Summary
The cellular signals controlling the formation of cardiomyocytes, vascular smooth muscle and endothelial cells from stem cell-derived mesoderm are poorly understood. To identify these signals, a mouse embryonic stem cell (ESC)-based differentiation assay was screened against a small molecule library resulting in a novel 1,4-dihydropyridine inducer of type II TGFβ receptor (TGFBR2) degradation-1 (ITD-1). ITD analogs enhanced proteasomal degradation of TGFBR2, effectively clearing the receptor from the cell surface and selectively inhibiting intracellular signaling (IC50 ~ 0.4-0.8μM). ITD-1 was used to evaluate TGFβ involvement in mesoderm formation and cardiopoietic differentiation, which occur sequentially during early development, revealing an essential role in both processes in ESC cultures. ITD-1 selectively enhanced the differentiation of uncommitted mesoderm to cardiomyocytes, but not to vascular smooth muscle and endothelial cells. Together, ITD-1 is the first selective TGFβ inhibitor and reveals an unexpected role for TGFβ signaling in controlling cardiomyocyte differentiation from multipotent cardiovascular precursors.
Keywords: Small molecule inhibitor; TGFβ; proteasomal degradation; embryonic stem cells; mesoderm; cardiogenesis; 1,4-dihydropyridine
Introduction
The ability to control stem cell cardiogenesis is critical to realize the promise of pluripotent stem cells as a source of cells for replacement therapies. Moreover, an improved understanding of the signals that regulate replication and differentiation of cardiac progenitors might reveal mechanisms that underlie the limited potential of the adult heart to replace muscle cells after injury and ultimately could lead to strategies for in vivo regeneration therapies (Sturzu and Wu, 2011). An important approach to defining the signals that drive stem cell cardiogenesis has been to mimic embryological mechanisms for mesoderm induction and cardiogenic patterning (Burridge et al., 2012). Although successful in revealing the underlying mechanisms of early differentiation events, little is known about the signals that drive later steps of cardiogenesis that may be key to achieving therapeutic regeneration.
Unbiased screening of small molecules in phenotypic assays can overcome some of the limitations of embryology studies and is thus an alternate approach to study gene, protein or pathway function in complex biological systems (Willems et al., 2011). Here, we describe a large scale, image-based screen to identify novel small molecule probes that would stimulate the specification of cardiac cells from uncommitted mesoderm in embryonic stem cells (ESCs). One of the most active compounds was a novel 1,4-dihydropyridine, which we named inducer of TGFβ type II receptor degradation (ITD). ITD and its analogs promote cardiomyocyte differentiation specifically via degradation of the TGFβ type II receptor (TGFBR2), revealing a role for TGFβ itself as a repressor of cardiomyocyte fate. Moreover, ITDs comprise the first selective TGFβ inhibitors that do not block the closely related Activin A signaling pathway, and represent novel reagents for exploring TGFβ function in various biological contexts such as embryonic development and models of disease.
Results
A cardiogenesis screen identifies a novel TGFβ selective inhibitor
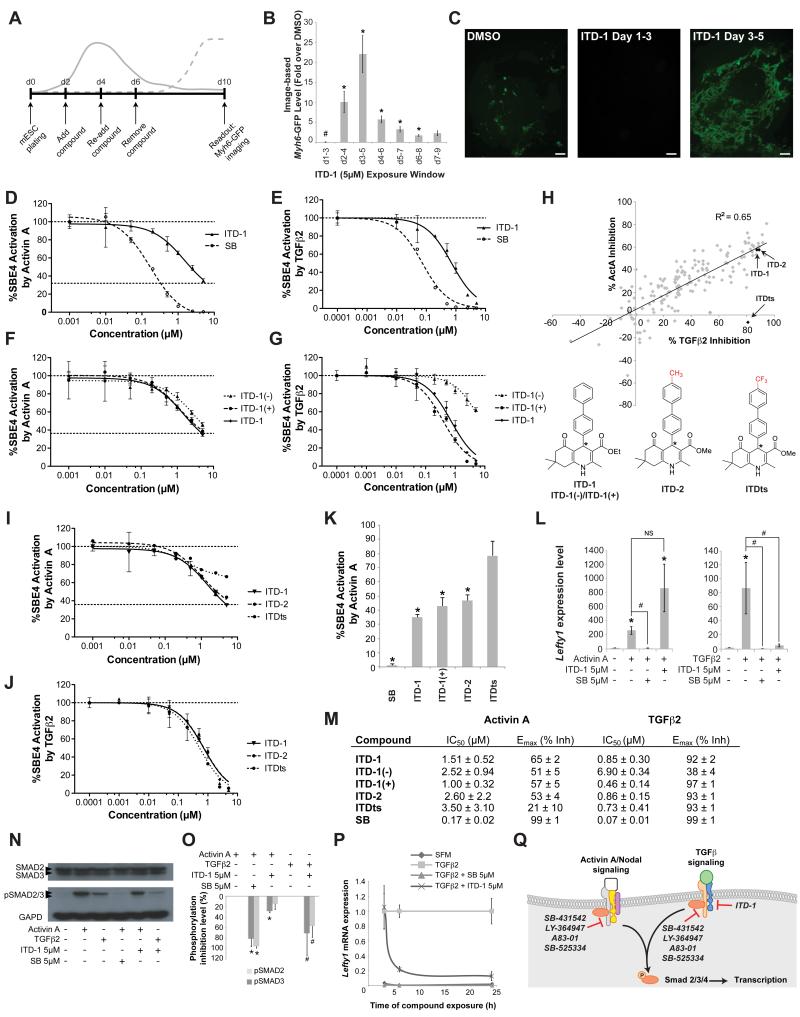
A mouse ESC (mESC) assay using an image-based Myh6-GFP reporter readout was screened between days 2 and 6 of differentiation, as uncommitted mesoderm (T/Bra+) cells become specified as cardiac. The assay identified a 1,4-dihydropyridine, which we named ITD-1 (inducer of TGFβ type II receptor degradation-1). ITD-1 optimally promoted cardiogenesis and beating cell clusters when added from day 3-5 of differentiation (Figure 1A-C and Movie S1). In contrast, ITD-1 completely abolished cardiogenesis when added before mesoderm induction, i.e. day 1-3 of differentiation (Figure 1B,C), suggesting a bi-phasic effect of ITD-1 during differentiation of mESC, inhibiting mesoderm early and inducing cardiac fate later.
Figure 1. High content screen in mESC identified a novel cardiogenic TGFβ selective inhibitor.
(A) The mESC screening assay that was used to identify compounds that affect cardiac fate at the mesoderm patterning stage. Solid line, mesoderm dynamics and dotted line, emerging cardiomyocytes.
