Abstract
microRNAs (miRNAs) and small interfering RNAs (siRNAs), which constitute two major classes of endogenous small RNAs in plants, impact a multitude of developmental and physiological processes by imparting sequence specificity to gene and genome regulation. Although lacking the third major class of small RNAs found in animals, Piwi-interacting RNAs (piRNAs), plants have expanded their repertoire of endogenous siRNAs, some of which fulfill similar molecular and developmental functions as piRNAs in animals. Research on plant miRNAs and siRNAs has contributed invaluable insights into small RNA biology, thanks to the highly conserved molecular logic behind the biogenesis and actions of small RNAs. Here, I review progress in the plant small RNA field in the past two years, with an emphasis on recent findings related to plant development. I do not recount the numerous developmental processes regulated by small RNAs; instead, I focus on major principles that have been derived from recent studies and draw parallels, when applicable, between plants and animals.
Keywords: argonaute, miRNA, siRNAs, piRNAs, cell-to-cell movement, germ line
Introduction
siRNAs derived from exogenous sources such as transgenes and viruses were first discovered in plants [1], and this discovery was instrumental in elucidating the molecular basis of RNA interference whereby siRNAs guide the sequence-specific cleavage of complementary mRNAs. Endogenous small RNAs, siRNAs and miRNAs, were discovered in plants in 2002 [2–5], and the past ten years have witnessed an explosion of our knowledge of these regulatory molecules. The major frameworks underlying the biogenesis of miRNAs and various types of siRNAs have been established. The modes of action of small RNAs in gene silencing at the transcriptional and posttranscriptional levels are being intensively dissected. The impacts of small RNAs in a multitude of biological processes including plant development are increasingly appreciated.
miRNAs are encoded by hundreds of MIR genes in the Arabidopsis genome and act in trans to repress the expression of target genes at primarily post-transcriptional levels (reviewed in [6]). The biogenesis of miRNAs entails a series of events that are mostly conserved in plants and animals, such as transcription, dicing, nuclear export, and loading of the miRNA into an argonaute (AGO) protein to form the miRNA-induced silencing complex (miRISC). Among the ten AGO proteins in Arabidopsis, AGO1 binds almost all miRNAs and serves as the major miRNA effector [7,8]. miRISCs repress the expression of target genes through mRNA cleavage [9], which requires the endonucleolytic (slicer) activities of AGO1 [7,8], and translational inhibition [10], for which the underlying mechanisms are poorly understood. The widespread and critical developmental functions of miRNAs are reflected by the pleiotropic developmental defects of miRNA biogenesis mutants [11–14] as well as ago1 mutants [15], and solidified by numerous studies demonstrating the roles of miRNAs throughout plant development including embryogenesis [16,17], organ formation, patterning and growth [16,18,19], developmental transitions [20,21], and reproduction [22,23].
The pioneering effort to use high throughput sequencing to profile endogenous small RNAs [24] led to the first glimpse of the complexity of endogenous siRNAs, which are represented by tens of thousands of distinct species, in plants (reviewed in [25]). This review will discuss recent, unexpected findings regarding the biogenesis and developmental functions of two types of endogenous siRNAs, heterochromatic siRNAs and trans-acting siRNAs (ta-siRNAs).
Heterochromatic siRNAs are derived from repetitive sequences and transposable elements and are recruited back to the source or homologous chromatin to trigger DNA methylation and transcriptional silencing. This process, also known as RNA-directed DNA methylation (RdDM), has been intensively dissected in the past decade in Arabidopsis such that major players in siRNA biogenesis, siRNA recruitment to chromatin, and DNA methylation have been uncovered (reviewed in [26]). Intriguingly, heterochromatic siRNAs share similar molecular functions with piRNAs in animals. piRNAs and their protein partners, the Piwi subclade of argonaute proteins, silence transposable elements in the germ line of C. elegans, Drosophila, zebrafish, and mice (reviewed in [27]). It was recently shown that piRNAs trigger DNA methylation in mice [28,29], but the underlying mechanisms are poorly understood.
