Abstract
Introduction
Circulating inflammatory markers may play an important role in cognitive impairment at older ages. Mice deficient for the chemokine (C-C motif) receptor 2 (CCR2) develop an accelerated Alzheimer-like pathology. CCR2 is also important in neurogenesis. To identify human gene transcripts most closely associated with Mini-Mental State Examination (MMSE) scores, we undertook a genome-wide and inflammation specific transcriptome screen in circulating leukocytes from a population-based sample.
Methods
We measured in vivo transcript levels by microarray analysis in 691 subjects (mean age 72.6 years) in the InCHIANTI study (Invecchiare in Chianti, aging in the Chianti area). We assessed expression associations with MMSE performance at RNA collection and prior 9-year change in MMSE score in linear regression models.
Results
In genome-wide analysis, raised CCR2 expression was cross-sectionally the most strongly associated transcript with lower MMSE score (beta=−0.16, p=5.1×10−6, false discovery rate (FDR; q=0.077). Amongst inflammatory transcripts, only CCR2 expression was associated with both MMSE score and accelerated decline in score over the preceding 9 years (beta=−0.16, p=5.1×10−6, q=0.003; and beta=−0.13, p=5.5×10−5, q=0.03, respectively). CCR2 expression was also positively associated with apolipoprotein E (ApoE) e4 Alzheimer disease risk haplotype.
Conclusions
We show for the first time that CCR2 expression is associated with lower MMSE scores in an older human population. Laboratory models of Ccr2-mediated β-amyloid removal and regulation of neurogenesis affecting cognitive function may be applicable in humans. CCR2-mediated pathways may provide a possible focus for intervention to potentiate protective reactions to Alzheimer pathology in older people, including for people with an adverse ApoE haplotype.
Introduction
Cognitive impairment and dementia at older ages commonly result from accumulating vascular and neurodegenerative pathology in the brain1 and are experienced by over half of adults aged 85 or over.2 Major risk factors for cognitive impairments in later life include the APOE e4 haplotype,3 but the underlying biological mechanisms are still unclear. Gene expression arrays offer a new approach to identifying the most important molecular mechanisms causing or responding to the pathologies underlying cognitive decline. The most accessible tissue for gene expression array analyses in large numbers of older people is circulating blood leukocytes, which are likely to be most sensitive to inflammatory and related mechanisms.
Chronic low-level inflammation has been proposed as a key mechanism underlying cognitive decline and dementia and has been implicated in the neuropathological cascade leading to late onset Alzheimer disease (LOAD).4 Inflammatory factors have been further implicated in cognitive impairment and dementia using mouse models.5 These include transgenic animals in which chemoattractant proteins or receptors such as Ccl2 or Ccr2 have been abolished.5 These genes, associated with the migration of phagocytic and inflammatory macrophages, have been associated with Alzheimer pathology and peripheral atherosclerosis.5–7 Ccr2-deficient mice show early accumulation of β-amyloid, with premature mortality.7 Blood-borne systemic factors, including CCR2 and CCL11, were also recently implicated in the negative regulation of neurogenesis and cognitive function in rodent studies.8
The importance of the chemokine (C-C motif) receptor 2 (CCR2) and related signaling in human age-related cognitive impairment is unclear. Maes et al. (2007) identified leukocyte expression differences between 14 Alzheimer disease and control individuals in a microarray experiment,9 and Grunblatt et al. identified five peripheral blood leukocyte genes whose mRNA correlated significantly with Mini-Mental State Examination (MMSE) scores in a smaller series of Alzheimer disease and control individuals.10 These studies present initial evidence that brain changes in pathologically specific dementia patients may be detectable using peripheral leukocyte gene expression, but it is currently not known if this also applies to age-associated cognitive decline.
In the current study, we used a genome-wide and inflammation-focused approach to identify the most strongly associated in vivo transcript levels in circulating leukocytes associated with MMSE score or rate of change in MMSE score in a general population sample of predominantly older people. The MMSE is a widely used measure of cognitive function in prospective epidemiological studies of elderly populations and is sensitive to moderate or severe cognitive declines, often due to dementia.11,12 To ensure population relevance, no exclusions for co-morbidity were made in the main analysis.
