Abstract
Autophagy is an essential process for the maintenance of cellular homeostasis in the heart under both normal and stress conditions. Autophagy is a key degradation pathway and acts as a quality control sensor. It protects myocytes from cytotoxic protein aggregates and dysfunctional organelles by quickly clearing them from cell. It also responds to changes in energy demand and mechanical stressors to maintain contractile function. The autophagic-lysosomal pathway responds to serum starvation to ensure that the cell maintains its metabolism and energy levels when nutrients run low. In contrast, excessive activation of autophagy is detrimental to cells and contributes to development of pathological conditions. A number of signaling pathways and proteins regulate autophagy. These include the AMPK/mTOR pathway, FoxO transcription factors, Sirt1, oxidative stress, Bcl-2 family proteins, and the E3 ubiquitin ligase Parkin. In this review, we will discuss how this diverse cast of characters regulates the important autophagic process in the myocardium.
Keywords: Autophagy, AMPK, mTOR, Beclin1, ULK1, Parkin, mitochondria
INTRODUCTION
Autophagy is an evolutionarily conserved catabolic process that is responsible for the degradation of cytoplasmic components via the lysosomal pathway 1. In the absence of stress, autophagy complements the function of the proteasome by degrading long-lived proteins. It also plays an important role in cellular quality control and is responsible for clearing protein aggregates and dysfunctional organelles that could become toxic to the cell. This quality control function is particularly important in post-mitotic cells such as myocytes and neurons that are not easily replaced. Autophagy increases under conditions of limited nutrients and degrades cytoplasmic material to provide the cell with amino acids and fatty acids. The breakdown of organelles and proteins ensures that the cell can maintain its metabolism and energy level when nutrients run low 2. Autophagy is also upregulated by many other stressors including opening of the mitochondrial permeability transition pore (mPTP) 3, ER stress 4, and increased production of reactive oxygen species (ROS) 5.
Autophagy has important functions in the myocardium and its dysregulation has been implicated in a wide variety of cardiovascular pathologies. For instance, Danon disease is a fatal cardiomyopathy caused by a defect in the fusion between autophagosomes and lysosomes that leads to an accumulation of autophagosomes in the myocytes 6, 7. Also, cardiac specific deletion of the autophagy-related gene 5 (Atg5), a critical autophagy protein, in the adult heart leads to disruption of autophagy with subsequent buildup of dysfunctional mitochondria and development of cardiac dysfunction 8. Similarly, deletion of Atg7 in skeletal muscle leads to accumulation of impaired mitochondria and a corresponding increase in intracellular ROS levels 9. Increased autophagy is commonly observed in the heart with acute and chronic ischemia, heart failure and dilated cardiomyopathy 5, 10–14.
Since cardiac myocytes are terminally differentiated and possess extremely limited regenerative capacity, rapid adaptive activation of autophagy in response to metabolic or mechanical stress is critical for the maintenance of normal cardiac function. Activation of autophagy will generate intracellular nutrients and energy required to survive the stress, and will also remove damaged organelles such as leaky mitochondria that can be harmful to the cell 15. Here, we review our current knowledge of metabolic and stress signaling pathways that regulate autophagy in the myocardium.
Initiation of Autophagy
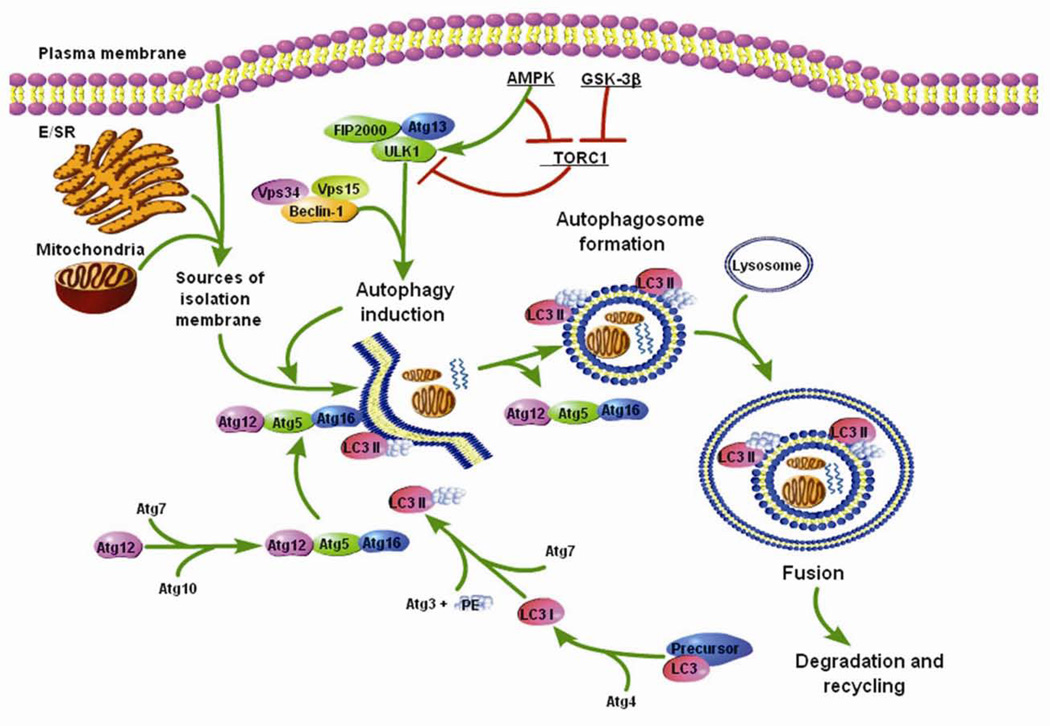
