Generating cells of different fate within a group of equipotent cells is the underlying principle of the development of complex multicellular organisms. Cell–cell signaling or environmental differences may cause two equipotent daughter cells to assume different fates. Alternatively, asymmetric distribution of cell fate determinants during cell division can generate two cells with different fates. In either case, developmental cues have to be interpreted correctly by the machinery that controls cell cycle progression and by the programs that induce cell specification. Despite great advances in identifying the proteins that govern cell cycle progression, little is known about how their activity is modulated by developmental signals. In this issue of the Proceedings, Quon et al. (1) provide new insights into understanding this fundamental problem. They identified an inhibitor of DNA replication whose activity is temporally and spatially regulated, thereby restricting its activity during development. However, these findings were not made, as one might have expected, in a eukaryotic organism, but in the eubacterium Caulobacter crescentus. The similarities in the regulation of this bacterial cell cycle regulator and cell fate determinant and that of eukaryotic cell cycle regulators and cell fate determinants will be discussed in this commentary.
Cell division in Caulobacter is asymmetric (2). A sessile stalked cell divides to give rise to two daughter cells, a stalked cell and a motile swarmer cell (Fig. 1A). The stalked cell is capable of immediately replicating its chromosome and subsequently dividing, whereas the swarmer cell must differentiate into a stalked cell before it can initiate DNA replication and cell division. The generation of a stalked cell and a swarmer cell during cell division and the initiation of DNA replication after the transition of a swarmer cell into a stalked cell are ultimately controlled by the CtrA protein (for cell cycle transcriptional regulator A; ref. 3). CtrA is a transcription factor of the response regulator superfamily that modulates transcription of genes important for flagellar biosynthesis, cell division, and DNA methylation (3). Although CtrA was implicated in the control of DNA replication (4), the paper by Quon et al. (1) reports that CtrA directly inhibits DNA replication in the swarmer cell. They show that CtrA inhibits DNA replication by binding to five distinct sites within the chromosomal replication origin. These sites overlap both a promoter within the origin whose transcription is required for initiation of chromosome replication and also binding sites for the DNA replication initiation factor DnaA. These findings demonstrate that CtrA is not only critically important for cell fate determination but is also an important cell cycle regulator.
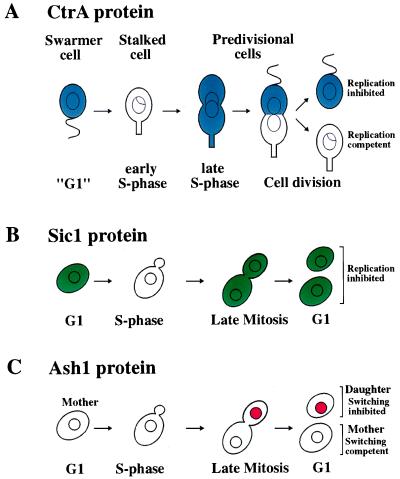
Figure 1.
Comparison of the temporal and spatial localization of CtrAp, Sic1p, and Ash1p. (A) To facilitate the comparison of CtrA accumulation with that of Sic1, the nomenclature of the eukaryotic cell cycle has been adapted. CtrA protein is present in replication incompetent swarmer cells (“G1”), where it inhibits the initiation of DNA replication. At the swarmer cell–stalked cell transition, CtrA is degraded and the cell initiates DNA replication. CtrA protein then accumulates during S phase, where it is uniformly distributed throughout the predivisional cell. Shortly before or at the time of chromosome partitioning and cell division, CtrA is cleared from the stalked cell but remains present in the swarmer cell portion of the predivisional cell. CtrA protein then persists in the swarmer cell. (B) Sic1 protein is present in G1 cells, where it inhibits cyclin-dependent kinases that trigger initiation of DNA replication. At the G1–S phase transition, Sic1 protein is degraded, allowing entry into S phase. The protein is absent throughout S phase and early mitosis, but accumulates during late mitosis. The protein then persists throughout the following G1 period. Because the localization of Sic1 within the cell is not known, the protein has been drawn to be distributed throughout the cell. (C) Ash1, like CtrA, is asymmetrically distributed between mother and daughter cells at the end of mitosis. Ash1 protein is absent in early stages of the cell cycle but accumulates toward the end of mitosis in the bud, the future daughter cell. It persists in the daughter cell throughout the G1 phase, inhibiting transcription of the HO endonuclease thereby preventing mating-type switching. As this daughter cell undergoes a new cell cycle, Ash1 protein is degraded. By the following G1 phase, when the daughter cell has become a mother cell, Ash1 protein has been eliminated, allowing the cell to switch its mating type.