(B-C) Myh6-GFP levels quantified by image analysis in mESC after treating with 5μM ITD-1 over different time windows, normalized to vehicle alone. Note cardiac fate suppression at d1-3 and promotion at later time windows. # p<0.05 for downregulation compared to DMSO vehicle, * p<0.05 for upregulation compared to DMSO (B). Error bars represent standard error of the mean (SEM). Representative day 10 Myh6-GFP images of the biphasic effect of ITD-1. Scale bar, 25μm (C).
(D-E) Inhibition of Smad4 response element-luciferase (SBE4-Luc) activity in HEK293T cells through a dose response of ITD-1 and the ACVR1/TGFBR1 kinase inhibitor SB-431542 (SB) in response to the TGFβ family members Activin A (D) and TGFβ2 (E).
(F-G) SBE4-Luc dose-response curves for ITD-1 and its enantiomers in presence of Activin A (F) or TGFβ2 (G).
(H) SAR analysis of over 200 ITD-1 analogs screened at 5μM against TGFβ2 and Activin A in the SBE4-Luc assay to identify compounds with high selectivity for TGFβ2. One confirmed compound (ITDts) and a structurally similar analog (ITD-2) are indicated with arrows. * chiral center.
(I-J) Dose response curves for ITDts, ITD-2 and ITD-1 against Activin A (I) and TGFβ2 (J) in the SBE4-Luc assay.
(K) Histogram plot representing the residual Activin A activity after treating with 5μM of the indicated compounds, normalized to Activin A alone (100%). * p<0.05 compared to DMSO vehicle.
(L) Functional inhibition of Activin A and TGFβ2 signaling by ITD-1, read out by Lefty1 mRNA levels in Cripto−/− mESC. * p<0.05 compared to no Activin A/TGFβ2 control, and # p<0.05 compared to Activin A/TGFβ2 alone and NS, not significant.
(M) Overview of IC50 values for Activin A/TGFβ2 inhibition and Emax values (shown as % inhibition) of key compounds in the SBE4-Luc assay. represented as average ± SEM
(N-O) Representative Western Blot for SMAD2/3, p-SMAD2/3 and GAPD in ITD-1 treated HEK293T cells after stimulation with TGFβ or Activin A (N). p-SMAD2/3 protein level quantification, normalized for GAPD and total SMAD2/3, plotted as % inhibition (O), * p<0.05 compared to Activin A, # p<0.05 compared to TGFβ2.
(P) Lefty1 mRNA time course analysis in a serum free Cripto−/− mESC assay after TGFβ2 treatment in the presence of ITD-1 or SB. SFM, serum free medium alone.
(Q) Schematic representation of the selectivity and targets of known small molecule inhibitors in respect to ITD-1 (see also Table S3). Error bars represent SEM.
Since dihydropyridines are well-known calcium channel blockers, we asked whether this mechanism was responsible for any of the ITD-1 activities, but no evidence was found for a role through calcium inhibition (Figure S1). To facilitate target identification of ITD-1, a panel of tyrosine kinase inhibitors was screened in the day 3-5 cardiogenic window, revealing that blocking Activin A and/or TGFβ signaling upregulated cardiogenesis significantly (Table S1). HEK293T cells were transfected with a Smad4 response element driving luciferase (SBE4-Luc) to test whether ITD-1 blocked Activin A/Nodal and/or TGFβ signaling, which utilize the same intracellular signaling cascade through Smad4. ITD-1 strongly inhibited TGFβ2 signaling with similar efficacy (92% vs. 99% respectively), but with lower potency compared to SB-431542, a ACVR1B/TGFBR1 kinase inhibitor (IC50 = 850nM vs. 70nM respectively), and was a weak and partial inhibitor of Activin A signals (Figure 1D,E,M). Toxicity was not responsible for the observed inhibitory activity of ITD-1 and other developmentally important pathways such as Wnt or BMP signaling were not inhibited by ITD-1 (Figure S2). To understand whether TGFβ2 and Activin A inhibition was through a shared target, we pursued a chemical biology approach asking whether or not we could generate ITD-1 analogs that would selectively target the TGFβ pathway and not the Activin A pathway. Optically pure (+)- and (−)-enantiomers of ITD-1 (chiral center at the 4-position) showed that the inhibitory effect was well separated stereochemically for TGFβ2, but not for Activin A, suggesting a highly selective mechanism for TGFβ2 inhibition, distinct from that of the weak Activin A inhibition (Figure 1F,G,M). This finding prompted a screen of ITD-1 analogs to increase the selectivity for TGFβ2 relative to Activin A. From over 200 analogs, one highly selective candidate, named ITDts (ITD TGFβ selective), retained TGFβ2 inhibition activity, but lost Activin A inhibition activity even at 5μM (Figure 1H-K,M). Interestingly, a structural homolog of ITDts, ITD-2, where only the 4-CF3 group was replaced by a 4-CH3 group, retained the weak activity against Activin A (Figure 1H-K,M).
We next evaluated whether ITD-1 is functionally selective in a defined stem cell system. We used Cripto−/− mESCs, which lack the essential co-receptor needed to respond to Nodal, to remove any confounding effects that endogenous Nodal might have if stimulated in response to exogenous Activin A or TGFβ. ITD-1 (5μM) effectively blocked induction of the Activin A/Nodal/TGFβ target gene Lefty1 in response to TGFβ2 but not to Activin A (Figure 1L). Since ITD-1 is functionally selective, we therefore used it in subsequent biological studies but confirmed key results with ITDts, which has poorer chemical stability in cell culture media.
ITD-1 blocks TGFβ signaling at the receptor level
The TGFβ signaling pathway was then probed at multiple levels to determine the point of inhibition. ITD-1 did not block the kinase activity of either type I (TGFBR1) or type II (TGFBR2) TGFβ receptors (Figure S3A,B), but ITD-1 potently blocked phosphorylation of the effector SMAD2/3 proteins induced by TGFβ2, and only minimally in response to Activin A (Figure 1N,O), corroborating the findings that ITD-1 is selective for TGFβ. Consequently, ITD-1 reduced transcriptional levels of Lefty1 in Cripto−/− mESCs, and needed ~3h more to inhibit Lefty1 expression compared to SB-431542 (Figure 1P). These results demonstrate that ITD-1 targets the TGFβ pathway at the receptor level but uses a different and more selective mechanism compared to kinase inhibitors such as SB-431542 (Figure 1Q, Table S2).