Materials and Methods
Study cohort
We used InCHIANTI (Invecchiare in Chianti, aging in the Chianti area), a population-based study of aging13 that has followed older persons over a 9-year period to assess ‘normal’ aging using both interviews (conducted at the participants home by experienced interviewers) and blood samples (in the study clinic, all patients fasted for 8 hr prior to collection). Peripheral blood samples for RNA extraction were collected from participants at the 9-year follow-up (2008/9). Cohort demographics are given in Table 1. Ethical approval was granted by the Instituto Nazionale Riposo e Cura Anziani institutional review board in Italy. Participants gave written informed consent to participate and for sample collection after having received an extensive description of the procedures, purposes, and potential risks of the study. RNA was extracted from each sample using the PAXgene Blood mRNA kit (Qiagen, Crawley, UK) according to the manufacturer's instructions. To ensure population relevance, no exclusions for co-morbidity were made in the main analysis.
Table 1.
Characteristics of the Study Cohort: Summary of the Population Statistics for the 691 Participants Eligible for Our Study
| Summary statistics | |
|---|---|
| Age (years) | n |
| 30–49 | 86 (12.5%) |
| 50–69 | 98 (14.2%) |
| 70–89 | 479 (69.3%) |
| 90–104 | 28 (4%) |
| Mean age at RNA collection | 72.6 (SD=15.3) |
| Gender | % |
| Men | 44.8 |
| Women | 55.2 |
| MMSE score | |
| Mean score at RNA collection | 25.58 (SD=5.66) |
| Mean change in MMSE (9 years) | −1.31 (SD=4.71) |
| Frequency of scores at RNA collection | % |
| 0–18 | 8.14 |
| 19–23 | 12.79 |
| 24–27 | 30.52 |
| 28–30 | 48.55 |
| Education | % |
| None | 13.31 |
| Elementary | 46.02 |
| Secondary | 13.46 |
| High school | 12.59 |
| University/Professional | 14.62 |
| Pack years smoked (lifetime) | % |
| None | 55.72 |
| 0.1–20 | 22.87 |
| 20–39 | 14.33 |
| 40+ | 7.09 |
MMSE, Mini-Mental State Examination; SD, standard deviation.
Whole-transcriptome scan
Whole-genome expression profiling of the samples was conducted using the Illumina Human HT-12 microarray (Illumina, San Diego, CA). Data processing was carried out using the Illumina and Beadstudio software (Illumina, San Diego, CA), as previously described.14 Baseline intensities were calculated as mean and standard deviation (SD) computed over all beads for a particular probe. Quality control (QC) steps included correction for local background effects, removal of outlier beads, computation of average bead signal, SD for each probe and gene, calculation of detection p values using negative controls present on the array, quantile normalization across arrays, check of outlier samples using a clustering algorithm, and checks of positive controls. Subject-level QC steps included removing individuals where the expression intensity was±3 SD from the mean. All microarray experiments and analyses complied with Minimum Information About a Microarray Experiment (MIAME) guidelines.15 Following microarray data QC steps, 16,571 transcripts gave reliable signals above background (p<0.01) in >5% of the sample population and were therefore eligible for analysis.
Assessment of cognitive function
Participant interviews and clinical assessment, including the MMSE of global cognitive function, were undertaken at baseline, and at years 3, 6, and 9 of follow-up. The MMSE is an assessment of global cognitive function widely used in both hospital and community settings as a dementia screening tool.16,17
Included sample and statistical analysis
From 733 blood samples collected, RNA quality and microarray QC steps resulted in loss of 35 participants from the analysis. From the remaining 698 participants, 7 were excluded due to absence of MMSE data, yielding 691 individuals. Three further individuals were dropped from the regression models due to incomplete leukocyte data. (See Table 1 for summary statistics of the cohort included in the analyses.) All of the remaining 688 individuals were included in the analyses, irrespective of other pathologies.
Associations between gene expression and MMSE score at RNA collection were analyzed using multiple regression models adjusted for the following potential confounding factors: Age in years (as a continuous variable), gender, highest level of education received (in five categories: none, elementary, secondary, high school, and university/professional), lifetime pack-years smoked (in four categories: none, 0.1–20 years, 20–39 years, and 40 plus years), blood leukocyte type (neutrophil, lymphocyte, monocyte, and eosinophil percentages) each as continuous variables, hybridization and amplification batch (in 10 and 14 categories, respectively), and study site. Participants lived in either a rural village (Greve) or an urban site (Bagno a Ripoli).
