Summary
Background
High plasma HDL cholesterol is associated with reduced risk of myocardial infarction, but whether this association is causal is unclear. Exploiting the fact that genotypes are randomly assigned at meiosis, are independent of non-genetic confounding, and are unmodified by disease processes, mendelian randomisation can be used to test the hypothesis that the association of a plasma biomarker with disease is causal.
Methods
We performed two mendelian randomisation analyses. First, we used as an instrument a single nucleotide polymorphism (SNP) in the endothelial lipase gene (LIPG Asn396Ser) and tested this SNP in 20 studies (20 913 myocardial infarction cases, 95 407 controls). Second, we used as an instrument a genetic score consisting of 14 common SNPs that exclusively associate with HDL cholesterol and tested this score in up to 12 482 cases of myocardial infarction and 41 331 controls. As a positive control, we also tested a genetic score of 13 common SNPs exclusively associated with LDL cholesterol.
Findings
Carriers of the LIPG 396Ser allele (2·6% frequency) had higher HDL cholesterol (0·14 mmol/L higher, p=8×10−13) but similar levels of other lipid and non-lipid risk factors for myocardial infarction compared with non-carriers. This difference in HDL cholesterol is expected to decrease risk of myocardial infarction by 13% (odds ratio [OR] 0·87, 95% CI 0·84–0·91). However, we noted that the 396Ser allele was not associated with risk of myocardial infarction (OR 0·99, 95% CI 0·88–1·11, p=0·85). From observational epidemiology, an increase of 1 SD in HDL cholesterol was associated with reduced risk of myocardial infarction (OR 0·62, 95% CI 0·58–0·66). However, a 1 SD increase in HDL cholesterol due to genetic score was not associated with risk of myocardial infarction (OR 0·93, 95% CI 0·68–1·26, p=0·63). For LDL cholesterol, the estimate from observational epidemiology (a 1 SD increase in LDL cholesterol associated with OR 1·54, 95% CI 1·45–1·63) was concordant with that from genetic score (OR 2·13, 95% CI 1·69–2·69, p=2×10−10).
Interpretation
Some genetic mechanisms that raise plasma HDL cholesterol do not seem to lower risk of myocardial infarction. These data challenge the concept that raising of plasma HDL cholesterol will uniformly translate into reductions in risk of myocardial infarction.
Funding
US National Institutes of Health, The Wellcome Trust, European Union, British Heart Foundation, and the German Federal Ministry of Education and Research.
Introduction
Cholesterol fractions such as LDL and HDL cholesterol are among the most commonly measured biomarkers in clinical medicine.1 Observational studies have shown that LDL and HDL cholesterol have opposing associations with risk of myocardial infarction, with LDL cholesterol being positively associated and HDL cholesterol being inversely associated.2,3 However, observational studies cannot distinguish between a causal role in the pathological process and a marker of the underlying pathophysiology. These two possibilities can be distinguished in human beings by changes of the cholesterol fractions in large-scale randomised trials or by studies of inherited DNA variation. For LDL cholesterol, the results of both randomised trials of LDL-cholesterol-lowering treatments4 and from human mendelian diseases5,6 are concordant and suggest that plasma LDL cholesterol is causally related to risk of myocardial infarction. However, the available evidence for the causal relevance of HDL cholesterol from randomised trials or mendelian diseases is scarce and inconsistent.7,8
If a particular plasma biomarker is directly involved in an underlying pathological process, then inherited variation changing plasma concentrations of this biomarker should affect risk of disease in the direction and magnitude predicted by the plasma concentrations. Referred to as mendelian randomisation,9–11 this analytical approach has been previously applied to plasma HDL cholesterol, albeit with restricted sample sizes, a small number of single nucleotide polymorphisms (SNPs) at a few genes, and with SNPs that affect multiple lipid fractions.8,12–15 Hence, these studies have not been able to resolve fully the possible causal relevance of HDL cholesterol concentrations for risk of myocardial infarction.
Recently, we have used the genome-wide association approach to identify SNPs that affect blood lipid concentrations.16,17 Additionally, through resequencing, we identified a loss-of-function coding SNP at the endothelial lipase gene (LIPG Asn396Ser) that affects plasma HDL cholesterol in isolation.18,19 Here, we use these SNPs in case-control studies and prospective cohort studies to test the hypothesis that genetically raised plasma HDL cholesterol might be protective for myocardial infarction.
Methods
Study design
The study design consisted of two components. First, using a case-control design, we tested lipid-associated SNPs individually for association with risk of myocardial infarction. Second, using a mendelian randomisation design, we tested two instruments: (1) a single SNP that related exclusively to plasma HDL cholesterol (a loss-of-function coding polymorphism at the endothelial lipase gene, LIPG Asn396Ser, rs61755018); and (2) a genetic score consisting of 14 common SNPs that exclusively associate with HDL cholesterol.
Study participants
Characteristics of cases of myocardial infarction and controls are shown in appendix p 19. Data for up to 19 139 cases of myocardial infarction and 50 812 myocardial-infarction-free controls were available from 30 studies. Characteristics of the participants in six prospective cohort studies are shown in the appendix p 20. 50 763 participants from six cohort studies were studied and, of these, 4228 developed an incident fatal or non-fatal myocardial infarction. All participants were of self-reported European or South Asian ancestry.