The study of autophagy in yeast has provided detailed knowledge about autophagic signaling pathways and advanced our general understanding of autophagy in the heart. At least sixteen different Atg gene products coordinate the formation of an autophagosome 16. When the cell receives a signal to initiate autophagy, an isolation membrane (also called phagophore) is formed (Figure 1). Although the origin of the membrane is still unclear, recent studies have found that sarcoplasmic/endoplasmic reticulum 17, mitochondrial outer membrane,18 and plasma membrane 19 can all serve as sources of the isolation membrane. Unc-51-like kinase (ULK1), which forms a complex with Atg13 and focal adhesion kinase-family interacting protein of 200 kD (FIP200), is a key regulator of the initiation of autophagy 20, 21. The initial phagophore formation (nucleation) requires assembly of the Beclin1 (Atg6 in yeast)-vacuolar sorting protein 34 (Vps34)-Vps15 complex 22. Beclin1 is regulated by the anti-apoptotic proteins Bcl-2 and Bcl-XL, which bind Beclin1 to inhibit activity and induction of autophagy 23–25. Subsequent expansion of the membrane is mediated by two ubiquitin-like conjugation systems, Atg12 and Atg8 (microtubule-associated protein 1 light chain 3 (LC3) in mammals) that together promote assembly of the Atg16L complex and the processing of LC3 26, 27. The isolation membrane elongates until the edges fuse around its target(s) forming a double-membrane structure called the autophagosome. The autophagosome then moves along the microtubules to the microtubule organizing center where it fuses with a lysosome 28. The contents are degraded by lysosomal digestive enzymes, and the breakdown products, including amino acids, lipids, nucleosides, and carbohydrates, are released into the cytosol where they can be used by synthetic and metabolic pathways.
Figure 1.
Schematic of the autophagic process. The plasma membrane, mitochondria, and E/SR serve as sources of the isolation membrane. The initiation of autophagy is regulated by the ULK1 protein complex. The initial phagophore formation requires Beclin1-Vps34-Vps15 complex and expansion of the membrane is mediated by two ubiquitin-like conjugation systems, Atg12 and LC3. The membrane elongates until the edges fuse around its target forming the autophagosome. The autophagosome then fuses with the lysosome and the content is degraded.
Autophagy and Energetic Balance
Induction of autophagy is critical for the maintenance of cardiac function under starved conditions 2, 29, and inhibition of autophagy results in reduced ATP levels and development of severe cardiac dysfunction 2. Moreover, autophagy plays an important role in salvaging myocytes during acute myocardial infarction (MI) by preserving amino acid and ATP levels. Autophagy was rapidly enhanced in the region bordering the infarction 30–32, and inhibiting autophagy prior to coronary ligation resulted in increased injury. In contrast, starvation of mice for 24 h prior to MI enhanced autophagy after occlusion of the left descending coronary artery and reduced infarct size compared to normally fed mice 30. Furthermore, amino acid and ATP levels were significantly reduced 4 h post-infarction in controls, but hearts of mice subjected to starvation prior to MI had preserved amino acid and ATP levels. Inhibition of autophagy abolished the protective effect of starvation 30. Clearly, this suggests that activation of autophagy during MI preserves ATP levels and maintains an adequate amino acid pool, which can be used for energy production and protein synthesis that required for the proper response to the ischemic insult.
The serine/threonine kinase mammalian target of rapamycin (mTOR) i s an important negative regulator of autophagy in mammalian cells 33, 34 and integrates intracellular signals such as growth factors, amino acids, glucose, and energy status 35. In the heart, mTORC1 plays an important role in regulating cellular homeostasis, and cardiac specific deletion of mTOR leads to development of a fatal dilated cardiomyopathy 36. mTOR exists in a multiprotein complex known as mTOR complex 1 (mTORC1) which includes Raptor, proline-rich Akt substrate of 40-kDa (PRAS40), and MTOR associated protein LST8 homolog (Mlst8) 37. It has been reported that mTORC1 suppresses autophagy by binding and phosphorylating the autophagy-initiating kinase ULK1 38. When inactivated, mTORC1 dissociates from ULK1, freeing it to initiate formation of the isolation membrane. Also, perfusion of isolated hearts with rapamycin, an inhibitor of mTOR and activator of autophagy, prior to I/R reduced infarct size suggesting that mTOR is an important regulator of essential cellular functions during ischemia 39, 40.