ITD-1 specifically targets TGFBR2 to the proteasome
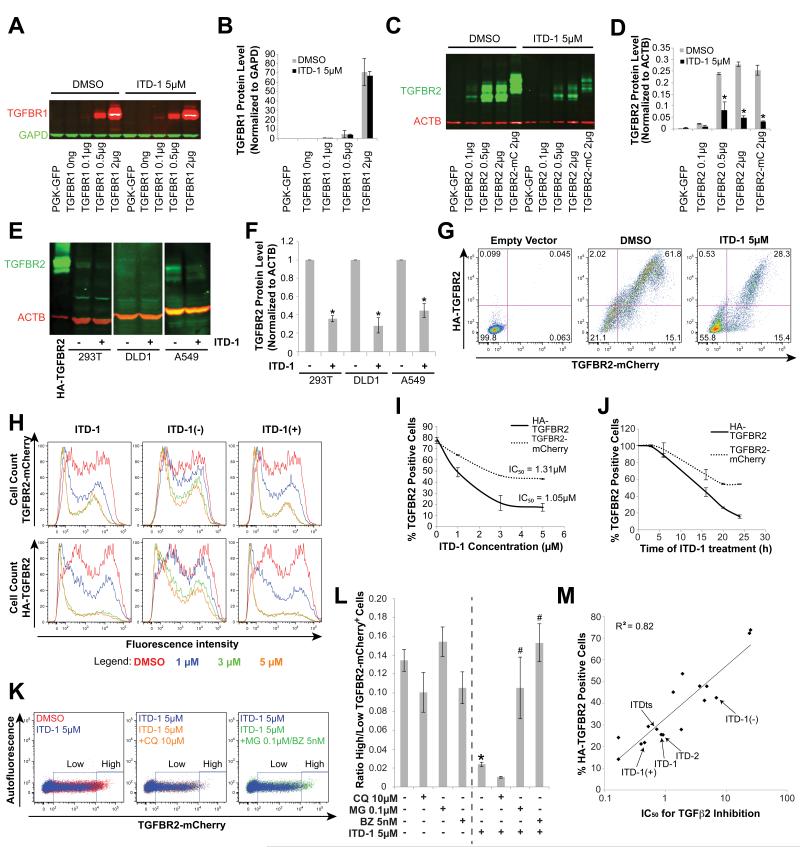
The above findings prompted us to investigate some of the dynamic processes of receptor internalization, degradation and recycling that regulate the ability of the receptor to signal (Chen, 2009). After binding the TGFβ ligand, TGFBR2 associates with TGFBR1, and the resulting ligand-receptor complex is internalized, as a requisite for signaling. Using an overexpressed extracellularly HA-tagged TGFBR2-mCherry fusion protein (HA-TGFBR2-mCherry), which is internalized in the presence of TGFβ2 ligand, we found that ITD-1 did not block immediate internalization of TGFBR2, in contrast to SB-431542, which abolished internalization (Figure S3C). We then asked if ITD-1 interfered with receptor recycling and/or degradation through the lysosome or proteasome (Di Guglielmo et al., 2003). ITD-1 did not affect TGFBR1 when overexpressed in HEK293T (Figure 2A,B), but strongly downregulated TGFBR2 protein levels (Figure 2C,D). ITD-1 similarly decreased endogenous TGFBR2 levels in several human cell lines, demonstrating that the ITD-1 effect was not an artifact of overexpression (Figure 2E-F and Figure S4A,B).
Figure 2. ITD-1 uniquely targets TGFBR2 to the proteasome.
(A-B) TGFBR1 protein levels in HEK293T cells, transiently transfected with different amounts of TGFBR1 plasmid and treated for 24h with 5μM ITD-1. A representative Western blot for TGFBR1 with GAPD as normalizing marker (A) and TGFBR1 protein level quantification, normalized for GAPD (B) are shown.
(C-D) Similarly, TGFBR2 and a TGFBR2-mCherry fusion (TGFBR2-mC) were overexpressed and detected by Western blot (C) and quantified relative to ACTB (D) after 24h of treatment with 5μM ITD-1.
(E-F) Endogenous protein levels of TGFBR2 in HEK293T, DLD1 and A549 cells, with or without 5μM ITD-1. HA-TGFBR2 is shown as blotting control (E). TGFBR2 protein level quantification, normalized for ACTB (F).
(G) A flow cytometry approach, using an extracellularly HA-tagged TGFBR2-mCherry fusion protein, quantified membrane associated (HA-TGFBR2) as well as total levels of TGFBR2 (TGFBR2-mCherry) upon ITD-1 treatment. Control and ITD-1 treated samples are shown.
(H-I) Flow cytometry analysis showing the dose dependent decrease of total (TGFBR2-mCherry) and extracellular TGFBR2 (HA-TGFBR2) for the indicated compounds. Representative histograms (H) and dose response curves based on the percentage of TGFBR2+ cells are shown (I).
(J) Time course analysis of ITD-1 on the percentage of TGFBR2+ cells assessed by flow cytometry with analysis for extracellular (HA-TGFBR2) and total TGFBR2 (TGFBR2-mCherry).
(K) Representative flow cytometry analysis for total TGFBR2 (TGFBR2-mCherry). ITD-1 reduced the TGFBR2-mCherry level per cell (high to low) compared to DMSO vehicle (blue versus red, left panel). Chloroquine (CQ) did not affect TGFBR2-mCherry levels on ITD-1 treated cultures (orange versus blue, middle panel), while MG132 (MG) or Bortezomib (BZ) treatment rescued the ITD-1 effect (green versus blue, right panel).
(L) Ratios of TGFBR2high cells over TGFBR2low cells for DMSO, ITD-1 and/or CQ/MG/BZ treatments, calculated as indicated in panel K. * p<0.05 over DMSO and # p<0.05 over ITD-1 alone.
(M) SAR analysis of highly active (IC50 <2μM), modestly active (IC50 = 2-5μM) and very weak to inactive ITD-1 analogs (IC50 >5μM) correlating SBE4-Luc TGFβ2 inhibition IC50 values with HA-TGFBR2 degradation. Key compounds are indicated with arrows. Error bars represent SEM.
To distinguish whether ITD-1 decreased cell surface or total TGFBR2 levels, we developed a flow cytometry assay with the HA-TGFBR2-mCherry vector that allowed the cell surface (extracellular HA-tag immunostaining) and total (mCherry fluorescence) TGFBR2 levels to be measured independently in the same experiment. Both HA-tag and mCherry levels declined in response to ITD-1, indicating that ITD-1 cleared TGFBR2 from the cell surface and targeted it for degradation (Figure 2G). The effect of ITD-1 was also observed on endogenous cell surface TGFBR2 and was selective, since ITD-1 did not affect other tyrosine kinase receptors (Figure S4C-F). ITD-1 diminished both the number of cells with TGFBR2 receptor, and the number of receptors per cell, in a dose-dependent manner (Figure 2H,I). ITD-1 downregulated HA-TGFBR2 and TGFBR2-mCherry with nearly the same potency (IC50 = 1.05 and 1.31μM, respectively), suggesting a common mechanism, although the maximal depletion of HA-TGFBR2 was greater (Emax = ~80% versus ~50%), consistent with the fact that ITD-1 acts by enhancing degradation rather than by blocking synthesis, which would affect cell surface and intracellular receptor pools equally (Figure 2I). At least 6h were needed for ITD-1 to reduce TGFBR2 levels, with a maximum reduction by 24h of treatment (Figure 2J).