Separate linear regression models were fitted for each of the full set of 16,571 probes that passed QC in the discovery dataset. We have expressed the effect sizes of the associations in standardized betas to aid in the interpretation (expression intensities vary from probe to probe, and the normalization procedures further affect the interpretability of coefficients). We controlled for the effect of multiple testing by measuring the statistical significance of each association using both the p value and the q value. The q value quantifies significance in terms of the false discovery rate (FDR) rather than the false positive rate,18 and forms a measure of how likely a particular p value is to represent a genuine association. Expression levels were taken to have a significant association with MMSE score at RNA collection, or cognitive decline from baseline 9 years previously, if the association achieved a nominal p value ≤0.05 and a q value<0.1. Our study was powered to detect expression differences of <0.25 SD for the 16,571 transcripts studied, allowing us to detect moderate expression differences between groups.
Inflammatory gene scan
The large number of transcripts tested in the whole genome scan yields a very stringent requirement for statistical significance, resulting in a substantial risk of type II (false negative) error. Because leukocytes are an inflammation-related tissue, we carried out a subanalysis focusing on a smaller subset of genes involved in inflammatory response only. The genes of interest were identified using the Molecular Signatures Database (MSigDB; www.broadinstitute.org/gsea/msigdb/index.jsp/),19 with search terms for inflammatory/immune-related gene pathways as defined by the Gene Ontology (GO) project (www.geneontology.org). A total of 44 inflammation-related gene-sets were returned, comprising 635 unique probe IDs, which equate to 425 unique genes with available data in our cohort (Supplemental Table 1 [Supplemental data available at www.liebertpub.com/rej). Linear regression models were carried out as described above. This study is powered to detect expression differences of <0.22 SD for the inflammation transcript set.
Real-time PCR validation
The expression of the CCR2 gene in a subcohort of 100 individuals selected at the extremes of the MMSE spectrum was validated by the use of a custom real-time PCR assay (probe and primer sequences available on request). Reaction mixes included 5μL of l 2×TaqMan universal master mix (no AMPerase) (Applied Biosystems, Foster City, CA), 30μL of dH2O, and 2 μL of cDNA template. PCR amplifications were performed on the ABI 7900HT platform (Applied Biosystems, Foster City, CA). Cycling conditions were 50°C for 2 min, 94.5°C for 10 min followed by 40 cycles of 97°C for 30 sec and 57.9°C for 1 min. The expression of each gene was measured in triplicate for each sample. Gene expression relative changes were quantified using the 2−DDCt method20 relative to the geometric mean of the GUSB, B2M, and PPIA endogenous controls. The correlation between quantifications achieved using microarray and by real-time PCR was then assessed using linear regression.
Gene Set Enrichment Analysis
We also performed gene set enrichment analysis (GSEA) to identify gene sets/pathways associated with cognitive function, according to the method of Subramanian et al.19 Molecular or biological function pathways were identified using GO gene sets from the MSigDB Gene set size; filtering excluded gene sets containing less than 15 or more than 500 genes. Genes were ranked according to the magnitude of their association with cognitive function, and representation of each gene set within this ranked list was analyzed. Gene sets significantly overrepresented at the top or bottom of the ranked list were taken to be significantly associated with cognitive function. A signal-to-noise metric was used to rank genes, and gene set enrichment scores were calculated using a weighted enrichment statistic. One thousand random permutations of the phenotype label were used to calculate the empirical p values of each pathway compared to the p values that would be ascertained by chance. Gene sets with a nominal p value<0.01 and q value<0.1 were considered associated with cognitive impairment or cognitive decline.
Penalized cubic regression spline
Using R package ‘mgcv’,21 we fitted an adjusted generalized additive model (GAM) using a smoothed penalized cubic regression spline; the smoothing parameter was chosen automatically by cross-validation. The same data and covariates that made up the original generalized linear regression (GLM) screen were used. GAM is applied here because it allows nonparametric fits, and thus if an aberrant relationship between expression and age had existed it could be highlighted.
Results
Cohort details
Participant characteristics are given in Table 1. The study sample had a mean age of 72.6 years (SD=15.3, range 30–104); 55.2% were female. At RNA collection, the mean MMSE score was 25.58 (SD=5.66), 21% of the sample had MMSE scores of 23 or less (8.1% with MMSE of 18 or less), and the mean change in MMSE scores over 9 years was −1.31 (SD=4.71).