The mTOR pathway is regulated by the 5’-AMP-activated protein kinase (AMPK) (Figure 1). AMPK is an intracellular energy sensor that is activated in response to changes in the AMP/ATP ratio. Activation of AMPK results in adaptive changes in growth and metabolism under conditions of low energy. AMPK stimulates autophagy by inhibiting mTOR through phosphorylation and activation of the tuberous sclerosis complex 2 (TSC2) 41 and Raptor 42. TSC2 exists in a complex with TSC1 in cells, and the TSC1/2 complex inhibits the mTOR activator Rheb 43. TSC2 acts as a GTPase activating protein towards Rheb and by stimulating the conversion of active Rheb-GTP into the inactive Rheb-GDP, the TSC1/2 complex inhibits mTOR 44. Raptor is an essential mTOR binding partner and is responsible for recruiting substrates to the mTORC1 complex 45, 46. Recently, it was reported that AMPK can directly activate autophagy by directly activating ULK1. Kim et al. discovered that under glucose starvation, AMPK promotes autophagy by directly activating ULK1 through phosphorylation of Ser 317 and Ser 777 38. In contrast, during nutrient sufficiency, high mTOR activity prevents ULK1 activation by phosphorylating ULK1 Ser757 and thus disrupting the interaction between ULK1 and AMPK. When nutrients are limited, as seen in myocardial ischemia, AMPK acts as a checkpoint by inhibiting cellular growth and inducing autophagy in cardiac myocytes. The AMPK-mTOR pathway is an important regulator of autophagy in response to glucose deprivation in neonatal myocytes as inhibition of AMPK reduced autophagy and increased cell death in these cells 13. Also, transgenic mice with cardiac specific expression of a dominant negative AMPK had attenuated induction of autophagy in response to ischemia in vivo 13. This suggests that AMPK-induced autophagy regulation via inhibition of mTOR plays an important role in the adaptation to ischemia.
Recently, glycogen synthase kinase-3 (GSK-3) was identified to be a regulator of the mTOR pathway in cardiac cells. GSK-3 is an important signaling molecule and is involved in regulating gene transcription, protein translation, and apoptosis, as well as hexose metabolism 47. GSK-3β is known to function downstream of phosphatidylinositol 3-kinase (PI3K) and Akt 48, and, in the heart, inhibition of GSK-3β has been reported to be cardioprotective 14, 48–50. Similar to AMPK, GSK-3β was found to inhibit mTOR signaling and activate autophagy via phosphorylation of TSC2 51,52. Moreover, Zhai et al. discovered that GSK-3β is a critical upstream regulator of mTOR during both ischemia and reperfusion in the heart 14.
Regulation of Autophagy by FoxO Transcription Factors and the NAD-dependent deacetylase SIRT1
Autophagy is also regulated by the FoxO (Forkhead box-containing protein, O subfamily) transcription factors 53. FoxO1 and FoxO3 are highly expressed in the heart 54 and regulate autophagy by activating transcription of the Atg genes 55. It was recently reported that glucose deprivation of cardiac myocytes induced translocation of FoxO1 and FoxO3 to the nucleus where they activated transcription of genes involved in autophagy 55,56. In addition, transgenic mice overexpressing FoxO3 in the heart showed increased levels of autophagy which correlated with the development of cardiac atrophy 57. In contrast, genetic deletion of FoxO3 resulted in development of cardiac hypertrophy 58. Both FoxO1 and FoxO3 have been reported to induce expression of the BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 (Bnip3) 59, 60 which is a potent inducer of autophagy.
Acetylation/deacetylation status of certain proteins is also linked to regulation of autophagy. The Sirtuin 1 (Sirt1) is a NAD-dependent deacetylase which is upregulated in response to starvation or caloric restriction 61, 62. Interestingly, Lee et al. found that starvation-induced autophagy was reduced in Sirt1−/− MEFs and that Sirt1 regulates autophagy via deacetylation of Atg5, Atg7 and Atg8 63. Another study recently reported that deacetylation of FoxO1 by Sirt1 was an essential step for autophagy in glucose-deprived cardiac myocytes as FoxO1 mutants that cannot be deacetylated by Sirt1 inhibited induction of autophagy 56.
Induction of Autophagy by Stress Signals
Many studies have reported that autophagy is upregulated in the heart in response to ischemia/reperfusion and hemodynamic stress 5, 10, 13. Multiple signaling pathways are activated in these hearts, and many of them are likely to contribute to the activation of autophagy. For instance, increased oxidative stress activates autophagy in cells, including cardiac myocytes 5, 64. Production of ROS played an important role in inducing autophagy during I/R in vivo, and the presence of an anti-oxidant significantly reduced the level of autophagy 5. Interestingly, mitochondria are a major source of ROS, which promotes activation of autophagy in cells. For instance, inhibition of electron transport chain complexes I or II induced ROS production and autophagy in cells, whereas the presence of a ROS scavenger decreased autophagy 65. Increased ROS could affect autophagy by directly acting on the autophagy proteins. Hydrogen peroxide was reported to oxidize and subsequently inactivate Atg4 during starvation. Atg4 is responsible for recycling of LC3 by cleaving phosphatidylethanolamine (PE) from PEconjugated LC3 and inhibition of Atg4 leads to accumulation of LC3-PE (LC3-II) and increased autophagosome formation 66. In addition, oxidative stress might indirectly activate autophagy by causing damage to organelles such as mitochondria that induces mitochondrial autophagy.