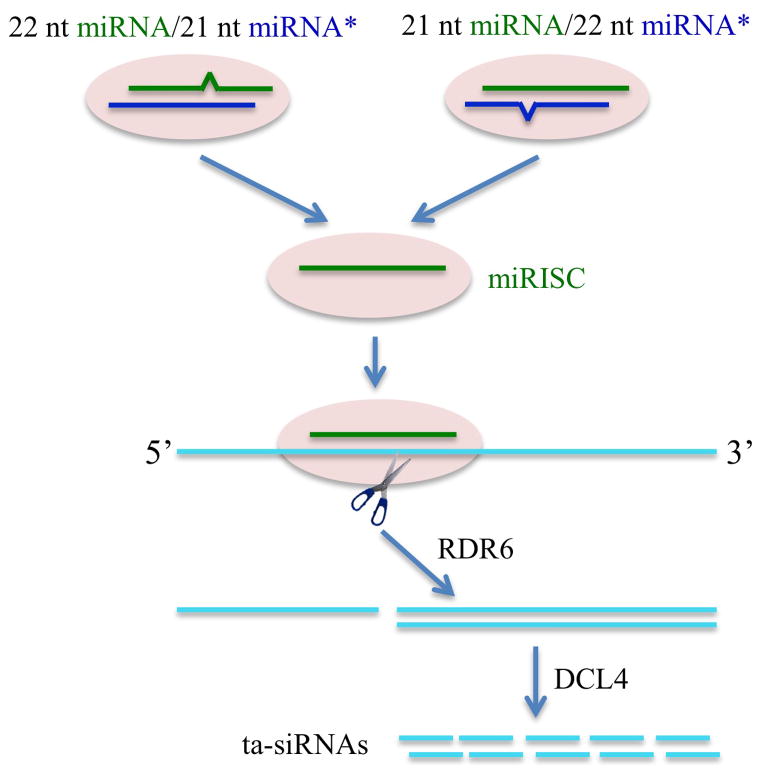
Ta-siRNAs are miRNA-triggered secondary siRNAs from miRNA-targeted noncoding transcripts [30–32] (Figure 1). Some of the secondary siRNAs generated at a particular locus are bound by an argonaute protein and regulate target transcripts other than their source transcripts, hence the name “trans-acting”. The miRNA-guided cleavage of a target transcript generates a fragment that serves as a template to synthesize double-stranded RNAs (dsRNAs) by RNA-DEPENDENT RNA POLYMERASE6 (RDR6). DICERLIKE4 (DCL4) processes the resulting dsRNAs into ta-siRNAs in succession from the end defined by miRNA-guided cleavage such that ta-siRNAs show strong phasing patterns, i.e., they are in a near perfect 21 nucleotide (nt) register from one another. Some ta-siRNAs, such as the tasiR-ARFs, so named because they target several AUXIN RESPONSE FACTOR (ARF) genes, are conserved in angiosperms and play critical roles in plant development [33–38].
Figure 1.
Biogenesis of trans-acting siRNAs (ta-siRNAs). A miRISC cleaves the target transcript into two fragments, one of which serves as a template for RNA-DEPENDENT RNA POLYMERASE6 (RDR6) to generate double-stranded RNAs (dsRNAs). The dsRNAs are diced into ta-siRNAs in a phased manner starting from the end defined by miRNA-guided cleavage. Recent studies show that the commitment to ta-siRNA biogenesis occurs already at the miRISC loading step. An asymmetric bulge (an unpaired nucleotide in one strand as indicated by the protrusion) in the miRNA/miRNA* duplex somehow endows the miRNA the capacity to generate ta-siRNAs.
Understanding ta-siRNA biogenesis
Ta-siRNAs are fascinating from the point of view of their biogenesis. All miRNAs do not trigger the production of ta-siRNAs. Thus, one wonders what features in the triggering miRNAs or the target transcripts endow them with this unique capacity to produce ta-siRNAs.
