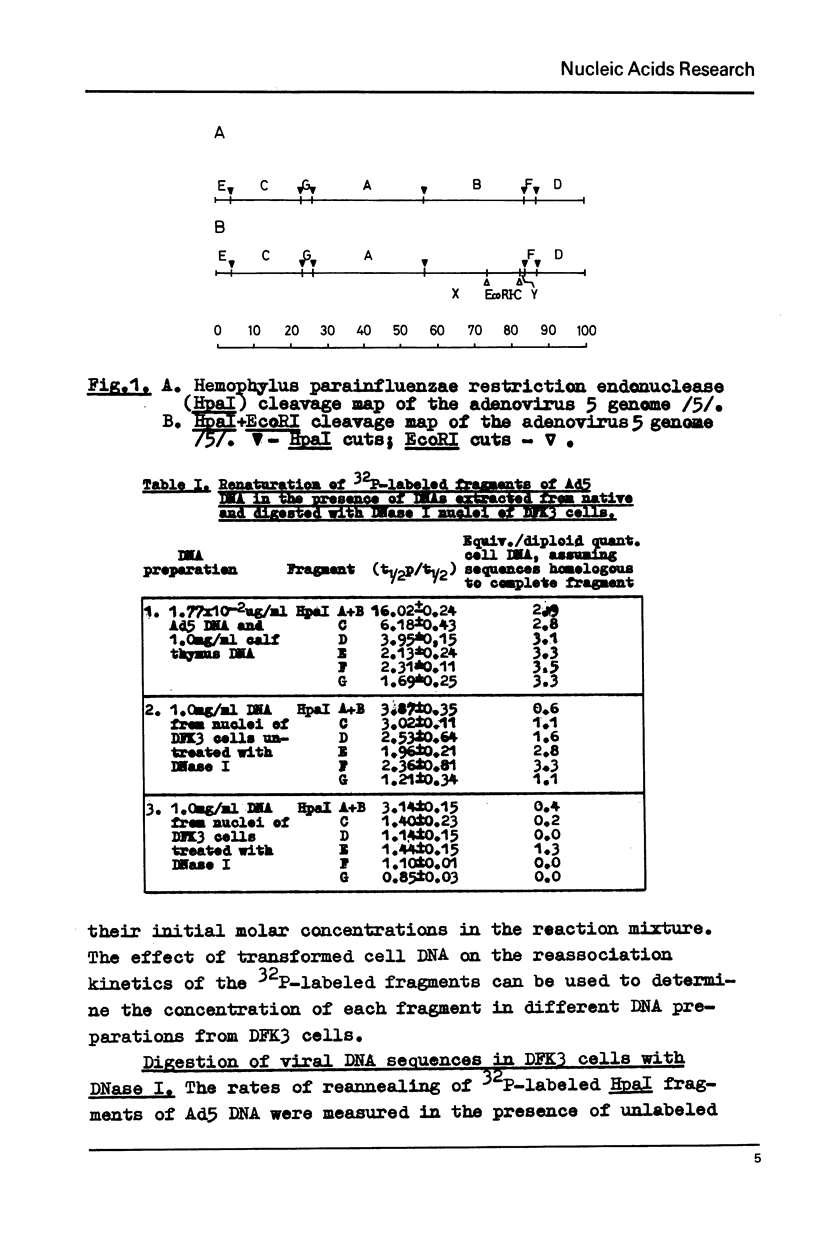
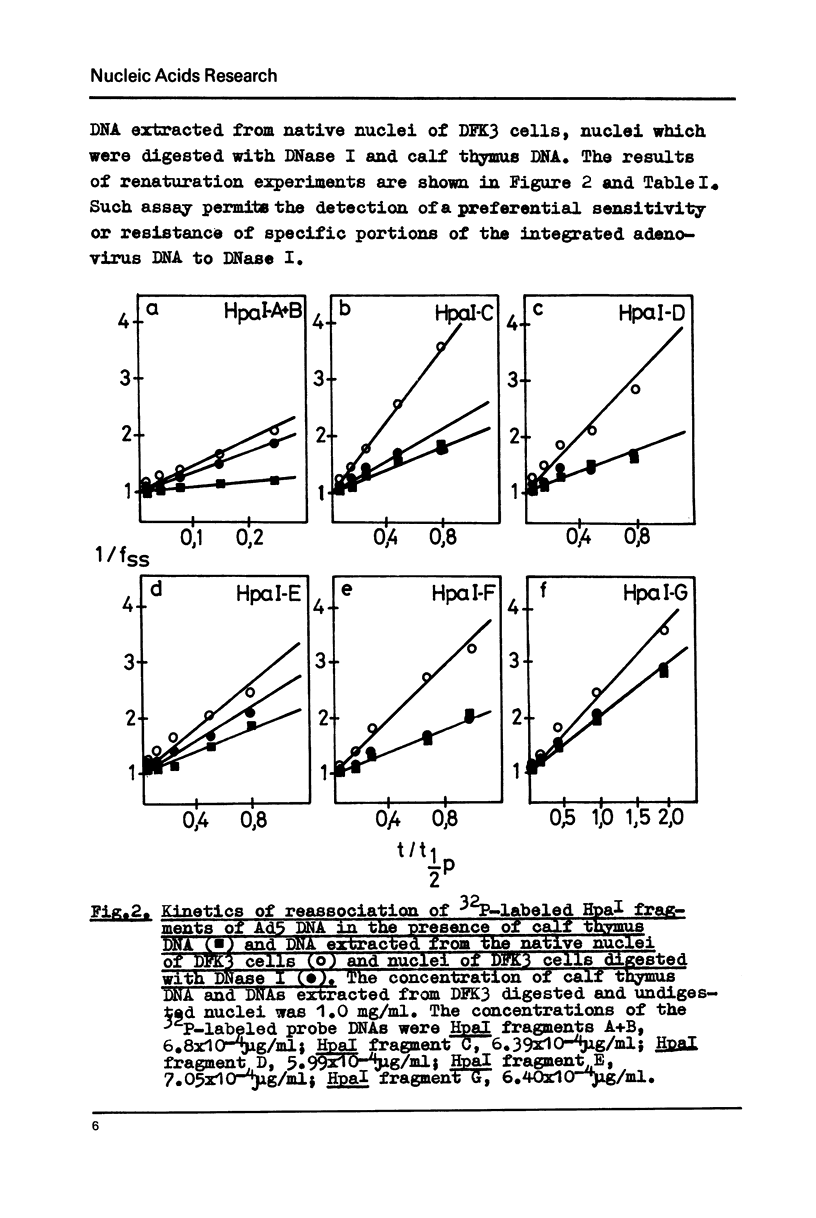

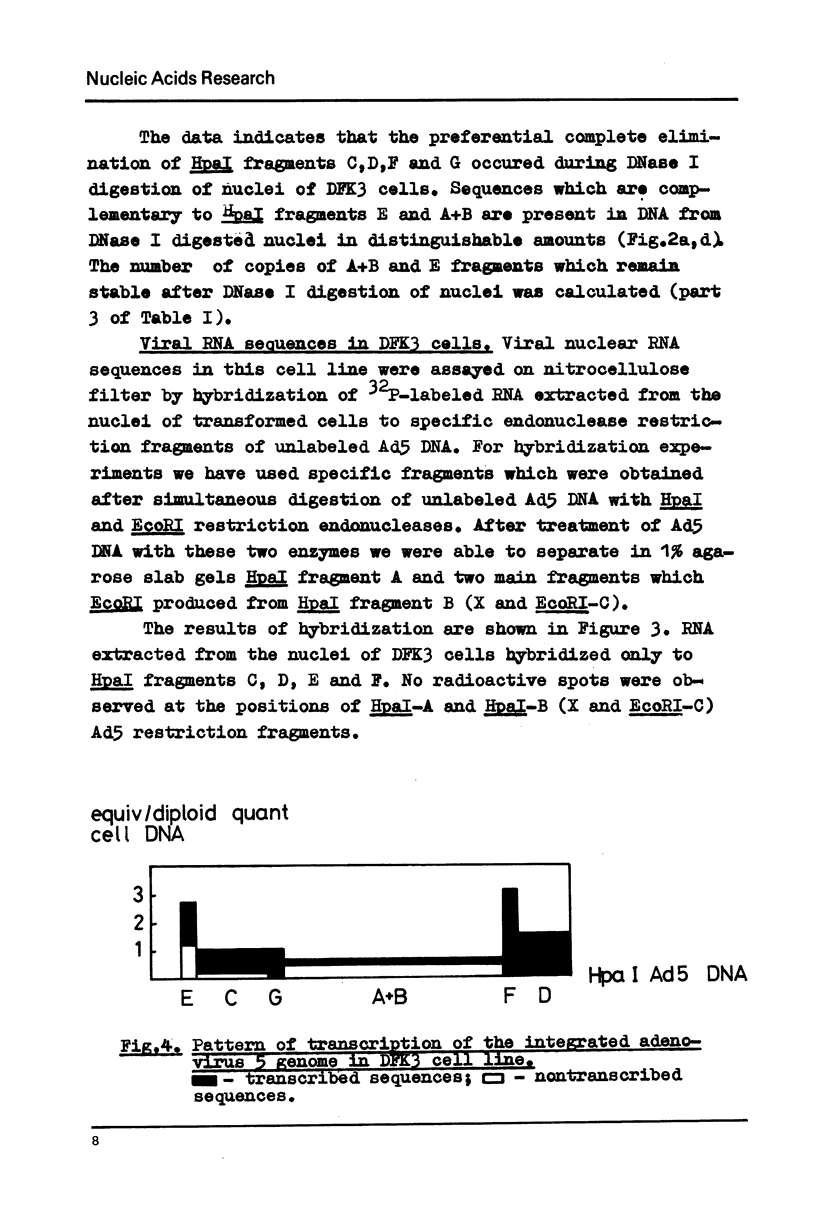



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachenheimer S., Darnell J. E. Hybridization of mRNA from adenovirus-transformed cells to segments of the adenovirus genome. J Virol. 1976 Jul;19(1):286–289. doi: 10.1128/jvi.19.1.286-289.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. doi: 10.1016/0092-8674(76)90118-5. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Flint J. The topography and transcription of the adenovirus genome. Cell. 1977 Feb;10(2):153–166. doi: 10.1016/0092-8674(77)90211-2. [DOI] [PubMed] [Google Scholar]
- Flint S. J., Gallimore P. H., Sharp P. A. Comparison of viral RNA sequences in adenovirus 2-transformed and lytically infected cells. J Mol Biol. 1975 Jul 25;96(1):47–68. doi: 10.1016/0022-2836(75)90181-3. [DOI] [PubMed] [Google Scholar]
- Flint S. J., Sambrook J., Williams J. F., Sharp P. A. Viral nucleic acid sequences in transformed cells. IV. A study of the sequences of adenovirus 5 DNA and RNA in four lines of adenovirus 5-transformed rodent cells using specific fragments of the viral genome. Virology. 