The mitochondrial permeability transition pore (mPTP) causes permeabilization of the inner mitochondrial membrane resulting in swelling of the inner membrane with subsequent rupture of the outer membrane 67. Opening of the mPTP have been reported to activate autophagy in mammalian cells 68–70. In contrast, starvation-induced autophagy was significantly reduced in the presence of cyclosporine A, an inhibitor of the mPTP 3. Cyclophilin D (CypD) is an essential component of the mPTP, and CypD-deficient mice are resistant to mPTP opening 71. This study found that fasting failed to induce autophagy in CypD-deficient mice while, transgenic mice overexpressing cypD in the heart showed enhanced levels of autophagy under normal and fasting conditions compared to wild type mice 3. This study suggests that the mPTP plays an important role in starvation-induced autophagy. In contrast, Quinsay et al. found that the BH3-only protein Bnip3 induced autophagy independent of the mPTP in cardiac myocytes 72. Although the mPTP plays a role in the induction of autophagy, it is unclear if loss of mitochondrial membrane potential due to pore opening alone is responsible for the induction of autophagy or if factor(s) released from the mitochondria activate the autophagic pathway.
The endoplasmic reticulum (ER) is important in regulating cytosolic Ca2+ homeostasis and proper folding of newly synthesized proteins 73. Defects in ER function lead to activation of the ER stress pathway and activation of autophagy 4 via down-regulation of mTOR signaling 74, monocyte chemotactic protein-1 induced protein (MCPIP) 75, and JNK/p38 and activating transcription factor 4 (ATF4)-dependent activation 76. Interestingly, treatment of hearts with low doses of tunicamycin or thapsigargin, two different inducers of ER stress, resulted in induction of autophagy and reductions in apoptosis and infarct size 77. This suggests that ER stress-mediated autophagy can serve a protective role in the heart.
Mitochondrial Autophagy in the Myocardium
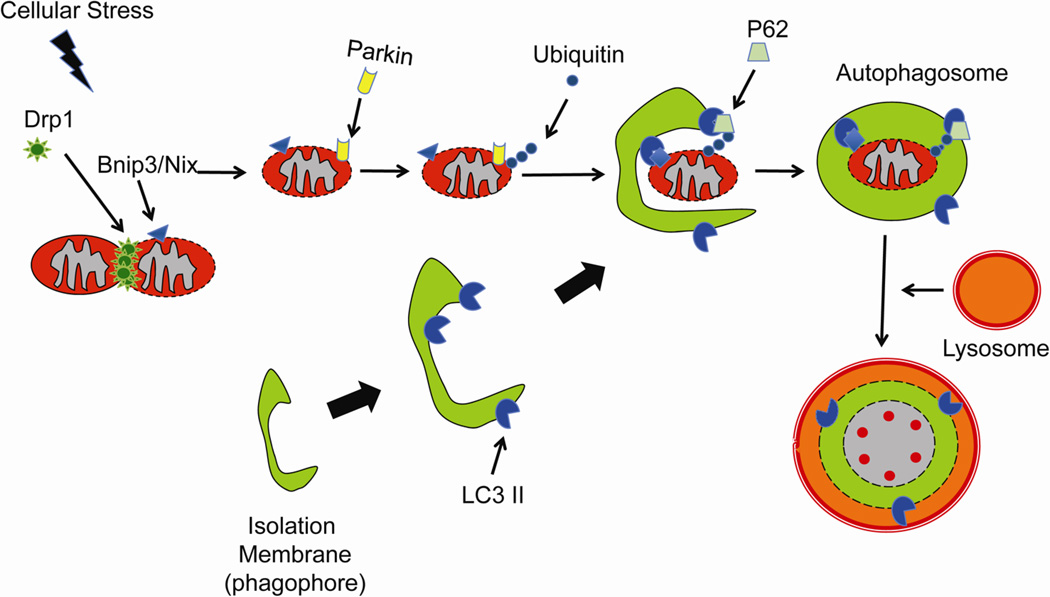
For a long time, autophagy was considered to be a non-selective process in which the autophagosomes randomly sequestered material in the cytosol for degradation. However, many studies have now demonstrated that autophagy can be selective and specifically remove damaged organelles and toxic protein aggregates (Figure 2). Bnip3 and Bnip3-like (Bnip3L/Nix) belong to the BH3-only proteins of the Bcl-2 family that are primarily localized to mitochondria in cells. Both Bnip3 and Nix are potent inducers of mitochondrial autophagy 78–80. Sandoval et al. found that Nix was required for mitophagy in erythroid cells 81. Interestingly, Nix was not required for the induction of general autophagy, but only for the elimination of mitochondria 79. Similarly, Bnip3 has been reported to induce mitochondrial autophagy in cardiac myocytes 72, 78, 82. Exactly how Nix and Bnip3 target mitochondria for autophagy is still unclear, but it has been shown that Nix-dependent loss of mitochondrial membrane potential (Δψm) was important in targeting the mitochondria to autophagosomes 81. In addition, Nix has been proposed to act as an autophagy receptor by interacting directly with the autophagy proteins LC3 and GABARAP 83. Bnip3 has also been reported to interact with LC3 84. However, the importance of Nix and Bnip3 as autophagy receptors is still unclear since mitophagy was restored when Nix−/− cells were treated with compounds that caused mitochondrial depolarization 81.