The activity and abundance of CtrA are under tight cell cycle as well as developmental control. Cell cycle regulation is manifested in changes in CtrA protein levels and activity during the cell cycle (temporal restriction), whereas developmental control is manifested in spatial restriction. Three layers of regulation restrict CtrA to the correct window of the cell cycle and the appropriate cell type (3, 4). First, transcriptional control ensures that CtrA is only transcribed in the predivisional cell after DNA replication has been initiated. Second, selective proteolysis clears CtrA protein from the stalked cell portion of the predivisional cell and from the swarmer cell as it develops into a stalked cell. Third, a phosphorylation event on the aspartic acid residue at position 51, which is required for CtrA to be active, occurs only in the swarmer cell and the swarmer portion of the predivisional cell. The combination of these controls leads to the dynamic changes in CtrA activity and protein levels observed during the life cycle of the bacterium (Fig. 1A). After the stalked cell has initiated DNA replication, CtrA transcription is initiated. The protein accumulates during the chromosome replication phase and is distributed equally between the stalked cell and swarmer cell portions of the predivisional cell. Shortly before or at the onset of cell division, CtrA is rapidly degraded in the stalked cell portion of the predivisional cell but remains stable and thus present at high levels in the swarmer portion of the cell, where the protein persists throughout the time the bacterium is locked in the swarmer cell fate. Concomitant with the transformation of the swarmer cell into a stalked cell, selective protein degradation clears CtrA protein from the cell allowing the initiation of DNA replication and stalked cell development.
The cell cycle regulation of CtrA protein levels exhibits similarities to that of the cyclin-dependent kinase inhibitor Sic1 in the budding yeast Saccharomyces cerevisiae (Fig. 1B), which, although indirect, is also an inhibitor of DNA replication. In eukaryotes, different types of cyclin-dependent kinases (CDKs, a complex between an activating regulatory cyclin subunit and a catalytic kinase subunit) trigger entry into the cell cycle, initiation of S phase, and entry into mitosis (5). The CDK inhibitor Sic1 inhibits the cyclin-dependent kinases that trigger entry into S phase and mitosis (6). As is the case for CtrA, transcriptional control and proteolysis (ubiquitin-dependent) restrict Sic1 protein to the correct window of the cell cycle. Sic1 is present only during late stages of mitosis and G1, where it acts as an inhibitor of cyclin-dependent kinases that trigger S phase and thus initiation of DNA replication. Like inactivation of CtrA at the swarmer-to-stalked cell transition, inactivation of Sic1 at the G1–S phase transition is a prerequisite for the initiation of DNA replication. When Sic1 is not degraded, cells fail to initiate DNA replication (7). Thus, in a remarkably parallel manner, cell cycle transitions in both eukaryotes and prokaryotes are regulated by selective protein degradation.
Stability and activity of CtrA, like that of eukaryotic cell cycle regulators, must be coordinated with all aspects of the cell cycle, including initiation of DNA replication, cell division, and morphogenesis. How these processes affect CtrA stability and activity is unclear. Protease activity or the accessibility of CtrA to the protease could be regulated. CtrA activity also is modulated by phosphorylation. Response regulator transcription factors such as CtrA are phosphorylated by histidine kinases that are involved in a wide variety of signal transduction systems. Thus, it is conceivable that CtrA is modulated by numerous histidine kinases, as is the case for the response regulator transcription factor Spo0A, which plays a critical role in spore formation in Bacillus subtilis (8). Thereby, CtrA may be capable of responding not to a single but several cell-intrinsic signals that reflect DNA replication, cell division, and morphogenesis.