To gain insight in the degradation mechanism of ITD-1, cells were treated with lysosome or proteasome inhibitors. ITD-1 reduced the levels of TGFBR2-mCherry fluorescence per cell, resulting in an increased number of TGFBR2-mCherrylow cells at the expense of TGFBR2-mCherryhigh cells (Figure 2K, left panel) and this effect was rescued by the proteasome inhibitors MG132 and Bortezomib (Figure 2K, right panel), but not by the lysosome inhibitor Chloroquine (CQ, Figure 2K, middle panel), as clearly demonstrated by the ratio of TGFBR2-mCherryhigh to TGFBR2-mCherrylow cells (Figure 2L). Additional support for induced degradation as the mechanism of ITD-1 action was the robust structure activity relationship (SAR) between TGFBR2 degradation and inhibition of TGFβ2 SBE4-Luc activity (R2>0.8) (Figure 2M). Since TGFBR2 was targeted to the proteasome, we examined ubiquitination of TGFBR2, but found no evidence of mono- or poly-ubiquitination (Figure S5). Taken together, the ITD class of molecules comprise selective TGFβ inhibitors that function by diverting TGFBR2 to the proteasome through an ubiquitin-independent mechanism.
Mesoderm induction in ESC requires TGFβ
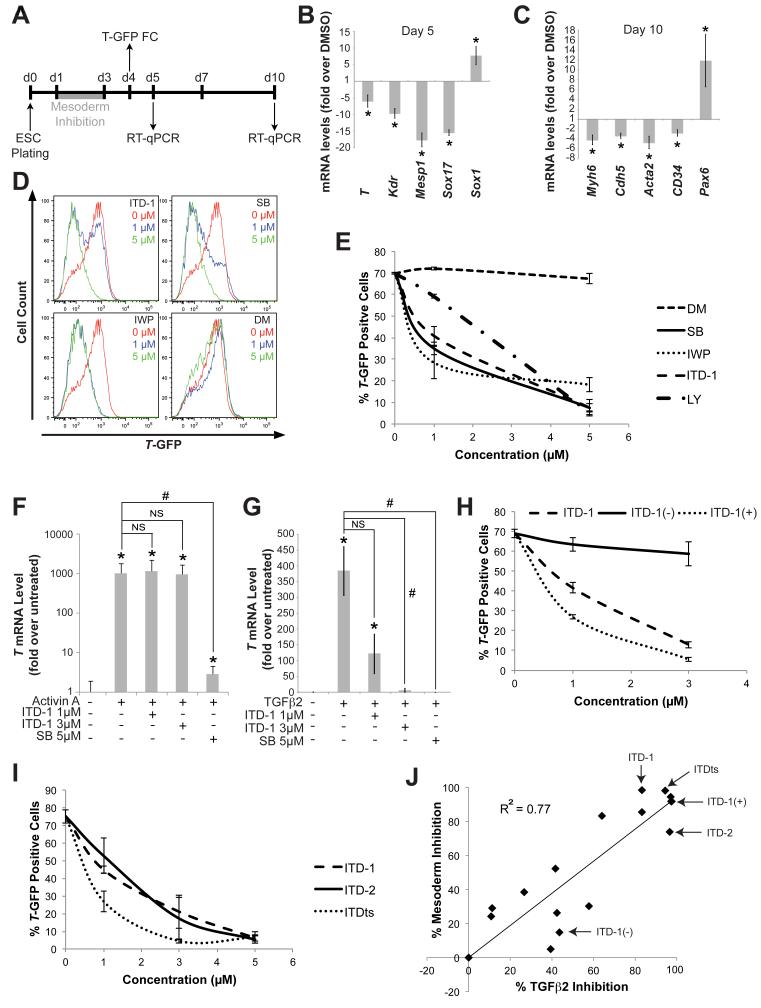
Inhibition of mesoderm formation by ITD-1 (Figure 1B,C) indicated that TGFβ was essential for this process, which was unexpected since prior studies had only implicated the TGFβ family member Nodal, Wnt and BMP (Burridge et al., 2012). Although TGFβ addition can mimic the native role of Nodal in generating mesoderm and heart cells in ESCs, it is not known to normally do so in either embryos or ESC cultures (Behfar et al., 2002). ITD-1 was therefore used to study the role of TGFβ in mesoderm induction. ESC cultures were exposed to ITD-1 from day 1 of differentiation and analyzed for germ layer segregation (Figure 3A-C). RT-qPCR analysis of mesoderm, endoderm, and ectoderm markers at day 5 of differentiation indicated that ITD-1 given at day 1 of differentiation induced ectoderm at the expense of mesoderm (Figure 3B). Consequently, on day 10 of differentiation, markers for mesoderm tissues such as heart, endothelium, smooth muscle and blood were all downregulated, while neural markers were upregulated (Figure 3C). A T-GFP mESC reporter line was then used to quantify mesoderm inhibition by ITD-1 and was compared to small molecule inhibitors of TGFβ and Activin A/Nodal (SB-431542 and LY-364947), Wnt (IWP) and BMP signaling (Dorsomorphin, DM), which are known to drive mesoderm (Figure 3D,E). Inhibition of Wnt, Activin A/TGFβ pathways diminished the number of mesoderm cells, similarly to ITD-1 (Figure 3D,E), suggesting that all three factors are involved. However, BMP inhibition did not affect mesoderm as documented previously (Yuasa et al., 2005). ITD-1 did not inhibit Wnt signaling (Figure S2B-E), and since ITD-1 retained weak activity against Activin A/Nodal signaling, it was evaluated in the Cripto−/− mESC assay, revealing that ITD-1 selectively blocked mesoderm induced by TGFβ but not by Activin A (Figure 3F,G). To confirm the specific involvement of TGFβ, the chemical tools described above were applied to correlate mesoderm inhibition with TGFβ signaling inhibition. Enantiomeric separation in the T-GFP assay was similar to TGFβ2 signaling inhibition (Figure 3H) and ITDts also reduced the number of T-GFP positive cells (Figure 3I). Moreover, the correlation between both activities was very strong as shown through SAR analysis (Figure 3J). The ITD class of molecules thus exposed an essential and specific role for TGFβ during mesoderm formation in ESC.