Figure 2.
Induction of mitochondrial autophagy in response to cellular stress. After Drp1-mediated mitochondrial fission, Parkin is recruited to dysfunctional mitochondria where it ubiquitinates proteins. The p62 adaptor protein binds to the ubiquitinated proteins and LC3 on the autophagosomes linking the two organelles. Alternatively, LC3 can directly interact with Bnip3 or Nix on the mitochondria.
The E3 ubiquitin ligase Parkin has also been identified as an important regulator of mitochondrial autophagy in cells. Parkin-mediated clearance of mitochondria plays an important role in cardiac preconditioning 85. Narendra et al. discovered that Parkin accumulated on depolarized mitochondria which promoted their removal by autophagy 86. Parkin was found to ubiquitinate proteins on dysfunctional mitochondria, which served to recruit HDAC6 and p62/SQSTM1, and promote assembly of the autophagy machinery at the impaired mitochondrion 87. Interestingly, p62 was not essential for Parkin-mediated mitophagy as p62 deficiency in cells did not prevent CCCP-mediated mitophagy 88. We recently found that Bnip3-mediated mitochondrial autophagy was dependent on Parkin in cardiac myocytes and that Parkin deficient myocytes had reduced induction of autophagy in response to Bnip3 overexpression 89. We also found that mitochondria had to undergo Drp1-mediated mitochondrial fission prior to Parkin-translocation and removal by autophagosomes. Also, Ding et al. found that CCCP-induced Parkin translocation was significantly reduced in Nix deficient MEFs 90. These studies suggest that Bnip3 and Nix cooperate with the Parkin pathway to clear dysfunctional mitochondria in cells.
Maladaptive role of Autophagy in the Myocardium
Most studies suggest that autophagy is a protective response activated by the cell during stress. However, it is now evident that constitutive and/or excessive autophagy can be detrimental to the heart. Recently, Schips et al. reported that constitutive activation of FoxO3 in mouse hearts led to increased autophagy and atrophy 57. These hearts reduced stroke volume and cardiac output, suggesting that constitutive activation of autophagy is harmful for the myocardium. Interestingly, this study discovered that the cardiac atrophy and dysfunction were reversible when FoxO3 expression was turned off 57. In addition, studies have reported that excessive autophagy contributes to pathological conditions in response to stress. Upregulation of Beclin1 and activation of autophagy during reperfusion after ischemia have been found to be detrimental to cardiac myocytes 13. Interestingly, Beclin1 heterozygous (Beclin1+/−) mice had reduced levels of autophagy and showed significantly smaller infarcts compared to wild type mice after I/R. This suggests that upregulation of Beclin1 contributes to excessive activation of autophagy which is detrimental to cells. Similarly, it was reported that reduced mTOR activity and enhanced autophagy during reperfusion contributed to I/R injury 14. Autophagy has also been reported to play a maladaptive role in a mouse model of pressure overload 10, 91. Zhu et al. found that Beclin1 heterozygous mice had decreased induction of autophagy and reduced hypertrophic growth in response to pressure over load, whereas overexpression of Beclin1 led to increased induction of autophagy and enhanced pathological hypertrophy compared to wild type mice 10. Collectively, these studies suggest that the duration and level of autophagy play an important role in determining whether autophagy will be protective or maladaptive in the heart.
Conclusion
Studies suggest that basal levels of autophagy are important for maintaining cellular homeostasis and for protecting cells against accumulation of toxic protein aggregates or dysfunctional organelles. It is also clear that enhancing autophagy can promote survival in response to stress, such as nutrient deprivation or hypoxia, by recycling macromolecules to maintain energy levels and removing damaged organelles such as mitochondria. In contrast, excessive levels of autophagy contribute to development of pathological conditions, most likely by removal of too many essential organelles and proteins. Manipulation of signaling pathways that regulate autophagy may represent a potential future therapeutic target to treat or prevent development of heart disease. A more thorough understanding of signaling pathways that regulate autophagy will be of great importance for future studies of the heart.