Genome-wide analysis
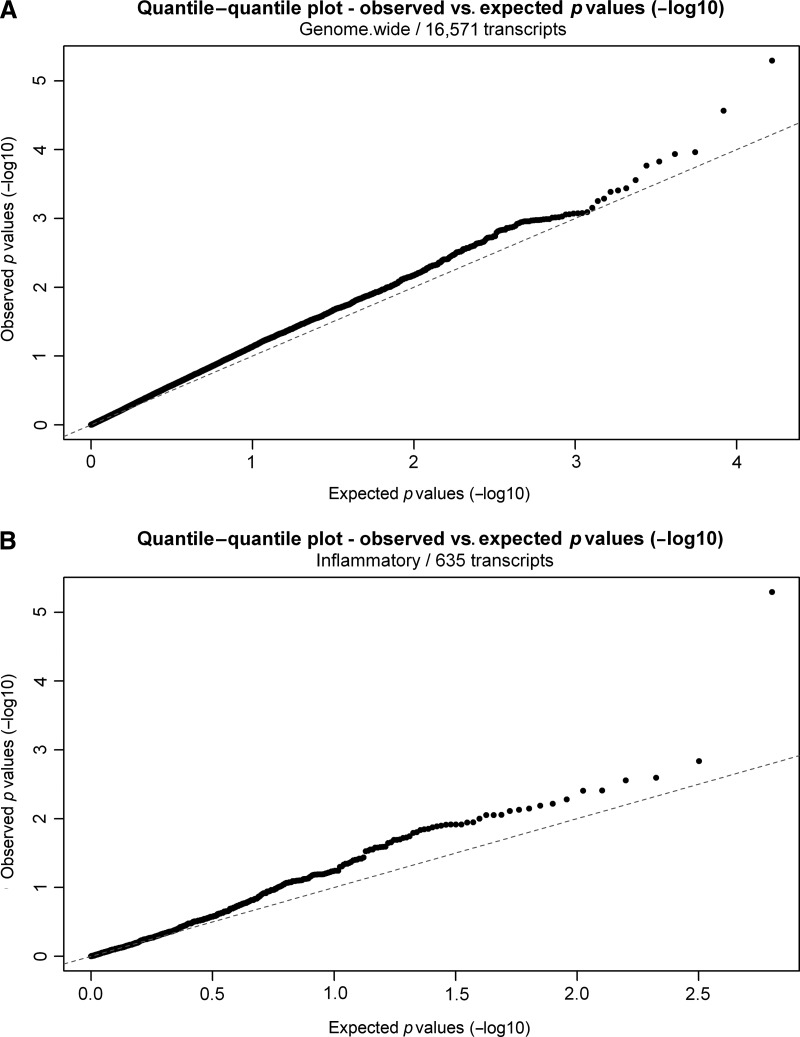
In genome-wide analysis, only one transcript (CCR2, measured via probe ilmn_1774761) showed a very near significant genome-wide association with MMSE score at RNA collection (beta=−0.16, p=5.1−10−6; q=0.076; Table 2). No probes were significantly associated (q<0.1) with change in MMSE score over the preceding 9 years (Table 2), although CCR2 was the most strongly associated transcript (beta=−0.13, p=5.5×10−5, q=0.70). Quantile–quantile (Q-Q) plots comparing the observed p values (−log10) with those that would be expected by chance alone (Fig. 1A) confirmed that large-scale disruption to gene expression levels was not a feature of MMSE score at RNA collection or change in score over the previous 9 years, although some small deviations from the expected pattern were noted.
Table 2.