Figure 3. ITD-1 inhibits mesoderm induction in mouse ESC.
(A) mESC differentiation timeline, showing the ITD-1 treatment window (grey bar) and the days of RT-qPCR or T-GFP flow cytometry (FC) analyses.
(B-C) Gene expression analysis of day 5 (B) and day 10 (C) samples after treating mESC with ITD-1 from day 1-3. Markers included mesoderm and endoderm (T, Kdr, Mesp1 and Sox17), neuroectoderm (Sox1), heart (Myh6), smooth muscle (Acta2), endothelium (Cdh5), blood (Cd34) and neuroectoderm (Pax6). * p<0.05 compared to DMSO vehicle.
(D-E) T-GFP flow cytometry analysis of ITD-1; SB-431542 (SB), Nodal/TGFβ signaling inhibitor; IWP, Wnt production inhibitor; and Dorsomorphin (DM), BMP signaling inhibitor treatments on mesoderm induction in T-GFP mESCs. (D) Representative histograms with three conditions are shown: DMSO vehicle (red), 1μM (blue) and 5μM (green) and (E) the quantification of T-GFP+ cells, with the inclusion of a second Nodal/TGFβ signaling inhibitor, LY-364947 (LY).
(F-G) T gene expression in Cripto−/− mESC to assess whether ITD-1 can block the ability of Activin A (F) or TGFβ2 (G) to induce mesoderm. * p<0.05 compared to no Activin A/TGFβ2 control, # p<0.05 compared to Activin A/TGFβ2 alone and NS, not significant.
(H-I) Flow cytometry quantification of T-GFP+ cells after treating with ITD-1 enantiomers (H) and TGFβ selective compounds (I).
(J) SAR profile spanning a wide activity range of ITD-1 analogs, correlating TGFβ2 inhibition (calculated from SBE4-Luc assays) with mesoderm inhibition (calculated from T-GFP flow cytometry analyses). Key compounds are indicated by arrows. Error bars represent SEM.
TGFBR2 degradation specifically promotes cardiac lineages in ESC
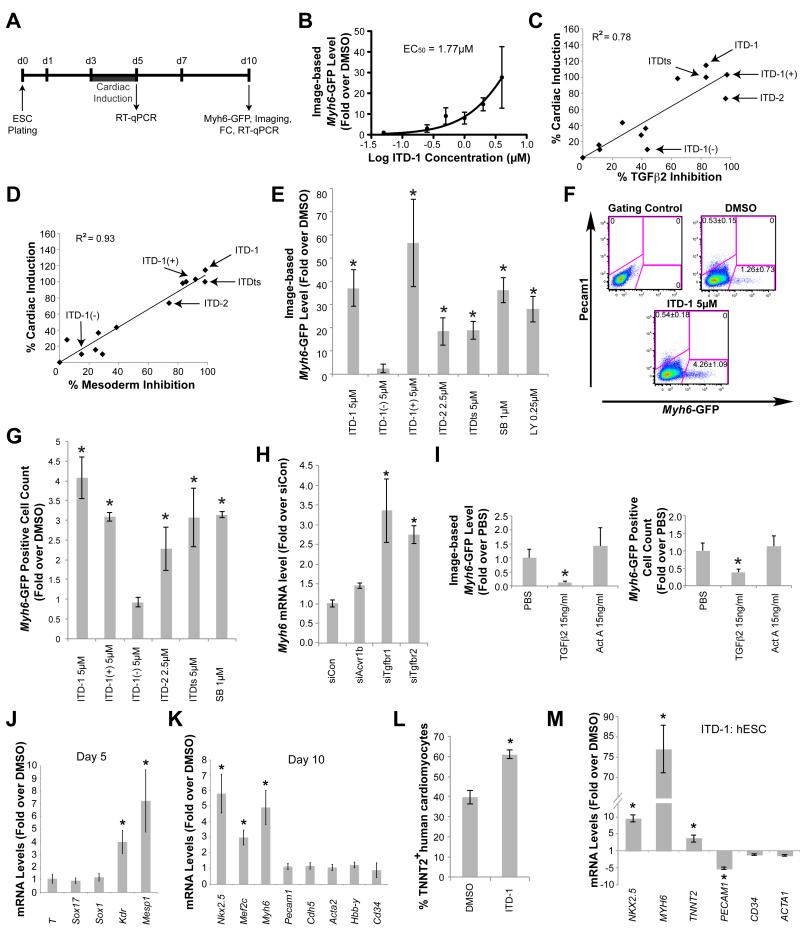
The pro-cardiac effect of ITD-1 between days 3-5 of mESC differentiation, suggested a specific and unappreciated role for TGFβ in regulating cardiac cell fate (Figure 1B,C and Figure 4A,B). SAR analyses showed a strong correlation between TGFβ inhibition and cardiomyocyte differentiation (R2 = 0.78), comparable to that between cardiogenesis and mesoderm inhibition (Figure 4C,D). Moreover, analysis of Myh6-GFP expression in mESC demonstrated that ITD-1, ITD-1(+), ITDts and ITD-2 all drove cardiomyogenesis as well as SB-431542 and LY-364947, whereas ITD-1(−) did not (Figure 4E-G), all consistent with an inhibitory effect of TGFβ on cardiomyocyte differentiation. Similarly, siRNA knockdown of Tgfbr1 or Tgfbr2 alone was sufficient to promote cardiac fate, while knockdown of Acvr1b and thus Activin signaling had no effect (Figure 4H), confirming a repressive role of TGFβ on cardiomyocyte development at this time. Conversely, addition of TGFβ2 and not Activin A inhibited cardiogenesis in the day 3-5 window of differentiation, bolstering the findings obtained with chemical- and siRNA-mediated knockdowns of TGFBR2 (Figure 4I).
Figure 4. ITD-1 promotes cardiogenesis via specific inhibition of TGFβ signaling.
(A) mESC cardiogenesis assay timeline, showing the day 3-5 ITD-1 treatment window (grey bar) that leads to cardiac induction and the times of RT-qPCR, Myh6-GFP imaging and flow cytometry (FC) analyses.
(B) ITD-1 dose response curve for cardiac induction in mESC, assessed by Myh6-GFP level quantification by image analysis, represented as a fold over DMSO vehicle alone.
(C-D) SAR correlation plots showing ITD-1 analogs for cardiac induction (calculated by image-based Myh6-GFP levels) and TGFβ2 inhibition (calculated from the SBE4-Luc assay) (C) and mesoderm inhibition (calculated from T-GFP flow cytometry analysis) (D) assays. Key compounds are indicated by arrows.