Acknowledgments
Funding
This manuscript was supported by NIH grants R01HL087023, R01HL101217, and R01HL092136.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Developmental cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Kanamori H, Takemura G, Maruyama R, Goto K, Tsujimoto A, Ogino A, Li L, Kawamura I, Takeyama T, Kawaguchi T, Nagashima K, Fujiwara T, Fujiwara H, Seishima M, Minatoguchi S. Functional significance and morphological characterization of starvation-induced autophagy in the adult heart. Am J Pathol. 2009;174:1705–1714. doi: 10.2353/ajpath.2009.080875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carreira RS, Lee Y, Ghochani M, Gustafsson AB, Gottlieb RA. Cyclophilin d is required for mitochondrial removal by autophagy in cardiac cells. Autophagy. 2010;6:462–472. doi: 10.4161/auto.6.4.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoyer-Hansen M, Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 5.Hariharan N, Zhai P, Sadoshima J. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. Antioxid Redox Signal. 2011;14:2179–2190. doi: 10.1089/ars.2010.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, Mora M, Riggs JE, Oh SJ, Koga Y, Sue CM, Yamamoto A, Murakami N, Shanske S, Byrne E, Bonilla E, Nonaka I, DiMauro S, Hirano M. Primary lamp-2 deficiency causes x-linked vacuolar cardiomyopathy and myopathy (danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in lamp-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 8.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 9.Wu JJ, Quijano C, Chen E, Liu H, Cao L, Fergusson MM, Rovira II, Gutkind S, Daniels MP, Komatsu M, Finkel T. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging. 2009;1:425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, Le V, Levine B, Rothermel BA, Hill JA. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tannous P, Zhu H, Nemchenko A, Berry JM, Johnstone JL, Shelton JM, Miller FJ, Jr., Rothermel BA, Hill JA. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan L, Vatner DE, Kim SJ, Ge H, Masurekar M, Massover WH, Yang G, Matsui Y, Sadoshima J, Vatner SF. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci U S A. 2005;102:13807–13812. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: Roles of amp-activated protein kinase and beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 14.Zhai P, Sciarretta S, Galeotti J, Volpe M, Sadoshima J. Differential roles of gsk-3beta during myocardial ischemia and ischemia/reperfusion. Circ Res. 2011;109:502–511. doi: 10.1161/CIRCRESAHA.111.249532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlieb RA, Gustafsson AB. Mitochondrial turnover in the heart. Biochim Biophys Acta. 2011;1813:1295–1301. doi: 10.1016/j.bbamcr.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juhasz G, Neufeld TP. Autophagy: A forty-year search for a missing membrane source. PLoS Biol. 2006;4:e36. doi: 10.1371/journal.pbio.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. Fip200, a ulk-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. Ulk1.Atg13.Fip200 complex mediates mtor signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang R, Zeh HJ, Lotze MT, Tang D. The beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the bcl-xl-beclin 1 peptide complex: Beclin 1 is a novel bh3-only protein. J Biol Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 24.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, Hickman JA, Geneste O, Kroemer G. Functional and physical interaction between bcl-x(l) and a bh3-like domain in beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, Korsmeyer S, Packer M, May HI, Hill JA, Virgin HW, Gilpin C, Xiao G, Bassel-Duby R, Scherer PE, Levine B. Exercise-induced bcl2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012;481:511–515. doi: 10.1038/nature10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the apg12p-apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. Lc3, gabarap and gate16 localize to autophagosomal membrane depending on form-ii formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 28.Huynh KK, Eskelinen EL, Scott CC, Malevanets A, Saftig P, Grinstein S. Lamp proteins are required for fusion of lysosomes with phagosomes. EMBO J. 2007;26:313–324. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanamori H, Takemura G, Goto K, Maruyama R, Ono K, Nagao K, Tsujimoto A, Ogino A, Takeyama T, Kawaguchi T, Watanabe T, Kawasaki M, Fujiwara T, Fujiwara H, Seishima M, Minatoguchi S. Autophagy limits acute myocardial infarction induced by permanent coronary artery occlusion. Am J Physiol Heart Circ Physiol. 2011;300:H2261–H2271. doi: 10.1152/ajpheart.01056.2010. [DOI] [PubMed] [Google Scholar]
- 31.Kanamori H, Takemura G, Goto K, Maruyama R, Tsujimoto A, Ogino A, Takeyama T, Kawaguchi T, Watanabe T, Fujiwara T, Fujiwara H, Seishima M, Minatoguchi S. The role of autophagy emerging in postinfarction cardiac remodelling. Cardiovasc Res. 2011;91:330–339. doi: 10.1093/cvr/cvr073. [DOI] [PubMed] [Google Scholar]
- 32.Zhang JL, Lu JK, Chen D, Cai Q, Li TX, Wu LS, Wu XS. Myocardial autophagy variation during acute myocardial infarction in rats: The effects of carvedilol. Chinese medical journal. 2009;122:2372–2379. [PubMed] [Google Scholar]