The 10 Probes Most Closely Associated with MMSE Score at Wave 9 and Change in MMSE Score (Baseline to Year 9), Ordered by False Discovery Rate q Value (n Observations=688)
| Probe ID | p value | Coefficient | Beta | 95% CIs | q value | Gene | |
|---|---|---|---|---|---|---|---|
| MMSE score at year 9 | |||||||
| ilmn_1774761 | 5.1E-06 | −0.0042 | −0.1616 | −0.0056 | −0.0027 | 0.076 | CCR2 |
| ilmn_2374362 | 2.7E-05 | −0.0015 | −0.1709 | −0.0021 | −0.0009 | 0.204 | FAM108B1 |
| ilmn_1791912 | 1.1E-04 | 0.0052 | 0.1475 | 0.0030 | 0.0074 | 0.356 | SIDT2 |
| ilmn_1758457 | 1.2E-04 | 0.0008 | 0.1473 | 0.0005 | 0.0011 | 0.361 | TBC1D16 |
| ilmn_1796094 | 1.5E-04 | −0.0044 | −0.1486 | −0.0063 | −0.0025 | 0.381 | CD36 |
| ilmn_1693949 | 1.7E-04 | 0.0011 | 0.1153 | 0.0006 | 0.0015 | 0.390 | UNQ1944 |
| ilmn_1724266 | 2.8E-04 | 0.0017 | 0.1382 | 0.0009 | 0.0025 | 0.418 | LYPD2 |
| ilmn_1772821 | 3.7E-04 | −0.0015 | −0.1561 | −0.0022 | −0.0008 | 0.430 | KIAA1671 |
| ilmn_2313434 | 3.9E-04 | 0.0013 | 0.1476 | 0.0007 | 0.0019 | 0.433 | TCP1 |
| ilmn_2393573 | 4.1E-04 | −0.0019 | −0.1073 | −0.0027 | −0.0010 | 0.434 | RASSF1 |
| Change in MMSE score (9 years) | |||||||
| ilmn_1774761 | 5.5E-05 | −0.0040 | −0.1305 | −0.0056 | −0.0024 | 0.700 | CCR2 |
| ilmn_2374362 | 1.5E-04 | −0.0015 | −0.1345 | −0.0022 | −0.0009 | 0.700 | FAM108B1 |
| ilmn_1796094 | 2.2E-04 | −0.0047 | −0.1146 | −0.0067 | −0.0026 | 0.700 | CD36 |
| ilmn_1761941 | 2.6E-04 | −0.0017 | −0.1135 | −0.0024 | −0.0009 | 0.700 | C4orf18 |
| ilmn_2313434 | 2.7E-04 | 0.0015 | 0.1273 | 0.0008 | 0.0022 | 0.700 | TCP1 |
| ilmn_1712684 | 3.1E-04 | −0.0011 | −0.1245 | −0.0015 | −0.0006 | 0.700 | FAM20C |
| ilmn_1866887 | 3.4E-04 | −0.0011 | −0.1290 | −0.0016 | −0.0006 | 0.700 | BX537605 |
| ilmn_1772821 | 5.6E-04 | −0.0016 | −0.1277 | −0.0023 | −0.0008 | 0.700 | KIAA1671 |
| ilmn_1703314 | 5.7E-04 | 0.0013 | 0.1167 | 0.0007 | 0.0020 | 0.700 | KLHL36 |
| ilmn_1789751 | 6.1E-04 | −0.0039 | −0.0985 | −0.0058 | −0.0020 | 0.700 | MFSD1 |
Models adjusted for: age, gender, highest education level, smoking-status, site, hybridization-batch, amplification-batch, leukocyte proportions (and MMSE-baseline score, in ‘change’ models).
MMSE, Mini-Mental State Examination; CI, confidence interval.
FIG. 1.
Quantile–quantile (Q-Q) plots for gene expression analysis of Mini-Mental State Examination (MMSE) score at RNA collection. (A) The Q-Q plot for the genome-wide analysis of MMSE at RNA collection is shown. The actual p values (−log10) obtained are given on the y axis, plotted against expected p values (−log10) given on the x axis. This graph shows potential deviations to the p value distribution that might be expected by chance. (B) The Q-Q plot for the focused analysis of inflammatory genes is shown. The actual p values (−log10) obtained are given on the y axis, plotted against expected p values (−log10) given on the x axis. This graph shows potential deviations to the p value distribution that might be expected by chance. The positive association with the CCR2 transcript is evident in both plots.