1976 Jul 15;72(2):456–470. doi: 10.1016/0042-6822(76)90174-4. [DOI] [PubMed] [Google Scholar]
- Flint S. J., Sharp P. A. Adenovirus transcription. V. Quantitation of viral RNA sequences in adenovirus 2-infected and transformed cells. J Mol Biol. 1976 Sep 25;106(3):749–774. doi: 10.1016/0022-2836(76)90263-1. [DOI] [PubMed] [Google Scholar]
- Graham F. L., Abrahams P. J., Mulder C., Heijneker H. L., Warnaar S. O., De Vries F. A., Fiers W., Van Der Eb A. J. Studies on in vitro transformation by DNA and DNA fragments of human adenoviruses and simian virus 40. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):637–650. doi: 10.1101/sqb.1974.039.01.077. [DOI] [PubMed] [Google Scholar]
- Lonberg-Holm K., Philipson L. Early events of virus-cell interaction in an adenovirus system. J Virol. 1969 Oct;4(4):323–338. doi: 10.1128/jvi.4.4.323-338.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson U., Sambrook J. Amount of viral DNA in the genome of cells transformed by adenovirus type 2. J Mol Biol. 1973 Jan;73(1):125–130. doi: 10.1016/0022-2836(73)90164-2. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Botchan M., Gallimore P., Ozanne B., Pettersson U., Williams J., Sharp P. A. Viral DNA sequences in cells transformed by simian virus 40, adenovirus type 2 and adenovirus type 5. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):615–632. doi: 10.1101/sqb.1974.039.01.075. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Gallimore P. H., Flint S. J. Mapping of adenovirus 2 RNA sequences in lytically infected cells and transformed cell lines. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):457–474. doi: 10.1101/sqb.1974.039.01.058. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Pettersson U., Sambrook J. Viral DNA in transformed cells. I. A study of the sequences of adenovirus 2 DNA in a line of transformed rat cells using specific fragments of the viral genome. J Mol Biol. 1974 Jul 15;86(4):709–726. doi: 10.1016/0022-2836(74)90348-9. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]