- 33.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, Rubinsztein DC. Inhibition of mtor induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of huntington disease. Nature genetics. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 34.Mousavi SA, Brech A, Berg T, Kjeken R. Phosphoinositide 3-kinase regulates maturation of lysosomes in rat hepatocytes. Biochem J. 2003;372:861–869. doi: 10.1042/BJ20021136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wullschleger S, Loewith R, Hall MN. Tor signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Zhang D, Contu R, Latronico MV, Zhang J, Rizzi R, Catalucci D, Miyamoto S, Huang K, Ceci M, Gu Y, Dalton ND, Peterson KL, Guan KL, Brown JH, Chen J, Sonenberg N, Condorelli G. Mtorc1 regulates cardiac function and myocyte survival through 4e-bp1 inhibition in mice. J Clin Invest. 2010;120:2805–2816. doi: 10.1172/JCI43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunlop EA, Tee AR. Mammalian target of rapamycin complex 1: Signalling inputs, substrates and feedback mechanisms. Cellular signaling. 2009;21:827–835. doi: 10.1016/j.cellsig.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 38.Kim J, Kundu M, Viollet B, Guan KL. Ampk and mtor regulate autophagy through direct phosphorylation of ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan S, Salloum F, Das A, Xi L, Vetrovec GW, Kukreja RC. Rapamycin confers preconditioning-like protection against ischemia-reperfusion injury in isolated mouse heart and cardiomyocytes. J Mol Cell Cardiol. 2006;41:256–264. doi: 10.1016/j.yjmcc.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 40.Wouters BG, Koritzinsky M. Hypoxia signalling through mtor and the unfolded protein response in cancer. Nature reviews. Cancer. 2008;8:851–864. doi: 10.1038/nrc2501. [DOI] [PubMed] [Google Scholar]
- 41.Inoki K, Zhu T, Guan KL. Tsc2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 42.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. Ampk phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mtor kinase. Curr Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 44.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, tuberin and hamartin, control mtor signaling by acting as a gtpase-activating protein complex toward rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 45.Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, Yonezawa K. The mammalian target of rapamycin (mtor) partner, raptor, binds the mtor substrates p70 s6 kinase and 4e-bp1 through their tor signaling (tos) motif. J Biol Chem. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- 46.Schalm SS, Fingar DC, Sabatini DM, Blenis J. Tos motif-mediated raptor binding regulates 4e-bp1 multisite phosphorylation and function. Current biology : CB. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 47.Grimes CA, Jope RS. The multifaceted roles of glycogen synthase kinase 3beta in cellular signaling. Progress in neurobiology. 2001;65:391–426. doi: 10.1016/s0301-0082(01)00011-9. [DOI] [PubMed] [Google Scholar]
- 48.Tong H, Imahashi K, Steenbergen C, Murphy E. Phosphorylation of glycogen synthase kinase-3beta during preconditioning through a phosphatidylinositol-3-kinase--dependent pathway is cardioprotective. Circ Res. 2002;90:377–379. doi: 10.1161/01.res.0000012567.95445.55. [DOI] [PubMed] [Google Scholar]
- 49.Tong H, Chen W, Steenbergen C, Murphy E. Ischemic preconditioning activates phosphatidylinositol-3-kinase upstream of protein kinase c. Circ Res. 2000;87:309–315. doi: 10.1161/01.res.87.4.309. [DOI] [PubMed] [Google Scholar]
- 50.Hirotani S, Zhai P, Tomita H, Galeotti J, Marquez JP, Gao S, Hong C, Yatani A, Avila J, Sadoshima J. Inhibition of glycogen synthase kinase 3beta during heart failure is protective. Circ Res. 2007;101:1164–1174. doi: 10.1161/CIRCRESAHA.107.160614. [DOI] [PubMed] [Google Scholar]
- 51.Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, Wang CY, He X, MacDougald OA, You M, Williams BO, Guan KL. Tsc2 integrates wnt and energy signals via a coordinated phosphorylation by ampk and gsk3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 52.Buller CL, Loberg RD, Fan MH, Zhu Q, Park JL, Vesely E, Inoki K, Guan KL, Brosius FC., 3rd A gsk-3/tsc2/mtor pathway regulates glucose uptake and glut1 glucose transporter expression. American journal of physiology. Cell physiology. 2008;295:C836–C843. doi: 10.1152/ajpcell.00554.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greer EL, Brunet A. Foxo transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 54.Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, Morrisey EE, Margulies KB, Cappola TP. Transcriptional genomics associates fox transcription factors with human heart failure. Circulation. 2006;114:1269–1276. doi: 10.1161/CIRCULATIONAHA.106.632430. [DOI] [PubMed] [Google Scholar]
- 55.Sengupta A, Molkentin JD, Yutzey KE. Foxo transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of foxo by sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schips TG, Wietelmann A, Hohn K, Schimanski S, Walther P, Braun T, Wirth T, Maier HJ. Foxo3 induces reversible cardiac atrophy and autophagy in a transgenic mouse model. Cardiovasc Res. 2011;91:587–597. doi: 10.1093/cvr/cvr144. [DOI] [PubMed] [Google Scholar]