(E) Quantification of image-based Myh6-GFP levels induced by indicated compounds and represented as fold over DMSO vehicle alone. SB, SB-431542; LY, LY-364947. * p<0.05 compared to DMSO vehicle.
(F) Representative flow cytometry analysis for Myh6-GFP+ cardiomyocytes and Pecam1+ endothelial cells after treatment with ITD-1. Numbers represent percent positive cells (average ± SEM).
(G) Fold increase in the number of Myh6-GFP+ cells quantified as in panel F for the indicated compounds. * p<0.05 compared to DMSO vehicle.
(H) The effect of siRNAs to Acvr1, Tgfbr1 and Tgfbr2 on Myh6 mRNA induction at day 10. * p<0.05 compared to siControl (siCon).
(I) Image based Myh6-GFP quantification (left panel) and flow cytometry quantification (right panel) of ESC cultures treated with TGFβ2 or Activin A from day 3-5, both represented as fold over PBS vehicle alone. * p<0.05 compared to PBS vehicle.
(J-K) Gene expression analysis of day 5 (B) and day 10 (C) samples after treatment of mESC with ITD-1 from day 3-5. Markers included mesoderm and endoderm (T, Kdr, Mesp1 and Sox17), neuroectoderm (Sox1), cardiac lineages (Nkx2.5, Mef2c, Myh6), endothelium (Pecam1, Cdh5), smooth muscle (Acta2) and blood progenitors (Hbb-y, Cd34). * p<0.05 compared to DMSO vehicle.
(L-M) hESC cultures were exposed to ITD-1 from day 1-5 and were analyzed by flow cytometry for TNNT2+ cardiomyocyte number on day 6 of differentiation (L) and were analyzed for gene expression indicative of cardiac (NKX2.5, MYH6, TNNT2), vascular endothelial (CD31), blood (CD34) and skeletal muscle (ACTA1) cells by RT-qPCR (M). * p<0.05 compared to DMSO vehicle. Error bars represent SEM.
To determine the developmental stages when TGFβ affected cardiac differentiation and whether other cardiovascular lineages were also affected, markers of the different germlayers, progenitors and lineages were examined at different time points by RT-qPCR. At day 5 of differentiation, mesoderm, endoderm and ectoderm markers were unaltered, and only the cardiac progenitor specific markers Kdr and Mesp1 were increased by ITD-1 (Figure 4J). Furthermore, only cardiac-specific markers were increased at day 10, whereas none of the vascular or hematopoietic markers were affected. TGFβ thus specifically repressed the formation of cardiomyocytes at this stage of differentiation (Figure 4K).
To understand the true magnitude of ITD-1 treatment towards cardiac induction from ESC, we asked whether TGFβ inhibition played a similarly specific role in promoting cardiomyocyte specification in a completely optimized cardiac differentiation protocol in human ESCs (hESCs). H9 cells were optimally differentiated to cardiomyocytes, with ITD-1 added during mesoderm patterning (day 1-5) and cultures were surveyed for cardiovascular markers by flow cytometry and RT-qPCR at day 6 of differentiation (Figure 4L,M). While SB completely blocked cardiogenesis at this stage because of Activin A dependence (data not shown), ITD-1 potently enhanced TNNT2+ cardiomyocyte yield by ~30% resulting in ~60% cardiomyocytes, and was also visible on the mRNA level (Figure 4L,M). A slight repression of vascular and hematopoietic markers was also observed (Figure 4M). These findings demonstrate that endogenous TGFβ regulates the yield of cardiomyocytes in hESCs, even under fully-optimized and defined media conditions.
In summary, ITD-1 and its analogs unraveled a novel conserved mechanism that exclusively directs cardiac fate in ESCs through temporal inhibition of TGFβ signaling.
Discussion
Screening of small molecules in a complex biological system through a phenotypic read-out can lead to the identification of novel probes of the biology of a cellular system. Such probes can then be linked to specific pathways or mechanisms, and may lead to the identification of novel drug targets (Ao et al., 2011; Willems et al., 2011). Therefore, we developed image-based screens in ESC to discover new pathways and/or mechanisms in cardiogenesis, in particular as a means to gain insight into endogenous regeneration. The molecule described here acts when uncommitted mesoderm cells become specified to a cardiac fate. ITD-1 treatment at this stage in an optimal hESC assay indicates that manipulation of endogenous TGFβ signaling is an important step to refine protocols that enhance cardiomyocyte differentiation, which can be highly variable in different human embryonic and induced pluripotent stem cell lines (Kattman et al., 2011).
1,4-dihydropyridines are well-known inhibitors of calcium channels (Edraki et al., 2009), but that mechanism was ruled out. By screening a panel of tyrosine kinase inhibitors that span a wide range of pathways, ITD-1 was found to inhibit the Activin A/TGFβ pathway specifically. Activin A and TGFβ act similarly in that they bind homologous receptors to form ligand-receptor complexes that activate an identical intracellular network of Smad2/3/4 proteins (Wharton and Derynck, 2009). Clear stereochemical separation for the strong TGFβ and no separation for the weak Activin A inhibitory activities, suggested that the molecular target responsible for effective TGFβ inhibition differs from that which accounts for the lower level of activity against Activin A. The identification of ITDts, which is highly selective for TGFβ, substantiates that idea. Pharmacological separation of the inhibitory effect on the two signaling pathways thus indicates that ITD-1 and analogs bind a molecular target that uniquely affects TGFβ signaling. To our knowledge, ITD-1 and ITDts are the first small molecule inhibitors that are highly selective for TGFβ relative to the Activin/Nodal signaling pathways.
A key observation of this study is that ITD-1 blocks TGFβ signaling by promoting degradation of TGFBR2. TGFβ receptor levels on the cell surface are dynamically regulated by vesicle-mediated ligand-triggered trafficking, recycling and lysosome degradation (Figure S6A, branch 1), as well as by direct proteasomal degradation (Figure S6A, branch 2) (Chen, 2009; Di Guglielmo et al., 2003). ITD-1 does not target the signaling/recycling/lysosome degradation loop. Instead, ITD-1 drives proteasomal degradation of TGFβ receptors as MG132 and Bortezomib rescued receptor degradation (Figure S6A, branch 2). However, previous studies described equal proteasomal degradation of both TGFBR1 and TGFBR2 through the ubiquitin ligase Smurf2 (Di Guglielmo et al., 2003). Our data on ITD-1 differs from this mechanism since TGFBR2, but not TGFBR1 levels are affected, and the process is ubiquitin-independent. Therefore, our findings suggest that there may be a third mechanism for the specific degradation of TGFBR2, which is enhanced by ITD-1 (Figure S6A, branch 3). Although the direct target of ITD-1 remains to be elucidated, several groups have reported different half lives for TGFBR1 and TGFBR2, consistent with the idea that distinct degradation processes may exist to clear these receptors from the cell surface (Wells et al., 1997). It therefore remains possible that ITD-1 directly binds TGFBR2 to drive its internalization and degradation. Interestingly, TGFBR2 appears to be exclusively downregulated in several human cancers, and in renal carcinomas this reduction has been attributed to increased proteasomal degradation (Fukasawa et al., 2010; Meng et al., 2011). ITD-1 might therefore be useful as a probe to understand how the altered dynamics of TGFBR2 trafficking contributes to cancer.