Statistical analysis
In myocardial infarction cases and controls, we tested each of 25 SNPs for association with myocardial infarction in up to 30 studies. These 25 SNPs represented the initial polymorphisms mapped for plasma HDL or LDL cholesterol concentrations with a genome-wide association approach.16 Each selected SNP has been associated with either HDL or LDL cholesterol at p<5×10−8. Genotyping details are provided in the appendix p 2. We undertook logistic regression with the outcome variable of myocardial infarction status, predictor variable of individual SNP genotype, and covariates of age, sex, and principal components of ancestry. We assumed a log-additive genetic model. Overall association for each SNP was evaluated with a fixed-effects inverse-variance-weighted meta-analysis.
Fatal or non-fatal myocardial infarction outcomes were ascertained in each of six prospective cohort studies as described in the appendix p 10. We constructed logistic regression models to examine the association of LIPG Asn396Ser genotype with myocardial infarction status, excluding participants who had had a previous myocardial infarction or ischaemic stroke. The predictor variable of LIPG Asn396Ser genotype was modelled in an additive model (ie, 0, 1, 2 copies of the 396Ser allele). Covariates in the model included age and sex. Overall association for each SNP was evaluated across the six studies with fixed-effects inverse-variance-weighted meta-analysis.
We estimated a predicted risk for LIPG Asn396Ser on the basis of the association of this SNP with plasma HDL cholesterol (appendix p 21) and the association of plasma HDL cholesterol with myocardial infarction (appendix p 22) in the population. Details are provided in the appendix p 2.
We undertook instrumental variable analysis using LIPG Asn396Ser in six prospective cohort studies as listed in the appendix p 23. We additionally did an instrumental variable analysis using multiple genetic variants as instruments.20 Details of the instrumental variable analysis methods are provided in the appendix p 4. We regarded a two-tailed p<0·05 as nominally significant. Heterogeneity statistics were calculated as described.21 SAS version 9.1, the R package, STATA, or PLINK software were used for all statistical analyses.22
Role of the funding source
The sponsors had no role in the conduct or interpretation of the study. The corresponding author had full access to all data in the study and had final responsibility for the decision to submit for publication.
Results
To validate the statistical framework and clinical samples, we first tested SNPs related to plasma LDL cholesterol in case-control studies (table 1). For nine of ten SNPs associated with LDL cholesterol, the allele correlated with increased LDL cholesterol was also associated with increased risk of myocardial infarction (p<0·05; table 1).
Table 1.
Association of myocardial infarction (MI) with single nucleotide polymorphisms (SNPs) previously found to relate to plasma LDL cholesterol
| Chromosome | Gene(s) of interest within or near associated interval | Major allele, minor allele (minor allele frequency)* | Modelled allele | Effect of modelled allele on plasma LDL cholesterol (mmol/L)* | Effect of modelled allele on plasma HDL cholesterol (mmol/L)* | Effect of modelled allele on plasma triglycerides (mmol/L)* | Sample size (MI cases/MI-free controls) | For modelled allele, observed change in MI risk (%; 95% CI) | For modelled allele, p value for association with MI | |
|---|---|---|---|---|---|---|---|---|---|---|
| rs646776 | 1p13 | CELSR2, PSRC1, SORT1† | T, C (0·21) | T | 0·20 | −0·03 | .. | 19 139/50 812 | 16% (12–19) | 4×10−16† |
| rs6511720 | 19p13 | LDLR† | G, T (0·10) | G | 0·23 | .. | 0·09 | 16 503/46 576 | 23% (17–30) | 4×10−12† |
| rs11206510 | 1p32 | PCSK9† | T, C (0·17) | T | 0·05 | .. | .. | 18 455/23 075 | 13% (9–16) | 1×10−9† |
| rs3798220 | 6q25 | LPA† | T, C (0·02) | C | 0·36‡ | .. | .. | 6658/5823 | 72% (39–211) | 4×10−7† |
| rs562338 | 2p24 | APOB† | G, A (0·20) | G | 0·14 | .. | .. | 19 139/50 812 | 8% (4–12) | 5×10−5† |
| rs6544713 | 2p21 | ABCG8† | C, T (0·32) | T | 0·13 | .. | .. | 14 818/45 454 | 8% (4–11) | 5×10−5† |
| rs7953249 | 12q24 | HNF1A† | A, G (0·44) | G | 0·07 | .. | .. | 19 139/50 812 | 5% (3–9) | 2×10−4† |
| rs10402271 | 19q13 | APOE-APOC1-APOC4-APOC2† | T, G (0·33) | G | 0·12 | .. | .. | 19 139/50 812 | 4% (1–7) | 0·007† |
| rs3846663 | 5q13 | HMGCR† | C, T (0·38) | T | 0·06 | .. | .. | 19 139/50 812 | 4% (1–7) | 0·01† |
| rs1501908 | 5q23 | TIMD4-HAVCR1 | C, G (0·37) | C | 0·06 | .. | .. | 18 310/49 897 | 3% (0–6) | 0·08 |
Data presented from a meta-analysis of seven cohorts (n up to 19 840) as presented in reference 16; the effect of each SNP on a lipid trait was modelled if the association of the SNP with a plasma lipid trait exceeded nominal significance (p<0·05).
Loci and SNPs that exceeded nominal significance (p<0·05) for association of modelled allele with MI; all modelled alleles increased LDL cholesterol.
The C allele at this LPA variant is related to increased plasma lipoprotein(a) as presented in reference 16.