The activity and protein levels of the cell cycle regulator and cell fate determinant CtrA are regulated not only temporally, but also spatially (Fig. 1A). Although asymmetric distribution of cell cycle regulators has not been reported in eukaryotes, asymmetric distribution of cell fate determinants is a mechanism commonly used in eukaryotes to generate daughter cells of different fate. Asymmetric distribution of the Numb and Prospero proteins plays a critical role in sensory organ development in Drosophila melanogaster (9). Asymmetric distribution of the Par proteins specifies cell fate during the early embryonic divisions in Caenorhabditis elegans (10). One of the best-studied examples of asymmetric segregation of cell fate determinants is mating-type switching in budding yeast (11). Mother cells, defined as cells that have budded in the previous cell cycle and thus have given birth to a daughter cell, switch their mating type. Daughter cells, on the other hand, do not switch their mating type. Mating-type switching is initiated by the Ho endonuclease, which is expressed only in mother cells. The potential transcriptional repressor Ash1 is ultimately responsible for repressing HO expression in the daughter cell (12, 13). The segregation of this cell fate determinant is similar to that of CtrA. During late mitosis, Ash1 accumulates in the future daughter cell, where it persists throughout the subsequent G1 period, preventing expression of HO (Fig. 1C; refs. 12 and 13). Ash1 is an unstable protein and thus decays as the daughter cell progresses through the cell cycle (14). By the following G1 phase, when the daughter cell has become a mother cell, the Ash1 protein has been eliminated. The mechanism whereby Ash1 is restricted to the daughter cells is very different from that of CtrA localization, however. ASH1 RNA is localized to the future daughter cell in a manner that depends on the actin cytoskeleton (14), whereas CtrA protein is cleared from the future stalked cell by selective proteolysis (4).
The finding that the cell fate and cell cycle regulator CtrA is asymmetrically regulated is a breakthrough in understanding how cell specification is controlled and how cell cycle progression is modulated by cell cycle and developmental cues in Caulobacter. It seems, however, that we have seen only the tip of the iceberg. As with every significant paper, the complex pattern of temporal and spatial control of CtrA activity poses as many questions about how asymmetry in gene expression and in cell cycle behavior is generated as it answers. What are the mechanisms that restrict CtrA proteolysis to the stalked cell portion of the predivisional cell? How is proteolysis activated as swarmer cells differentiate into stalked cells? Similar questions can be posed concerning the identity and regulation of the histidine kinase(s) that activate CtrA in the swarmer cell but not the stalked cell. Answering these questions will provide key insights into the complex regulatory networks that integrate developmental cues with the cell cycle and differentiation programs in both bacteria and eukaryotes.
References
- 1.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brun Y V, Marczynski G, Shapiro L. Annu Rev Biochem. 1994;63:419–450. doi: 10.1146/annurev.bi.63.070194.002223. [DOI] [PubMed] [Google Scholar]
- 3.Quon K C, Marczynski G T, Shapiro L. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 4.Domian I J, Quon K C, Shapiro L. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 5.Nigg E A. BioEssays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- 6.Mendenhall M D M. Science. 1993;259:216–219. doi: 10.1126/science.8421781. [DOI] [PubMed] [Google Scholar]
- 7.Schwob E, Böhm T, Mendenhall M D, Nasmyth K. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 8.Ireton K, Grossman A. EMBO J. 1994;13:1566–1573. doi: 10.1002/j.1460-2075.1994.tb06419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knoblich J A, Jan L Y, Jan Y N. Nature (London) 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- 10.Guo S, Kemphues K J. Curr Opin Genet Dev. 1996;6:408–415. doi: 10.1016/s0959-437x(96)80061-x. [DOI] [PubMed] [Google Scholar]
- 11.Nasmyth K. Curr Opin Genet Dev. 1993;3:286–294. doi: 10.1016/0959-437x(93)90036-o. [DOI] [PubMed] [Google Scholar]
- 12.Bobola N, Jansen R-P, Shin T-H, Nasmyth K. Cell. 1996;84:699–710. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- 13.Sil A, Herskowitz I. Cell. 1996;84:711–721. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 14.Long R M, Singer R H, Meng X, Gonzalez I, Nasmyth K, Jansen R-P. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]