Through its high selectivity for TGFβ, ITD-1 to revealed a biphasic role of TGFβ signaling in ESC cardiogenesis (Figure S6B). When applied early in the differentiation process, ITD-1 prevented mesoderm formation and enhanced neuroectodermal fates. A direct role for TGFβ in mesoderm induction was unanticipated as Nodal (and Activin A) and Wnt are thought to be the principal effectors of the mesoderm and neuroectoderm fate choice in ESCs and mouse embryos (Naito et al., 2006; Perea-Gomez et al., 2002). TGFβ is expressed in differentiating ESCs and embryos, and can induce mesoderm if provided exogenously, but Tgfbr1 and Tgfbr2 null embryos do form mesoderm (Larsson et al., 2001; Oshima et al., 1996). Thus, our data provide the first evidence for the requirement of TGFβ in mesoderm/neuroectoderm induction in mESCs and potentially in embryos, although compensatory and/or other mechanisms may allow mesoderm formation in Tgfbr1 and Tgfbr2 mutant embryos.
Additionally, ITD-1 stimulated cardiomyocyte differentiation from uncommitted mesodermal progenitors in mouse and human ESCs, indicating that TGFβ represses cardiac fate at this developmental stage. Interestingly, inhibition of the TGFβ signaling pathway at this time did not increase vascular smooth muscle or endothelial markers or cell number, further substantiating the idea that ITD-1 specifically stimulated cardiac differentiation from a cardiovascular progenitor (Figure S6B, light blue cell). In addition, TGFβ inhibition continues to enhance cardiogenesis at considerably later stages (>day 8) by enhancing proliferation of immature cardiomyocytes (Kitamura et al., 2007). In embryos, TGFβ ligands are expressed in the newly forming cardiac crescent where cardiomyocyte committed cells arise and may thus naturally contribute to the proper apportioning of cardiomyocyte versus other cardiovascular lineages (Dickson et al., 1993; Kitamura et al., 2007).
In summary, a phenotypic screen of ESC cardiogenesis yielded ITD-1, which, to our knowledge, is the first selective TGFβ inhibitor. ITD-1 acts by stimulating clearance of TGFBR2 from the cell surface and subsequent proteasomal degradation. The chemical biology approach revealed that TGFβ plays a critical, biphasic role in the formation of cardiovascular derivatives from ESC, first by promoting mesoderm induction and subsequently by inhibiting cardiomyocyte differentiation specifically. Moreover, ITD-1 is a potentially valuable reagent, not only to study ESC differentiation, but also to probe the role of TGFβ signaling in pathological processes such as cancer, fibrosis, or adult cardiovascular disease.
Materials and methods
Embryonic stem cell culture and differentiation assays
mESC cell lines CGR8 carrying a Myh6-GFP, J1 carrying a T-GFP reporter and Cripto−/− ES cells were maintained as described in supplemental methods.
For screening, Myh6-GFP ESCs were seeded and differentiated in 384-well plates. About 17000 compounds from the DIVERSet library (Chembridge) were added from day 2-6 of differentiation. ITD-1, SB-431542 (Sigma), LY-364947 (Cayman Chemical) and tyrosine kinase inhibitor panel (EMD/Millipore) treatments were performed as indicated. Myh6-GFP positive cells were imaged on an InCell 1000 System (GE Healthcare) and GFP levels from images were quantified with Cyteseer (Vala Sciences).
Mesoderm inhibition assays with T-GFP and Myh6-GFP cells were performed as suspension cultures in serum containing media. Embryoid bodies (EB) formed in differentiation media were exposed to ITDs, SB-431542, LY-364947, Dorsomorphin (Tocris) or IWP (EMD/Millipore) at day 1 of differentiation.
For the Cripto−/− mESC assay, cells were transferred to serum free conditions and were treated at day 2 of EB formation with Wnt3a, 15 ng/ml TGFβ2 (EMD/Millipore), 10 ng/ml BMP4 (R&D Systems) or 15 ng/ml Activin A (R&D Systems) in the presence of indicated compounds.
H9 hESC were differentiated as EBs in StemPro34 (GIBCO) in ultra-low attachment plates. EBs were optimally differentiated with BMP4, Activin A, bFGF (Sigma), VEGF (R&D Systems) and DKK1 (R&D Systems) as described (Kattman et al., 2011), with addition of ITD-1 from day 1-5.
Reverse transcription quantitative PCR (RT-qPCR)
cDNA samples, synthetized from total RNA with the Quantitect RT kit (Qiagen), were run on a LightCycler 480 (Roche) using LC480 Sybr Green master mix (Roche). Primers are available at http://www.rtprimerdb.org/ and primer ID numbers are listed in supplemental methods.
SBE4-Luciferase assays
HEK293T cells, grown in DMEM-high glucose with 1% FBS, were transfected with SBE4-Luciferase plasmid and CMV-Renilla-Luciferase using Lipofectamine 2000 (Invitrogen). After 12h of adhesion, cells were induced with either TGFβ2 (15ng/ml) or Activin A (15ng/ml) and inhibitors were added simultaneously. Luminescence was measured through the Dual-Glo kit (Promega).
Receptor degradation assays
TGFBR1, extracellularly HA-tagged TGFBR2, HA-TGFBR2-mCherry fusions, pcDNA3.1 or PGK-GFP plasmids were transfected in HEK293T as above and treated >6h later with ITD-1 for the indicated time points. Chloroquine (Sigma), Bortezomib (Selleck Chemicals) or MG132 (Sigma) were added 3h after ITD-1 treatment.
Flow cytometry
Cultures were dissociated with enzyme free cell dissociation buffer (GIBCO) (or trypsin for cardiomyocytes) and analyzed on a FACSCanto or LSRFortessa (BD Biosciences). For intracellular stains, cells were permeabilized with saponin (Sigma) before staining. Single cells were stained for 30′ with indicated antibodies, which are listed in the supplemental methods. FlowJo (Treestar) was used for data analysis.