Having established that SNPs related to plasma LDL cholesterol consistently affected risk of myocardial infarction, we applied the same methodological framework in the same samples to test the hypothesis that genetic modulation of HDL cholesterol would affect risk of myocardial infarction. Of 15 loci related to plasma HDL cholesterol, at six loci (LPL, TRIB1, APOA1-APOC3-APOA4-APOA5 cluster, CETP, ANGPTL4, and GALNT2) the allele correlated with raised HDL cholesterol was also associated with decreased risk of myocardial infarction (p<0·05; table 2). Of note, at the HNF4A locus, the HDL-cholesterol-raising allele was surprisingly associated with increased risk of myocardial infarction (p=0·0009).
Table 2.
Association of myocardial infarction (MI) with single nucleotide polymorphisms (SNPs) previously found to relate to plasma HDL cholesterol
| Chromosome | Gene(s) of interest within or near associated interval | Major allele, minor allele (minor allele frequency)* | Modelled allele | Effect of modelled allele on plasma HDL cholesterol (mmol/L)* | Effect of modelled allele on plasma triglycerides (mmol/L)* | Effect of modelled allele on plasma LDL cholesterol (mmol/L)* | Sample size (MI cases/MI-free controls) | For modelled allele, observed change in MI risk (%; 95% CI) | For modelled allele, p value for association with MI | |
|---|---|---|---|---|---|---|---|---|---|---|
| rs17482753 | 8p21 | LPL† | G, T (0·10) | T | 0·08 | −0·24 | .. | 19 139/50 812 | −12% (−16 to −7) | 4×10−7† |
| rs17321515 | 8q24 | TRIB1† | A, G (0·45) | G | 0·02 | −0·11 | −0·05 | 19 139/50 812 | −7% (−9 to −4) | 2×10−6† |
| rs6589566 | 11q23 | APOA1-APOC3-APOA4-APOA5† | A, G (0·07) | A | 0·05 | −0·27 | −0·09 | 18 310/49 897 | −10% (−15 to −5) | 8×10−5† |
| rs4846914 | 1q42 | GALNT2† | A, G (0·40) | A | 0·02 | −0·03 | .. | 19 139/50 812 | −3% (−6 to −1) | 0·02† |
| rs2967605 | 19p13 | ANGPTL4† | C, T (0·16) | C | 0·05 | −0·07 | .. | 13 595/16 423 | −5% (−10 to −1) | 0·03† |
| rs3764261 | 16q13 | CETP† | C, A (0·32) | A | 0·10 | .. | −0·03 | 16 503/46 576 | −4% (−7 to 0) | 0·04† |
| rs61755018 (Asn396Ser) | 18q21 | LIPG | A, G (0·015) | G | 0·14‡ | .. | .. | 17 165/49 077 | −6% (−18 to 9) | 0·41 |
| rs17145738 | 7q11 | MLXIPL | C, T (0·11) | T | 0·03 | −0·15 | .. | 19 139/50 812 | −1% (−4 to 3) | 0·61 |
| rs3890182 | 9q31 | ABCA1 | G, A (0·14) | G | 0·03 | .. | 0·05 | 19 139/50 812 | −1% (−5 to 4) | 0·76 |
| rs2338104 | 12q24 | MMAB, MVK | G, C (0·46) | G | 0·03 | .. | .. | 19 139/50 812 | 0% (−3 to 3) | 0·85 |
| rs471364 | 9p22 | TTC39B | T, C (0·12) | T | 0·03 | .. | .. | 15 693/47 098 | 0% (−5 to 5) | 0·97 |
| rs2271293 | 16q22 | LCAT | G, A (0·11) | A | 0·03 | .. | .. | 19 139/50 812 | 4% (−1 to 8) | 0·10 |
| rs174547 | 11q12 | FADS1-FADS2-FADS3 | T, C (0·33) | T | 0·03 | −0·06 | .. | 19 139/50 812 | 3% (−1 to 6) | 0·11 |
| rs1800588 | 15q22 | LIPC | C, T (0·22) | T | 0·05 | 0·07 | .. | 17 917/49 514 | 4% (0 to 7) | 0·04 |
| rs16988929 | 20q13 | HNF4A | C, T (0·01) | T | 0·01 | .. | .. | 17 041/20 137 | 31% (12 to 54) | 9×10−4 |
Data presented from a meta-analysis of seven cohorts (n up to 19 840) as presented in reference 16; the effect of each SNP on a lipid trait was modelled if the association of the SNP with a plasma lipid trait exceeded nominal significance (p<0·05).
Loci and SNPs that exceeded nominal significance (p<0·05) for association of modelled allele with MI; all modelled alleles increased HDL cholesterol.
Effect size presented is from the Atherosclerosis Risk in Communities Study.
All six SNPs associated with both HDL cholesterol and myocardial infarction had additional effects on plasma LDL cholesterol or triglycerides, or both (p<5×10−8 for the additional effects on LDL cholesterol or triglycerides). For example, at APOA1-APOC3-APOA4-APOA5 rs6589566, the allele associated with increased HDL cholesterol also relates to reduced LDL cholesterol and reduced triglycerides. The pleiotropic effects of such SNPs undermine the ability to define a causal role for HDL cholesterol, independent of effects on LDL cholesterol or triglycerides.