The biogenesis of the four canonical and well-characterized ta-siRNAs in Arabidopsis involves three miRNAs and four types of TRANS ACTING SIRNA (TAS) loci. The TAS loci are transcribed into long noncoding RNAs, which are cleaved by one of three miRNAs to trigger ta-siRNA production [30,31,39,40]. The three triggering miRNAs are miR173, miR390, and miR828. How are these miRNAs different from other miRNAs that do not trigger ta-siRNA production? miR390 is unique in that it is specifically bound by AGO7, one of ten AGO proteins with the distinctive ability to trigger tasiR-ARF production from two TAS3 loci [41,42]. However, miR173 and probably miR828, are bound by AGO1 as are most miRNAs [43] and it has been unknown what distinguishes these two miRNAs from other AGO1-bound miRNAs in terms of ta-siRNA biogenesis. Several recent findings described below have begun to shed light on this problem.
It is now apparent that the stringently defined ta-siRNAs represent a small fraction of secondary siRNAs generated from miRNA-targeted transcripts, which can be either non-protein coding or protein-coding transcripts. Until a function is ascribed to the secondary siRNAs from a particular locus, the secondary siRNAs are referred to as phasiRNAs because they are in 21 nt (or sometimes 24 nt depending on the DICERLIKE protein that generates them) register from one another. Through analysis of high throughput sequencing data of small RNAs for features of phasiRNAs (phased secondary siRNAs from miRNA target transcripts), recent studies [44–49] have uncovered many phasiRNA loci in Arabidopsis, rice, the legume Medicago, and three Solanaceae species. These studies provided insights into the features that endow a miRNA the ability to trigger phasiRNA biogenesis.
The size of the triggering miRNA is one such distinguishing feature [44,45]. In Arabidopsis, more phasiRNA-generating loci have been identified (in addition to the previously known TAS1-4 loci) as well as miRNAs other than the previously known miR173, miR390, and miR828 that can trigger the production of phasiRNAs. Intriguingly, all miRNAs that have the ability to induce phasiRNA production, with the exception of miR390, are 22 nt in length, as opposed to 21 nt, the length for most Arabidopsis miRNAs produced by DCL1. The production of 22 nt miRNAs by DCL1 is due to the presence of an asymmetric bulge (an unpaired nucleotide) in the miRNA/miRNA* duplex, the product of DCL1 (Figure 1). Deletion of the asymmetric bulge in the miRNA precursors results in 21 nt miRNAs and abolishes the ability of the miRNAs to trigger phasiRNA biogenesis. Conversely, artificially changing a miRNA from 21 nt to 22 nt imparts the ability to trigger phasiRNA biogenesis. In rice, Medicago, and Solanaceae species, hundreds to thousands of loci produce phasiRNAs, and the triggering miRNAs are also 22 nt long [46,47,49], suggesting that the underlying mechanism is conserved across plants.
While the 22 nt size of miRNAs is sufficient for triggering secondary siRNA biogenesis, it may not be necessary. When siRNAs from high throughput sequencing were mapped to miRNA-targeted transcripts, one third of the transcripts were found to yield secondary siRNAs, although they were not necessarily phased [48]. Intriguingly, the presence of an asymmetric bulge in the miRNA/miRNA* duplex was sufficient for the miRNA to trigger secondary siRNA production [48]. In fact, the miRNA does not have to be 22 nt long – a 21 nt miRNA that has a 22 nt miRNA* is able to cause secondary siRNA production. It is hypothesized that the path to secondary siRNA production is already determined at the miRISC loading step (Figure 1).
Features of an mRNA that contribute to ta-siRNA biogenesis were also recently examined. The miR173 target site was introduced into a GFP reporter gene and the position of the target site was found to influence the efficiency of ta-siRNA biogenesis [50]. Interestingly, the optimal position of the target site is within 10 nt downstream of the stop codon; placing the target site within the coding region or in the 3′ UTR further away from the stop codon drastically reduces ta-siRNA biogenesis without affecting miRNA-guided cleavage. Ribosome profiling experiments in animals show strong pauses of ribosomes at the stop codons of many genes [51,52]. These observations raise the possibility that stalled ribosomes promote ta-siRNA biogenesis. Although the TAS loci that give rise to ta-siRNAs are thought to encode noncoding transcripts, the transcripts do harbor short open reading frames. In fact, many transcripts previously annotated as long noncoding RNAs in mammals are bound by ribosomes in vivo [52].