Western blotting
Cells were washed in cold PBS, collected with enzyme free dissociation buffer and lysed with ice cold RIPA buffer supplemented with protease and phosphatase inhibitors (Sigma). Lysates were run on 10% SDS-tris glycine gels (Invitrogen) and transferred to 45μm PVDF membranes, which were blocked and stained in 5% w/v skim milk in TBST. Detection was performed with the ECL Plus detection kit (Abcam) or with an Odyssey system (LICOR). Antibodies are listed in the supplemental methods.
Statistic analysis of samples
All data is represented as the mean with error bars indicating SEM for at least 3 biological replicates, p-values were obtained by a Student’s t-test. Dose response curve fitting and EC50/IC50 calculation using the (log)agonist vs. normalized response equation for induction and the (log)inhibitor vs. normalized response equation for inhibition were done in Prism 5 (GraphPad Software). Toxic doses were removed from EC50 or IC50 analysis, as judged by Renilla luciferase levels, Alamar Blue cell viability assays or microscopy.
Supplementary Material
Highlights.
Highly selective small molecule inhibitor of TGFβ degrades TGFBR2 specifically
TGFβ signaling is essential for mesoderm establishment in mouse ESC
Degradation of TGFBR2 promotes cardiac fate specifically in mouse and human ESC
Chemical biology approach to correlate or separate biological activities
Acknowledgements
The authors would like to thank Fabio Cerignoli and Karl Willert for running calcium transient assays and Wnt TOPflash assays, respectively. This work was supported by CIRM T2-00004 and AHA fellowship to EW, German Research Foundation Grant SCHA 1663/1-1 to DS NIH HL088293 and MINECO to JCIB, CIRM Seed RS-00169-1 and T Foundation to JC, CIRM RC1-000132 and NIH HL059502 to MM and NIH STTR R41-HL108714 to ChemRegen Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ao A, Hao J, Hong CC. Regenerative chemical biology: current challenges and future potential. Chem Biol. 2011;18:413–424. doi: 10.1016/j.chembiol.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behfar A, Zingman LV, Hodgson DM, Rauzier JM, Kane GC, Terzic A, Puceat M. Stem cell differentiation requires a paracrine pathway in the heart. FASEB J. 2002;16:1558–1566. doi: 10.1096/fj.02-0072com. [DOI] [PubMed] [Google Scholar]
- Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG. Endocytic regulation of TGF-beta signaling. Cell Res. 2009;19:58–70. doi: 10.1038/cr.2008.315. [DOI] [PubMed] [Google Scholar]
- Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol. 2003;5:410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]
- Dickson MC, Slager HG, Duffie E, Mummery CL, Akhurst RJ. RNA and protein localisations of TGF beta 2 in the early mouse embryo suggest an involvement in cardiac development. Development. 1993;117:625–639. doi: 10.1242/dev.117.2.625. [DOI] [PubMed] [Google Scholar]
- Edraki N, Mehdipour AR, Khoshneviszadeh M, Miri R. Dihydropyridines: evaluation of their current and future pharmacological applications. Drug Discov Today. 2009;14:1058–1066. doi: 10.1016/j.drudis.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Fukasawa H, Yamamoto T, Fujigaki Y, Misaki T, Ohashi N, Takayama T, Suzuki S, Mugiya S, Oda T, Uchida C, et al. Reduction of transforming growth factor-beta type II receptor is caused by the enhanced ubiquitin-dependent degradation in human renal cell carcinoma. Int J Cancer. 2010;127:1517–1525. doi: 10.1002/ijc.25164. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-Specific Optimization of Activin/Nodal and BMP Signaling Promotes Cardiac Differentiation of Mouse and Human Pluripotent Stem Cell Lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kitamura R, Takahashi T, Nakajima N, Isodono K, Asada S, Ueno H, Ueyama T, Yoshikawa T, Matsubara H, Oh H. Stage-specific role of endogenous Smad2 activation in cardiomyogenesis of embryonic stem cells. Circ Res. 2007;101:78–87. doi: 10.1161/CIRCRESAHA.106.147264. [DOI] [PubMed] [Google Scholar]
- Larsson J, Goumans MJ, Sjostrand LJ, van Rooijen MA, Ward D, Leveen P, Xu X, ten Dijke P, Mummery CL, Karlsson S. Abnormal angiogenesis but intact hematopoietic potential in TGF-beta type I receptor-deficient mice. Embo J. 2001;20:1663–1673. doi: 10.1093/emboj/20.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W, Xia Q, Wu L, Chen S, He X, Zhang L, Gao Q, Zhou H. Downregulation of TGF-beta receptor types II and III in oral squamous cell carcinoma and oral carcinoma-associated fibroblasts. BMC Cancer. 2011;11:88. doi: 10.1186/1471-2407-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, Kikuchi A, Komuro I. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci U S A. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima M, Oshima H, Taketo MM. TGF-beta receptor type II deficiency results in defects of yolk sac hematopoiesis and vasculogenesis. Dev Biol. 1996;179:297–302. doi: 10.1006/dbio.1996.0259. [DOI] [PubMed] [Google Scholar]
- Perea-Gomez A, Vella FD, Shawlot W, Oulad-Abdelghani M, Chazaud C, Meno C, Pfister V, Chen L, Robertson E, Hamada H, et al. Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev Cell. 2002;3:745–756. doi: 10.1016/s1534-5807(02)00321-0. [DOI] [PubMed] [Google Scholar]
- Sturzu AC, Wu SM. Developmental and regenerative biology of multipotent cardiovascular progenitor cells. Circ Res. 2011;108:353–364. doi: 10.1161/CIRCRESAHA.110.227066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RG, Yankelev H, Lin HY, Lodish HF. Biosynthesis of the type I and type II TGF-beta receptors. Implications for complex formation. J Biol Chem. 1997;272:11444–11451. doi: 10.1074/jbc.272.17.11444. [DOI] [PubMed] [Google Scholar]
- Wharton K, Derynck R. TGFbeta family signaling: novel insights in development and disease. Development. 2009;136:3691–3697. doi: 10.1242/dev.040584. [DOI] [PubMed] [Google Scholar]
- Willems E, Lanier M, Forte E, Lo F, Cashman J, Mercola M. A Chemical Biology Approach to Myocardial Regeneration. J Cardiovasc Transl Res. 2011;4:340–350. doi: 10.1007/s12265-011-9270-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa S, Itabashi Y, Koshimizu U, Tanaka T, Sugimura K, Kinoshita M, Hattori F, Fukami S, Shimazaki T, Ogawa S, et al. Transient inhibition of BMP signaling by Noggin induces cardiomyocyte differentiation of mouse embryonic stem cells. Nat Biotechnol. 2005;23:607–611. doi: 10.1038/nbt1093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.