- 58.Ni YG, Berenji K, Wang N, Oh M, Sachan N, Dey A, Cheng J, Lu G, Morris DJ, Castrillon DH, Gerard RD, Rothermel BA, Hill JA. Foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation. 2006;114:1159–1168. doi: 10.1161/CIRCULATIONAHA.106.637124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. Foxo3 controls autophagy in skeletal muscle in vivo. Cell metabolism. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Xu P, Das M, Reilly J, Davis RJ. Jnk regulates foxo-dependent autophagy in neurons. Genes & development. 2011;25:310–322. doi: 10.1101/gad.1984311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates sirt1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 62.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the sirt1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 63.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the nad-dependent deacetylase sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dai DF, Johnson SC, Villarin JJ, Chin MT, Nieves-Cintron M, Chen T, Marcinek DJ, Dorn GW, 2nd, Kang YJ, Prolla TA, Santana LF, Rabinovitch PS. Mitochondrial oxidative stress mediates angiotensin ii-induced cardiac hypertrophy and galphaq overexpression-induced heart failure. Circ Res. 2011;108:837–846. doi: 10.1161/CIRCRESAHA.110.232306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Y, McMillan-Ward E, Kong J, Israels SJ, Gibson SB. Mitochondrial electron-transport-chain inhibitors of complexes i and ii induce autophagic cell death mediated by reactive oxygen species. J Cell Sci. 2007;120:4155–4166. doi: 10.1242/jcs.011163. [DOI] [PubMed] [Google Scholar]
- 66.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Halestrap AP, Clarke SJ, Javadov SA. Mitochondrial permeability transition pore opening during myocardial reperfusion--a target for cardioprotection. Cardiovasc Res. 2004;61:372–385. doi: 10.1016/S0008-6363(03)00533-9. [DOI] [PubMed] [Google Scholar]
- 68.Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 69.Arrington DD, Van Vleet TR, Schnellmann RG. Calpain 10: A mitochondrial calpain and its role in calcium-induced mitochondrial dysfunction. American journal of physiology. Cell physiology. 2006;291:C1159–C1171. doi: 10.1152/ajpcell.00207.2006. [DOI] [PubMed] [Google Scholar]
- 70.Teckman JH, An JK, Blomenkamp K, Schmidt B, Perlmutter D. Mitochondrial autophagy and injury in the liver in alpha 1-antitrypsin deficiency. American journal of physiology. Gastrointestinal and liver physiology. 2004;286:G851–G862. doi: 10.1152/ajpgi.00175.2003. [DOI] [PubMed] [Google Scholar]
- 71.Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, Robbins J, Molkentin JD. Loss of cyclophilin d reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 72.Quinsay MN, Thomas RL, Lee Y, Gustafsson AB. Bnip3-mediated mitochondrial autophagy is independent of the mitochondrial permeability transition pore. Autophagy. 2010;6:855–862. doi: 10.4161/auto.6.7.13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Berridge MJ. The endoplasmic reticulum: A multifunctional signaling organelle. Cell calcium. 2002;32:235–249. doi: 10.1016/s0143416002001823. [DOI] [PubMed] [Google Scholar]
- 74.Qin L, Wang Z, Tao L, Wang Y. Er stress negatively regulates akt/tsc/mtor pathway to enhance autophagy. Autophagy. 2010;6:239–247. doi: 10.4161/auto.6.2.11062. [DOI] [PubMed] [Google Scholar]
- 75.Younce CW, Kolattukudy PE. Mcp-1 causes cardiomyoblast death via autophagy resulting from er stress caused by oxidative stress generated by inducing a novel zinc-finger protein, mcpip. Biochem J. 2010;426:43–53. doi: 10.1042/BJ20090976. [DOI] [PubMed] [Google Scholar]
- 76.Rzymski T, Milani M, Singleton DC, Harris AL. Role of atf4 in regulation of autophagy and resistance to drugs and hypoxia. Cell Cycle. 2009;8:3838–3847. doi: 10.4161/cc.8.23.10086. [DOI] [PubMed] [Google Scholar]
- 77.Petrovski G, Das S, Juhasz B, Kertesz A, Tosaki A, Das DK. Cardioprotection by endoplasmic reticulum stress-induced autophagy. Antioxid Redox Signal. 2011;14:2191–2200. doi: 10.1089/ars.2010.3486. [DOI] [PubMed] [Google Scholar]
- 78.Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, Gottlieb RA, Gustafsson AB. Response to myocardial ischemia/reperfusion injury involves bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 79.Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, Ney PA. Nix is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J, Ney PA. Role of bnip3 and nix in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee Y, Lee HY, Hanna RA, Gustafsson AB. Mitochondrial autophagy by bnip3 involves drp1-mediated mitochondrial fission and recruitment of parkin in cardiac myocytes. Am J Physiol Heart Circ Physiol. 2011;301:H1924–H1931. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dotsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rikka S, Quinsay MN, Thomas RL, Kubli DA, Zhang X, Murphy AN, Gustafsson AB. Bnip3 impairs mitochondrial bioenergetics and stimulates mitochondrial turnover. Cell Death Differ. 2011;18:721–731. doi: 10.1038/cdd.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by parkin and p62/sqstm1. PLoS One. 2011;6:e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and hdac6-dependent mitophagy. J Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS, Kimura M, Sato S, Hattori N, Komatsu M, Tanaka K, Matsuda N. P62/sqstm1 cooperates with parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee Y, Lee HY, Hanna RA, Gustafsson AB. Mitochondrial autophagy by bnip3 involves drp1-mediated mitochondrial fission and recruitment of parkin in cardiac myocytes. Am J Physiol Heart Circ Physiol. 2011;301:H1924–H1931. doi: 10.1152/ajpheart.00368.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ding WX, Ni HM, Li M, Liao Y, Chen X, Stolz DB, Dorn GW, 2nd, Yin XM. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales CR, Kong Y, Rothermel BA, Gillette TG, Hill JA. Histone deacetylase (hdac) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci U S A. 2011;108:4123–4128. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]