Together, these findings reveal that the mechanisms underlying ta-siRNA biogenesis are far more complex than summarized by the current molecular framework (Figure 1) and new insights into small RNA biology are expected to emerge from future studies in this area.
Cell-to-cell and long distance movements of small RNAs
Most plant cells are connected with neighboring cells by plasmodesmata (PD), through which nutrients and biomolecules such as certain RNAs and proteins can move from cell to cell. Once the molecules enter the phloem, they can be transported systemically in the plant. However, the PD is restrictive in terms of what goes through and cell-to-cell movement of biomolecules is a regulated process. Given that miRNAs and some siRNAs play crucial roles in plant development, whether small RNAs can traffic between cells and the extent of their movement in the plant would impact how they exert their patterning roles.
RNA silencing in plants has long been known to be systemic. Grafting experiments in the 1980s showed that silencing triggers exert long distance effects in the plant [53,54]. Expression of a silencing trigger in the phloem companion cells in Arabidopsis leads to the silencing of an endogenous gene within approximately 15 cell layers from the source of the silencing trigger. siRNA duplexes have been implicated as the agents that move between cells to cause non-cell autonomous RNA silencing [55,56]. By grafting two genotypes differing in the ability to generate siRNAs and employing high throughput sequencing to detect siRNAs in recipient tissues, endogenous heterochromatic siRNAs were shown to travel systemically and to direct epigenetic changes in recipient cells [56,57].
The two types of small RNAs with large impacts in plant development, miRNAs and ta-siRNAs, have also been implicated to move between cells. miR390, which affects many aspects of plant development through the tasiR-ARFs, accumulates in a broader domain than that of its gene promoter activity or its precursor accumulation, suggesting that miR390 moves across a few cell layers. miR165/166 acts in meristem maintenance, vasculature patterning, and leaf polarity specification. In the root, transcription of the gene is activated specifically in one of the concentric rings that make up the radial axis of the root, the endodermis. However, the miRNA represses the expression of its target gene PHABULOSA (PHB) within the central cylinder, suggesting that the miRNA moves inward across a few layers [58,59]. Intriguingly, the expression of PHB in the central cylinder occurs in a gradient with the highest expression being the farthest away from the endodermis [58,59]. This suggests that miR165/166 forms a gradient emanating from the endodermis and regulates its targets in a concentration-dependent manner. In a mutant with a reduced PD aperture, expression of PHB within the central cylinder is highly increased, suggesting that the movement of miR165/166 is compromised [60]. The tasiR-ARFs also appear to move across cell layers. While the TAS3 gene is transcribed in the epidermal layer to produce the precursors to the tasiR-ARFs, the tasiR-ARFs are found in a gradient emanating from the epidermal layer on the adaxial side of leaf primordia towards the abaxial side [61]. Interestingly, ta-siRNAs have a greater range of cell-to-cell spread than miRNAs. When identical small RNAs were produced only in phloem companion cells with either a miRNA or a ta-siRNA backbone, the non-cell autonomous silencing effects were much greater for small RNAs produced from the ta-siRNA backbone [62].
In summary, siRNAs and miRNAs that impact plant development can move across a few cell layers. This makes it possible for small RNAs to form gradients across cell layers and, in the case of the tasiR-ARFs, such gradients have been detected [61]. If small RNAs regulate their targets in a concentration-dependent manner, it is possible that these small RNAs serve as morphogens in development.
Non-canonical functions of argonaute proteins
Most characterized eukaryotic organisms have an AGO gene family consisting of two or more members with the paralogs assuming distinct binding preferences for different types of small RNAs. Despite the functional divergence between AGO members, AGO proteins are generally known to mediate the activities of the small RNAs that they are associated with. However, recent studies in Arabidopsis uncovered a non-canonical function of AGO10, a function that has strong developmental implications.