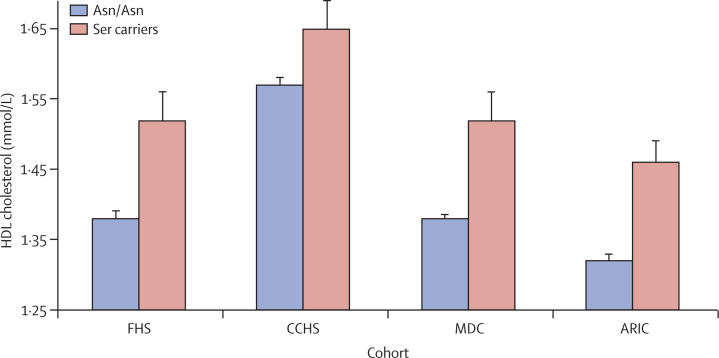
To evaluate plasma HDL cholesterol specifically, we undertook mendelian randomisation, a form of instrumental variable analysis.23 We identified a variant that affected only plasma HDL cholesterol without changing other lipid or non-lipid cardiovascular risk factors. In the LIPG gene, roughly 2·6% of individuals carry a serine substitution at aminoacid 396 (substituted for wild-type asparagine). Carrier status for 396Ser was associated with significant increases in HDL cholesterol in each of four prospective cohort studies, with the effect size ranging from 0·08 mmol/L to 0·28 mmol/L per copy of the Ser allele (figure 1, appendix p 21; p=0·002 in FHS, p=0·02 in CCHS, p=5×10−6 in MDC, and p=7×10−7· in ARIC).
Figure 1.
Plasma HDL cholesterol concentrations in carriers versus non-carriers of the Ser allele at the LIPG Asn396Ser polymorphism
Error bars show standard error. FHS=Framingham Heart Study. CCHS=Copenhagen City Heart Study. MDC=Malmo Diet and Cancer Study. ARIC=Atherosclerosis Risk in Communities Study.
In a meta-analysis including all four studies, carrier status for 396Ser was associated with an increase of roughly 0·29 SD units in HDL cholesterol (p=8×10−13). There was no evidence of heterogeneity across the four studies (I2=0·58; Cochran's heterogeneity p=0·07). LIPG Asn396Ser was not significantly associated with other risk factors for myocardial infarction including plasma LDL cholesterol, triglycerides, systolic blood pressure, body-mass index, risk of type 2 diabetes, fasting glucose, plasma C-reactive protein, waist-to-hip ratio, fibrinogen, and small LDL particle concentration (p>0·05 for each; appendix pp 24–26). Therefore, LIPG Asn396Ser satisfied the three main criteria for mendelian randomisation analysis—ie, that the genotype should be associated with an intermediate biomarker (figure 1), should not be associated with confounding factors (appendix pp 24–46), and should exert its effect on the clinical outcome only through the specific intermediate biomarker (appendix p 22).24
Under the model that plasma HDL cholesterol causally relates to risk of myocardial infarction, individuals with an inherited increase in HDL cholesterol (eg, those carrying the LIPG 396Ser allele) are expected to have reduced risk of myocardial infarction. On the basis of the associations between SNPs and HDL cholesterol, and HDL cholesterol and myocardial infarction, we estimated that carriage of LIPG 396Ser should decrease risk of myocardial infarction by 13% (odds ratio [OR] 0·87, 95% CI 0·84–0·91).
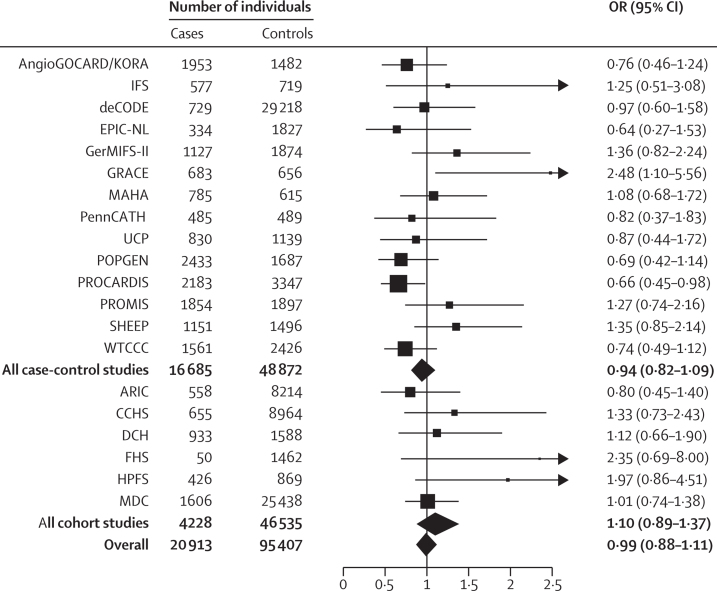
To establish whether LIPG 396Ser carriers are indeed protected from risk of myocardial infarction, we studied the association of LIPG Asn396Ser with incident myocardial infarction in 50 763 participants from six prospective cohort studies, the gold standard with respect to epidemiological study design. Of these participants, 4228 developed a first myocardial infarction event. LIPG Asn396Ser was not associated with myocardial infarction in any of the six studies (figure 2). Combining these studies in a meta-analysis, LIPG Asn396Ser allele was not associated with myocardial infarction (OR 1·10, 95% CI 0·89–1·37, p=0·37; figure 2). There was no evidence for heterogeneity across the six cohorts (I2=0·17; Cochran's heterogeneity p=0·31).
Figure 2.
Association of LIPG Asn396Ser with myocardial infarction in 116320 participants from 20 studies
In each study, the HDL-cholesterol-raising serine allele was modelled.