AGO10 is most closely related to AGO1 among the ten AGO proteins in Arabidopsis. Phenotypes of loss-of-function ago10 mutants indicate a critical role of AGO10 in stem cell regulation. While the shoot apical meristem (SAM), which harbors stem cells, continues to generate organs to result in an elaborate shoot structure in wild type, in ago10 mutants, the SAM terminates in a pin-like structure [63,64], suggesting that AGO10 is required for stem cell maintenance in the SAM. In floral meristems, stem cells undergo programed termination so that a fixed number of floral organs are produced. AGO10 promotes the termination of stem cell maintenance in the floral meristems [65]. In both developmental contexts, AGO10 exerts its functions through miR165/166 [65,66]. AGO10 binds many miRNAs [65], but it has the highest preference for miR165/166 such that this miRNA accounts for 90% of AGO10-associated miRNAs in vivo [66]. AGO10 also exhibits higher affinity for miR165/166 than AGO1 [66]. Although AGO10 has miR165/166-guided slicer activity in vitro, it does not mediate the activities of the miRNA in vivo [65,66]. On the contrary, ago10 mutants have reduced expression of the type III HD-Zip genes, targets of miR165/166 [65–67], suggesting that AGO10 negatively affects the activities of this miRNA. These observations lead to the model that AGO10, which accumulates in a highly restricted pattern in the SAM and organ primordia [63,64], sequesters miR165/166 from AGO1, which is expressed ubiquitously [63], to prevent repression of miR165/166 targets in the AGO10 expression domain (Figure 2).
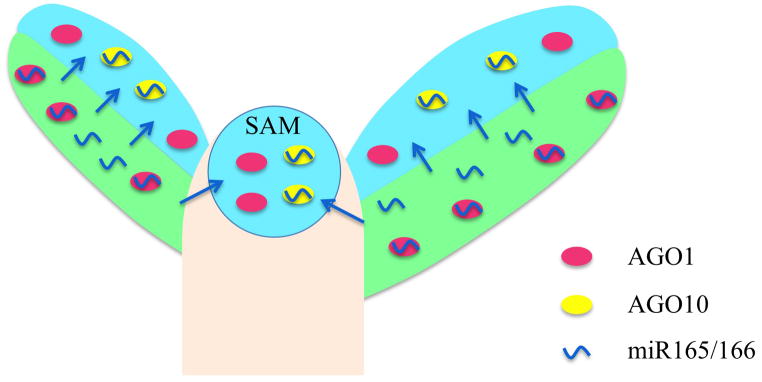
Figure 2.
AGO10 competes with AGO1 for binding to miR165/166 and sequesters the miRNA to prevent it from acting on its targets in the AGO10 expression domain. The apex of an Arabidopsis plant is diagrammed with the shoot apical meristem (SAM) in the center and two emerging leaf primordia on the flanks. Early in development, the adaxial (top; light blue) and abaxial (bottom; light green) sides of the leaf primordia are specified such that the two sides differ in structures and morphology in the adult leaves. In the leaf primordia, miR165/166 is enriched in the abaxial domain to restrict the expression of its target genes to the adaxial domain. AGO10 is expressed in the adaxial domain of leaf primordia and in the SAM (represented by the light blue regions), whereas AGO1 is ubiquitously expressed. AGO10 competes with AGO1 for binding to miR165/166 to prevent it from repressing its targets. This mechanism is probably necessitated by the fact that miR165/166 can potentially move across a few cell layers (represented by the arrows) to enter into the adaxial domain and the SAM; such that restriction of miR165/166 to the abaxial domain cannot be achieved solely by transcriptional regulation.
Why is such a sequestration mechanism necessary? Is transcriptional restriction of MIR165/166 expression not sufficient to limit miR165/166 activity spatially? One speculation is that this has to do with the movement of miRNAs, which precludes transcriptional regulation alone from establishing sharp boundaries of miRNA accumulation. Perhaps sequestration of miRNAs in specific spatial domains together with regulated transcription of MIR genes delineates sharp boundaries for miRNA activities (Figure 2).
Endogenous siRNAs in germ line specification and gamete formation
miRNAs and some endogenous siRNAs such as the tasiR-ARFs exert obvious development functions. Heterochromatic siRNAs, which are thought to act mainly to maintain genome stability by silencing repetitive sequences and transposable elements, are not known to have developmental functions. Arabidopsis mutants that largely eliminate heterochromatic siRNAs do not exhibit obvious developmental defects. Recent studies have changed this view and uncovered an intriguing parallel between plant siRNAs and animal piRNAs in germ line specification and gamete formation.
Unlike animals that set aside a germ line early during embryogenesis, plants specify their germ line late in development in both the female and male reproductive organs. Within each of the many developing ovules in Arabidopsis, one cell in the subepidermal layer becomes the female archespore, or megaspore mother cell (MMC), which is the female germ line. The MMC undergoes meiosis to produce four spores, one of which undergoes three rounds of mitosis to generate an eight-celled structure, the female gametophyte, with the egg being one of the cells. Loss of function of AGO9, which encodes a member of the AGO4-clade that associates with heterochromatic siRNAs, results in ovules with multiple MMCs [68,69]. In maize, loss of function of the AGO4-clade member ago104 causes defects in chromosome condensation and chromosome segregation during meiosis of the MMC [70]. Therefore, heterochromatic siRNAs are probably required for proper female germ line specification and gamete formation. However, the above-mentioned phenotypes of the Arabidopsis and maize mutants are not fully penetrant, implying that additional mechanisms are at work.
The redundant players may in part be ta-siRNAs. In Arabidopsis, mutants defective in ta-siRNA biogenesis, such as rdr6 and sgs3, also exhibit partially penetrant female germ line defects similar to ago9 mutants [69]. In rice and maize, an intriguing class of phasiRNAs with strong accumulation in flowers has been discovered. Two miRNAs that are 22 nt in length and conserved in rice and maize trigger the production of phasiRNAs from nearly 1000 genomic loci in rice [46]. One speculation is that the phasiRNAs promote germ line specification or meiosis. A rice argonaute gene, MEL1, is specifically expressed in male and female archesporal cells. The mel1 mutants exhibit meiosis defects [71], although it is not known what small RNA species are bound by MEL1.
An intriguing parallel between endogenous siRNAs in plants and piRNAs in animals has emerged. Both types of small RNAs guide sequence-specific DNA methylation in transcriptional gene silencing. Both have roles in germ line specification or gamete formation. It is still unknown whether the RNA-directed DNA methylation and the germ line functions of the small RNAs are related.
Concluding remarks
miRNAs and certain endogenous siRNAs have long been known to impact developmental patterning in plants. Recent studies have provided a glimpse of the strategies behind the patterning roles of small RNAs. The ability to move between cells allows small RNAs to form a gradient across a number of cell layers to potentially influence patterning in a concentration-dependent manner. When such a gradient is undesirable, mechanisms that inhibit the activities of small RNAs help enable their sharp spatial restriction. Recent studies have also uncovered the massive production of phasiRNAs from numerous genomic loci in many plant species and implicated these siRNAs and heterochromatic siRNAs in germ line specification and/or meiosis. The intriguing parallel between endogenous siRNAs in plants and piRNAs in animals suggests that a conserved and fundamental logic underlying germ line development in eukaryotes awaits discovery.
Highlights.