We also studied LIPG Asn396Ser in case-control studies involving an additional 16 685 cases of myocardial infarction and 48 872 controls and noted that this SNP was not associated with myocardial infarction (OR 0·94, 95% CI 0·82–1·09, p=0·41; table 2, figure 2), with no evidence of heterogeneity across the 14 case-control studies (I2=0·34; Cochran's heterogeneity p=0·11). Finally, we used meta-analysis to combine the evidence from both the prospective studies and case-control studies (116 320 participants; 20 913 cases and 95 407 controls). In all available samples, LIPG Asn396Ser remained not associated with risk of myocardial infarction (OR 0·99, 95% CI 0·88–1·11, p=0·85; figure 2). There was no evidence for heterogeneity across all 20 studies (I2=0·30; Cochran's heterogeneity p=0·10).
We formally estimated the magnitude of an association of genetically raised HDL cholesterol with myocardial infarction, using LIPG Asn396Ser as the instrument. The mendelian randomisation estimate was computed from the ratio of the coefficient of the association between genotype and disease to that of the association between genotype and plasma HDL cholesterol; this instrumental variable estimate reflects the potential causal effect of plasma HDL cholesterol on the risk of myocardial infarction.
Table 3 presents the instrumental variable estimate for the association of plasma HDL cholesterol with risk of myocardial infarction in each of six prospective cohort studies. In each study, genetically raised plasma HDL cholesterol was not associated with risk of myocardial infarction. In meta-analysis, genetically raised plasma HDL cholesterol was also not associated with risk of myocardial infarction (OR 1·02 per 0·03 mmol/L [1 mg/dL] increase in HDL cholesterol, 95% CI 0·95–1·09, p=0·64; OR 1·28 per 0·39 mmol/L [15 mg/dL] increase in HDL cholesterol, 95% CI 0·46–3·61, p=0·64).
Table 3.
Instrumental variable analysis estimate of the association of genetically raised HDL cholesterol and risk of myocardial infarction using LIPG Asn396Ser as an instrument
|
Observational epidemiology |
Genetically raised |
|||
|---|---|---|---|---|
| Odds ratio (95% CI) per 0·03 mmol/L (1 mg/dL) increase in plasma HDL cholesterol | p value | Odds ratio (95% CI) per 0·03 mmol/L (1 mg/dL) increase in plasma HDL cholesterol | p value | |
| Cohort | ||||
| Atherosclerosis Risk in Communities Study | 0·97 (0·96–0·98) | 7×10−18 | 0·96 (0·86–1·07) | 0·44 |
| Copenhagen City Heart Study | 0·98 (0·98–0·99) | 6×10−13 | 1·09 (0·95–1·26) | 0·21 |
| Malmo Diet and Cancer Study, Cardiovascular Cohort | 0·97 (0·96–0·98) | 1×10−6 | 0·82 (0·66–1·01) | 0·06 |
| Framingham Heart Study | 0·96 (0·94–0·98) | 4×10−6 | 1·17 (1·00–1·37) | 0·06 |
| Health Professionals Follow-up Study | .. | .. | 1·84 (0·39–8·62) | 0·16 |
| Danish Diet, Cancer, and Health Study | .. | .. | 1·05 (0·79–1·41) | 0·71 |
| Meta-analysis of cohort studies | ||||
| Per 0·03 mmol/L (1 mg/dL) increase in plasma HDL cholesterol | 0·98 (0·97–0·98) | 4×10−36 | 1·02 (0·95–1·09) | 0·64 |
| Per 0·39 mmol/L (15 mg/dL) increase in plasma HDL cholesterol | 0·70 (0·66–0·74) | 4×10−36 | 1·28 (0·46–3·61) | 0·64 |
Statistical power for instrumental variable analysis could be increased if multiple genetic variants in combination are used as instruments, according to a recent proposal.20 From our genome-wide association study of plasma lipid traits involving more than 100 000 individuals,17 we noted that 13 common SNPs had statistical evidence at genome-wide levels of significance (p<5×10−8) for plasma LDL cholesterol and no evidence for association with triglycerides (p>0·01) or HDL cholesterol (p>0·01). We constructed a genetic score for LDL cholesterol combining the LDL-cholesterol-raising alleles at each of these 13 SNPs (appendix p 27).25 We also noted that 14 common SNPs had statistical evidence at genome-wide levels of significance (p<5×10−8) for plasma HDL cholesterol and no evidence for association with triglycerides (p>0·01) or LDL cholesterol (p>0·01). We constructed a genetic score for HDL cholesterol combining the HDL-cholesterol-raising alleles at each of these 14 SNPs (appendix p 28). Each SNP was given a weight based on the degree of change in LDL or HDL cholesterol as estimated in roughly 100 000 individuals.17
We tested the association of genetic scores for LDL and HDL cholesterol separately for association with myocardial infarction in up to 53 146 cases and controls from the CARDIoGRAM study.26 From observational epidemiology, an increase of 1 SD in usual LDL cholesterol was associated with raised risk of myocardial infarction (OR 1·54, 95% CI 1·45–1·63; appendix p 22). In a mendelian randomisation analysis, a 1 SD increase in LDL cholesterol due to genetic score was also associated with risk of myocardial infarction (OR 2·13, 95% CI 1·69–2·69, p=2×10−10; table 4). From observational epidemiology, a 1 SD rise in usual HDL cholesterol was associated with lowered risk of myocardial infarction (OR 0·62, 95% CI 0·58–0·66; appendix p 22). However, in mendelian randomisation analysis, a 1 SD increase in HDL cholesterol due to genetic score was not associated with risk of myocardial infarction (OR 0·93, 95% CI 0·68–1·26, p=0·63; table 4).
Table 4.
Estimate of the association of genetically raised LDL cholesterol or HDL cholesterol and risk of myocardial infarction using multiple genetic variants as instruments
| Odds ratio (95% CI) per SD increase in plasma lipid based on observational epidemiology* | Odds ratio (95% CI) per SD increase in plasma lipid conferred by genetic score† | |
|---|---|---|
| LDL cholesterol | 1·54 (1·45–1·63) | 2·13 (1·69–2·69), p=2×10−10 |
| HDL cholesterol | 0·62 (0·58–0·66) | 0·93 (0·68–1·26), p=0·63 |
Observational epidemiology estimates derived from more than 25 000 individuals from prospective cohort studies as shown in the appendix p 22.
LDL genetic score consisting of 13 single nucleotide polymorphisms (SNPs) as shown in the appendix p 27; HDL genetic score consisting of 14 SNPs as shown in the appendix p 28.
Discussion
For a biomarker directly involved in disease pathogenesis, we expect a genetic variant that modulates the biomarker to likewise confer risk of disease. We tested this hypothesis for two plasma biomarkers: LDL and HDL cholesterol. SNPs affecting LDL cholesterol were consistently related to risk of myocardial infarction. However, we unexpectedly found that LIPG Asn396Ser, a genetic variant that specifically and substantially increases plasma HDL cholesterol, did not reduce risk of myocardial infarction. A genetic score combining 14 variants exclusively related to HDL cholesterol also showed no association with risk of myocardial infarction (Panel).
Panel. Research in context.
Systematic review
Electronic searches of Medline and PubMed, supplemented by hand searches of reference lists of other review articles, identified reports from three large mendelian randomisation studies for plasma HDL cholesterol.7,12,15 In each of these previous reports, genetically increased plasma HDL cholesterol was not associated with risk of ischaemic heart disease.
Interpretation
The present study tested a naturally occurring loss-of-function variant in the endothelial lipase gene and, with this new instrument, we confirm that genetically raised plasma HDL cholesterol is not associated with risk of myocardial infarction. The study further extends previous work by testing an instrument consisting of 14 common variants exclusively associated with plasma HDL cholesterol. A genetic score consisting of these 14 variants was not associated with risk of myocardial infarction. These results show that some ways of raising HDL cholesterol might not reduce risk of myocardial infarction in human beings. Therefore, if an intervention such as a drug raises HDL cholesterol, we cannot automatically assume that risk of myocardial infarction will be reduced.
These results challenge several established views about plasma HDL cholesterol. First, these data question the concept that raising of plasma HDL cholesterol should uniformly translate into reductions in risk of myocardial infarction. We raise the strong possibility that a specific means of raising of HDL cholesterol in human beings—namely, inhibition of endothelial lipase—will not reduce risk of myocardial infarction. In animal models, inhibition or deletion of the endothelial lipase gene increases HDL cholesterol concentrations,27 but there has been debate as to the consequent effect on atherosclerosis. One report suggested that mice deleted for Lipg on an Apoe knockout genetic background have decreased aortic atherosclerosis,28 but a subsequent study showed no effect of Lipg deletion on aortic atherosclerosis.29
Second, these findings emphasise the potential limitation of plasma HDL cholesterol as a surrogate measure for risk of myocardial infarction in intervention trials. The data presented here using mendelian randomisation are consistent with results from completed randomised controlled trials focused on raising plasma HDL cholesterol. Hormone replacement therapy raised plasma HDL cholesterol but did not lower risk of myocardial infarction.30 In the Atherothrombosis Intervention in Metabolic Syndrome with Low HDL Cholesterol/High Triglyceride and Impact on Global Health Outcomes (AIM-HIGH) trial,31 the addition of long-acting niacin to background simvastatin increased HDL cholesterol and lowered triglycerides but did not lower risk of cardiovascular events.
Of note, at the cholesterol ester transfer protein (CETP) gene, we did find that common genetic variation reduces risk of myocardial infarction by 4%, a result in line with an earlier meta-analysis.32 However, the CETP variant both increases HDL cholesterol and lowers LDL cholesterol17 in a manner similar to pharmacological inhibitors of CETP.33 As such, whether the protection afforded by the CETP variant is due to the change in HDL or LDL cholesterol is difficult to distinguish.
Third, biomarkers that assay HDL function might overcome some limitations of standard HDL cholesterol assays. However, a challenge will remain—namely, to prove that new functional HDL biomarkers reflect a causal association with myocardial infarction rather than an indirect one, as seems to be the case with plasma HDL cholesterol. For example, using a new in-vitro measure involving mouse macrophages and human serum, Khera and colleagues34 showed an inverse correlation between a specific functional property of HDL, cholesterol efflux capacity, and coronary artery disease status. The present study suggests that a fruitful approach to the causal evaluation of such functional measures in human beings might be large-scale study of relevant inherited DNA variation of HDL function.
There are inherent limitations to the mendelian randomisation study design. Naturally occurring genetic variation could be a useful instrument to assess causality provided that several requirements have been satisfied.35,36 First, one needs suitable genetic variants for the study of the modifiable exposure of interest (in our case, plasma HDL cholesterol). Although many loci are associated with plasma HDL cholesterol, we found LIPG Asn396Ser to be particularly informative because it is specifically associated with substantial increases in HDL cholesterol. Additionally, we evaluated a set of 14 common genetic variants that also exclusively affected HDL cholesterol. Both instruments, LIPG Asn396Ser and the genetic score, produced similar results.