Small RNAs move cell-to-cell and systemically in plants
Some plant miRNAs have the capacity to trigger the production of secondary siRNAs to impact plant development
AGO10 acts in plant development through sequence-specific small RNA sequestration
Heterochromatic siRNAs in plants and piRNAs in mammals silence transposable elements by guiding DNA methylation
Heterochromatic siRNAs in plants and piRNAs in animals impact germ line specification and meiosis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 2.Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mette MF, van der Winden J, Matzke M, Matzke AJ. Short RNAs can identify new candidate transposable element families in Arabidopsis. Plant Physiology. 2002;130:6–9. doi: 10.1104/pp.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes & Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X. Small RNAs and their roles in plant development. Ann Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci U S A. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 10.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke JH, Tack D, Findlay K, Van Montagu M, Van Lijsebettens M. The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J. 1999;20:493–501. doi: 10.1046/j.1365-313x.1999.00623.x. [DOI] [PubMed] [Google Scholar]
- 13.Lu C, Fedoroff N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell. 2000;12:2351–2366. doi: 10.1105/tpc.12.12.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ray S, Golden T, Ray A. Maternal effects of the short integument mutation on embryo development in Arabidopsis. Dev Biol. 1996;180:365–369. doi: 10.1006/dbio.1996.0309. [DOI] [PubMed] [Google Scholar]
- 15.Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. Embo J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grigg SP, Canales C, Hay A, Tsiantis M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005;437:1022–1026. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- 17.Nodine MD, Bartel DP. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes & Dev. 2010;24:2678–2692. doi: 10.1101/gad.1986710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 20.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–759. doi: 10.1016/j.cell.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Millar AA, Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell. 2005;17:705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu MF, Tian Q, Reed JW. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 2006;133:4211–4218. doi: 10.1242/dev.02602. [DOI] [PubMed] [Google Scholar]
- 24.Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- 25.Chen X. Small RNAs - secrets and surprises of the genome. Plant J. 2010;61:941–958. doi: 10.1111/j.1365-313X.2009.04089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Rev Genetics. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juliano C, Wang J, Lin H. Uniting germline and stem cells. the function of Piwi proteins and the piRNA pathway in diverse organisms. Ann Rev Genetics. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 29.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes & Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 32.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes & Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 34.Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 35.Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 36.Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz Gutierrez-Nava M, Poethig SR. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nogueira FT, Madi S, Chitwood DH, Juarez MT, Timmermans MC. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes & Dev. 2007;21:750–755. doi: 10.1101/gad.1528607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Gao X, Li L, Shi X, Zhang J, Shi Z. Overexpression of Osta-siR2141 caused abnormal polarity establishment and retarded growth in rice. Journal of Experimental Botany. 2010;61:1885–1895. doi: 10.1093/jxb/erp378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes & Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunter C, Sun H, Poethig RS. The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr Biol. 2003;13:1734–1739. doi: 10.1016/j.cub.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 42.Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 transacting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 43.Montgomery TA, Yoo SJ, Fahlgren N, Gilbert SD, Howell MD, Sullivan CM, Alexander A, Nguyen G, Allen E, Ahn JH, et al. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci U S A. 2008;105:20055–20062. doi: 10.1073/pnas.0810241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA. 2010;107:15269–15274. doi: 10.1073/pnas.1001738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45**.Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nature Struc & Mol Biol. 2010;17:997–1003. doi: 10.1038/nsmb.1866. Studies in references 44 and 45 uncover the 22 nt length as a sufficient feature in miRNAs for triggering the production of ta-siRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson C, Kasprzewska A, Tennessen K, Fernandes J, Nan GL, Walbot V, Sundaresan V, Vance V, Bowman LH. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 2009;19:1429–1440. doi: 10.1101/gr.089854.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47*.Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. MicroRNA regulation of plant innate immune receptors. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. Studies in references 47 and 49 show that several 22 nt miRNAs target highly conserved regions in defense-related NB-LRR-encoding genes to trigger the production of ta-siRNAs in Medicago and several Solanaceae species. The ta-siRNAs can potentially regulate 60% of all approximately 540 NB-LRR genes in Medicago. This suggests that miRNAs serve as master regulators of gene families through ta-siRNA production. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48**.Manavella PA, Koenig D, Weigel D. Plant secondary siRNA production determined by microRNA-duplex structure. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2461–2466. doi: 10.1073/pnas.1200169109. This study reveals that an asymmetric bulge in the miRNA/miRNA* duplex is sufficient for the miRNA to trigger secondary siRNA production from target transcripts. The triggering miRNA does not have to be 22 nt in length. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, Gonzalez AJ, Yan Z, Kitto SL, Grusak MA, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes & Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50*.Zhang C, Ng DW, Lu J, Chen ZJ. Roles of target site location and sequence complementarity in trans-acting siRNA formation in Arabidopsis. Plant J. 2012;69:217–226. doi: 10.1111/j.1365-313X.2011.04783.x. The study reveals that the position of the miRNA-binding site relative to the stop codon in an mRNA affects the ability of the mRNA to be channeled into ta-siRNA production. [DOI] [PubMed] [Google Scholar]
- 51.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palauqui JC, Elmayan T, Pollien JM, Vaucheret H. Systemic acquired silencing. transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. Embo J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voinnet O, Vain P, Angell S, Baulcombe DC. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]
- 55.Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nature genetics. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- 56.Dunoyer P, Schott G, Himber C, Meyer D, Takeda A, Carrington JC, Voinnet O. Small RNA duplexes function as mobile silencing signals between plant cells. Science. 2010;328:912–916. doi: 10.1126/science.1185880. [DOI] [PubMed] [Google Scholar]
- 57.Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 58.Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vaten A, Thitamadee S, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59**.Miyashima S, Koi S, Hashimoto T, Nakajima K. Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development. 2011;138:2303–2313. doi: 10.1242/dev.060491. The studies in references 58 and 59 show that miR165/166 is produced in one cell layer and traffics across a few cell layers to result in graded expression of its target gene. [DOI] [PubMed] [Google Scholar]
- 60.Vaten A, Dettmer J, Wu S, Stierhof YD, Miyashima S, Yadav SR, Roberts CJ, Campilho A, Bulone V, Lichtenberger R, et al. Callose Biosynthesis Regulates Symplastic Trafficking during Root Development. Dev Cell. 2011;21:1144–1155. doi: 10.1016/j.devcel.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 61.Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC. Pattern formation via small RNA mobility. Genes & Dev. 2009;23:549–554. doi: 10.1101/gad.1770009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Felippes FF, Ott F, Weigel D. Comparative analysis of non-autonomous effects of tasiRNAs and miRNAs in Arabidopsis thaliana. Nucleic Acids Res. 2011;39:2880–2889. doi: 10.1093/nar/gkq1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- 64.Moussian B, Schoof H, Haecker A, Jurgens G, Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. Embo J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65*.Ji L, Liu X, Yan J, Wang W, Yumul RE, Kim YJ, Dinh TT, Liu J, Cui X, Zheng B, et al. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genetics. 2011;7:e1001358. doi: 10.1371/journal.pgen.1001358. This work shows that AGO10 binds many miRNAs including miR165/166 and exhibits miR165/166-guided slicer activity in vitro. However, in vivo, AGO10 promotes, instead of inhibits, the expression of miR165/166 target genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66**.Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou LW, Barefoot A, Dickman M, Zhang X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145:242–256. doi: 10.1016/j.cell.2011.03.024. This study reveals a special preference or affinity of AGO10 for miR165/166. AGO10 competes with AGO1 for binding to this miRNA and sequesters the miRNA to prevent AGO1-mediated repression of miR165/166 target genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Q, Yao X, Pi L, Wang H, Cui X, Huang H. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and leaf polarity establishment by repressing miR165/166 in Arabidopsis. Plant J. 2008 doi: 10.1111/j.1365-313X.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- 68.Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell. 2010;22:321–334. doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69**.Olmedo-Monfil V, Duran-Figueroa N, Arteaga-Vazquez M, Demesa-Arevalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–632. doi: 10.1038/nature08828. This work reveals a previously unknown role of AGO9, which binds heterochromatic siRNAs, and of RDR6 and SGS3, which produce ta-siRNAs, in female germ line formation. Loss of function mutations in these genes result in the formation of multiple female archespores. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70*.Singh M, Goel S, Meeley RB, Dantec C, Parrinello H, Michaud C, Leblanc O, Grimanelli D. Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell. 2011;23:443–458. doi: 10.1105/tpc.110.079020. This study shows that maize ago104, a member of the AGO4-clade that associates with heterochromatic siRNAs, is required for the meiosis of the female archespore to produce gametes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell. 2007;19:2583–2594. doi: 10.1105/tpc.107.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]