Second, reliable genotype-to-intermediate-phenotype and intermediate-phenotype-to-disease effect estimates are needed. To obtain as precise estimates as possible, we derived SNP-to-lipid effect estimates from analysis of a large sample involving more than 24 000 participants. Estimates of plasma lipid to myocardial infarction were derived from meta-analysis of four large cohort studies involving more than 25 000 participants.
Third, there must not be pleiotropic effects of the genetic variants of interest. We cannot exclude all potential pleiotropic effects of the LIPG Asn396Ser SNP; however, we have assessed but did not detect pleiotropic effects on other cardiovascular risk factors including LDL cholesterol, small LDL particle concentration, triglycerides, body-mass index, systolic blood pressure, plasma C-reactive protein, and type 2 diabetes status.
Finally, the absence of association of individual SNPs with myocardial infarction could be due to low statistical power. However, for the crucial SNP in the mendelian randomisation study for plasma HDL cholesterol, we had sufficient power. In this study, LIPG Asn396Ser has been tested in 20 913 myocardial infarction cases and 95 407 myocardial-infarction-free participants. We had 90% power to detect the expected 13% reduction in risk of myocardial infarction for the LIPG Asn396Ser variant (at a two-sided α of 0·05).
In summary, our results showed that polymorphisms related to plasma LDL cholesterol were consistently associated with risk of myocardial infarction, whereas this was not the case for variants related to plasma HDL cholesterol. A polymorphism in the endothelial lipase gene and a genetic score of 14 common SNPs that specifically raised HDL cholesterol were not associated with myocardial infarction, suggesting that some genetic mechanisms that raise HDL cholesterol do not lower risk of myocardial infarction. Hence, interventions (lifestyle or pharmacological) that raise plasma HDL cholesterol cannot be assumed ipso facto to lead to a corresponding benefit with respect to risk of myocardial infarction.
This online publication has been corrected. The corrected version first appeared at thelancet.com on June 1, 2012
Acknowledgments
Acknowledgments
This Article is dedicated to Leena Peltonen, who passed away on March 11, 2010. A full listing of the acknowledgments is provided in the appendix p 12.
Contributors
BFV, DAltshuler, and SK contributed to study design. BFV, GMP, MO-M, RF-S, MB, MKJ, GH, HH, ELD, TJ, HS, NJS, RClarke, JCH, JFT, ML, GThorleifsson, CN-C, KM, JP, DS, LC, AFRS, SA, JCE, TM, JS, MS, KB, NM, DG, PPM, CCP, UT, GThorgeirsson, BG, PIWdeB, SR, CWiller, JE, PD, JD, SB, RR, RM, HW, ASH, KO, ER, EB, AT-H, LAC, MPR, OM, PMM, DAltshuler, DS, RE, KS, CJO, VS, DJR, LP, SMS, DArdissino, SK, and all collaborators contributed to data collection and did research. BFV, GMP, MO-M, RF-S, MB, MJK, GH, ELD, JCH, JFT, ML, GThorleifsson, KM, JP, DS, LD, ASurti, JCE, TM, MS, NM, CCP, BG, PIWdeB, SR, CWijmenga, SMS, DArdissino, and SK contributed to data analysis and interpreted results. BFV, GMP, DAltshuler, and SK wrote the report. HS, NJS, RClarke, CN-C, KM, JP, JCE, TM, JS, DG, PPM, EB, MPR, OM, DS, RE, CJO, VS, DJR, SMS, DAltshuler, and SK revised and reviewed the final report.
Conflicts of interest
KS, UT, HH, GThorleifsson, and GThorgeirsson are employees of or own stock options in deCODE Genetics, or both. SK serves on a scientific advisory board for Merck and has received research grants from Pfizer, Shire Therapeutics, and Alnylam Pharmaceuticals. HS serves on scientific advisory boards for Merck, Servier, and AstraZeneca and received lecture fees from Pfizer, Novartis, and Boehringer Ingelheim. The collection of clinical and sociodemographic data in the Dortmund Health Study was supported by the German Migraine & Headache Society (DMKG) and by unrestricted grants of equal share from AstraZeneca, Berlin Chemie, Boots Healthcare, GlaxoSmithKline, McNeil Pharma (formerly Woelm Pharma), MSD Sharp & Dohme, and Pfizer to the University of Muenster. VM, DW, CK, and MW are full-time employees of GlaxoSmithKline. All other authors declare that they have no conflicts of interest.
Supplementary Material
References
- 1.Expert Panel on Detection, Evaluation and Treatment of High Blood Cholesterol in Adults Executive summary of the third report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III) JAMA. 2001;285:2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 2.Prospective Studies Collaboration Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55 000 vascular deaths. Lancet. 2007;370:1829–1839. doi: 10.1016/S0140-6736(07)61778-4. [DOI] [PubMed] [Google Scholar]
- 3.Di Angelantonio E, Sarwar N, Perry P. Major lipids, apolipoproteins, and risk of vascular disease. JAMA. 2009;302:1993–2000. doi: 10.1001/jama.2009.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cholesterol Treatment Trialists' (CTT) Collaborators Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90 056 participants in 14 randomised trials of statins. Lancet. 2005;366:1267–1278. doi: 10.1016/S0140-6736(05)67394-1. [DOI] [PubMed] [Google Scholar]
- 5.Rader DJ, Cohen J, Hobbs HH. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J Clin Invest. 2003;111:1795–1803. doi: 10.1172/JCI18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 7.Barter PJ, Caulfield M, Eriksson M. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med. 2007;357:2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 8.Frikke-Schmidt R, Nordestgaard BG, Stene MC. Association of loss-of-function mutations in the ABCA1 gene with high-density lipoprotein cholesterol levels and risk of ischemic heart disease. JAMA. 2008;299:2524–2532. doi: 10.1001/jama.299.21.2524. [DOI] [PubMed] [Google Scholar]
- 9.Gray R, Wheatley K. How to avoid bias when comparing bone marrow transplantation with chemotherapy. Bone Marrow Transplant. 1991;7(suppl 3):9–12. [PubMed] [Google Scholar]
- 10.Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1986;327:507–508. doi: 10.1016/s0140-6736(86)92972-7. [DOI] [PubMed] [Google Scholar]
- 11.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 12.Johannsen TH, Kamstrup PR, Andersen RV. Hepatic lipase, genetically elevated high-density lipoprotein, and risk of ischemic cardiovascular disease. J Clin Endocrinol Metab. 2009;94:1264–1273. doi: 10.1210/jc.2008-1342. [DOI] [PubMed] [Google Scholar]
- 13.Kathiresan S, Melander O, Anevski D. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 14.Willer CJ, Sanna S, Jackson AU. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haase CL, Tybjaerg-Hansen A, Ali Qayyum A, Schou J, Nordestgaard BG, Frikke-Schmidt R. LCAT, HDL cholesterol and ischemic cardiovascular disease: a mendelian randomization study of HDL cholesterol in 54,500 individuals. J Clin Endocrinol Metab. 2012;97:E248–E256. doi: 10.1210/jc.2011-1846. [DOI] [PubMed] [Google Scholar]
- 16.Kathiresan S, Willer CJ, Peloso GM. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teslovich TM, Musunuru K, Smith AV. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.deLemos AS, Wolfe ML, Long CJ, Sivapackianathan R, Rader DJ. Identification of genetic variants in endothelial lipase in persons with elevated high-density lipoprotein cholesterol. Circulation. 2002;106:1321–1326. doi: 10.1161/01.cir.0000028423.07623.6a. [DOI] [PubMed] [Google Scholar]
- 19.Edmondson AC, Brown RJ, Kathiresan S. Loss-of-function variants in endothelial lipase are a cause of elevated HDL cholesterol in humans. J Clin Invest. 2009;119:1042–1050. doi: 10.1172/JCI37176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer TM, Lawlor DA, Harbord RM. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. 2011 doi: 10.1177/0962280210394459. published online Jan 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K. PLINK: a tool set for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:3. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 24.Ding EL, Song Y, Manson JE. Sex hormone-binding globulin and risk of type 2 diabetes in women and men. N Engl J Med. 2009;361:1152–1163. doi: 10.1056/NEJMoa0804381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehret GB, Munroe PB, Rice KM. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schunkert H, Konig IR, Kathiresan S. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishida T, Choi S, Kundu RK. Endothelial lipase is a major determinant of HDL level. J Clin Invest. 2003;111:347–355. doi: 10.1172/JCI16306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishida T, Choi SY, Kundu RK. Endothelial lipase modulates susceptibility to atherosclerosis in apolipoprotein-E-deficient mice. J Biol Chem. 2004;279:45085–45092. doi: 10.1074/jbc.M406360200. [DOI] [PubMed] [Google Scholar]
- 29.Ko KW, Paul A, Ma K, Li L, Chan L. Endothelial lipase modulates HDL but has no effect on atherosclerosis development in apoE-/- and LDLR-/- mice. J Lipid Res. 2005;46:2586–2594. doi: 10.1194/jlr.M500366-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Hulley S, Grady D, Bush T. Randomized trial of estrogen plus progestin for secondary prevention of coronary heart disease in postmenopausal women. Heart and Estrogen/progestin Replacement Study (HERS) Research Group. JAMA. 1998;280:605–613. doi: 10.1001/jama.280.7.605. [DOI] [PubMed] [Google Scholar]
- 31.Boden WE, Probstfield JL, Anderson T. Niacin in patients with low HDL cholesterol levels receiving intensive statin therapy. N Engl J Med. 2011;365:2255–2267. doi: 10.1056/NEJMoa1107579. [DOI] [PubMed] [Google Scholar]
- 32.Thompson A, Di Angelantonio E, Sarwar N. Association of cholesteryl ester transfer protein genotypes with CETP mass and activity, lipid levels, and coronary risk. JAMA. 2008;299:2777–2788. doi: 10.1001/jama.299.23.2777. [DOI] [PubMed] [Google Scholar]
- 33.Cannon CP, Shah S, Dansky HM. Safety of anacetrapib in patients with or at high risk for coronary heart disease. N Engl J Med. 2010;363:2406–2415. doi: 10.1056/NEJMoa1009744. [DOI] [PubMed] [Google Scholar]
- 34.Khera AV, Cuchel M, de la Llera-Moya M. Cholesterol efflux capacity, high-density lipoprotein function, and atherosclerosis. N Engl J Med. 2011;364:127–135. doi: 10.1056/NEJMoa1001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 36.Nitsch D, Molokhia M, Smeeth L, DeStavola BL, Whittaker JC, Leon DA. Limits to causal inference based on Mendelian randomization: a comparison with randomized controlled trials. Am J Epidemiol. 2006;163:397–403. doi: 10.1093/aje/kwj062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.