Specific analysis of inflammation-related transcripts
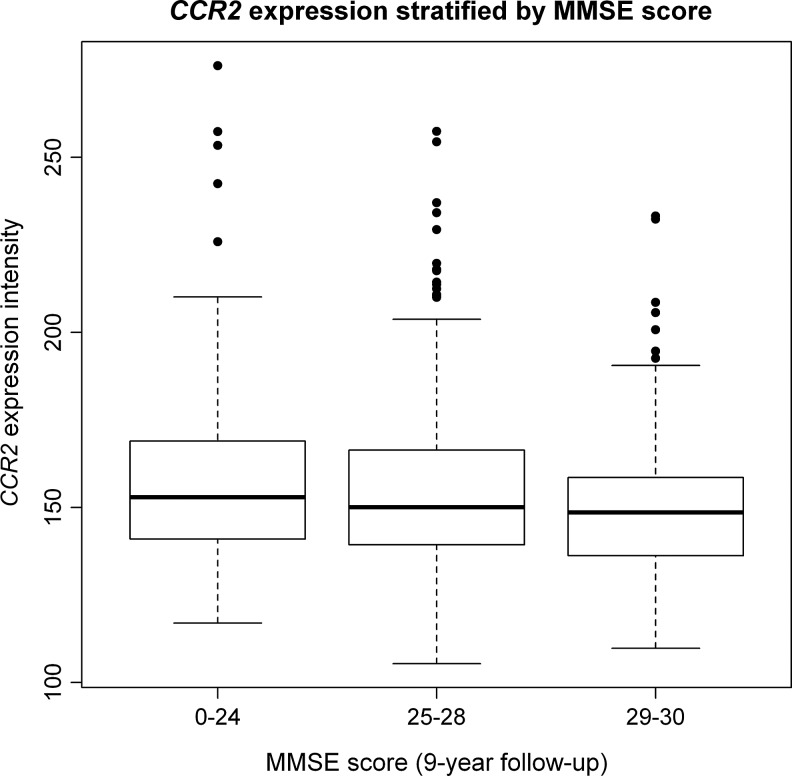
In a targeted analysis on inflammation-related transcripts only, the CCR2 transcript was associated with MMSE score at RNA collection (beta=−0.16, p=5.1×10−6, q=0.003; Table 3) and also with cognitive decline score over the previous 9 years, using a FDR of q<0.1 (beta=−0.13, p=5.5×10−5, q=0.03; Table 3). As in the genome-wide analysis, other large-scale alterations in the expression of inflammatory genes were not present (after accounting for multiple testing) for either cognitive function or decline, although one large effect (CCR2) was evident on the Q-Q plot (Fig. 1B). Figure 2 shows the unadjusted relationship between CCR2 and MMSE score; the trend is apparent even prior to adjustment for leukocyte proportions and other known confounders.
Table 3.
The 10 Inflammation-Related Probes Most Closely Associated with MMSE Score at Wave 9 and Change in MMSE Score (Baseline to Year 9), Ordered by False Discovery Rate q Value (n Observations=688)
| Probe ID | p value | Coefficient | Beta | 95% CIs | q value | Gene | |
|---|---|---|---|---|---|---|---|
| MMSE score at year 9 | |||||||
| ilmn_1774761 | 5.1E-06 | −0.0042 | −0.1616 | −0.0056 | −0.0027 | 0.003 | CCR2 |
| ilmn_2366212 | 1.5E-03 | 0.0083 | 0.1267 | 0.0040 | 0.0125 | 0.232 | CD79B |
| ilmn_1677440 | 2.6E-03 | −0.0055 | −0.1194 | −0.0084 | −0.0025 | 0.262 | ATP6AP2 |
| ilmn_2276996 | 2.8E-03 | −0.0009 | −0.1071 | −0.0014 | −0.0004 | 0.266 | CCR2 |
| ilmn_1785439 | 3.9E-03 | 0.0042 | 0.1239 | 0.0018 | 0.0066 | 0.279 | CD79B |
| ilmn_1710017 | 3.9E-03 | 0.0045 | 0.1302 | 0.0019 | 0.0070 | 0.279 | CD79B |
| ilmn_1764396 | 5.3E-03 | −0.0011 | −0.0725 | −0.0017 | −0.0004 | 0.288 | HDAC4 |
| ilmn_1763875 | 6.1E-03 | 0.0022 | 0.0842 | 0.0009 | 0.0035 | 0.291 | ABCF1 |
| ilmn_1738767 | 6.5E-03 | −0.0012 | −0.0780 | −0.0019 | −0.0005 | 0.293 | PLP2 |
| ilmn_1747227 | 7.1E-03 | 0.0008 | 0.1412 | 0.0003 | 0.0013 | 0.295 | ADORA1 |
| Change in MMSE score (9 years) | |||||||
| ilmn_1774761 | 5.5E-05 | −0.0040 | −0.1305 | −0.0056 | −0.0024 | 0.033 | CCR2 |
| ilmn_2366212 | 2.1E-03 | 0.0087 | 0.1047 | 0.0041 | 0.0134 | 0.305 | CD79B |
| ilmn_1710017 | 2.2E-03 | 0.0052 | 0.1013 | 0.0024 | 0.0079 | 0.310 | CD79B |
| ilmn_1677440 | 2.9E-03 | −0.0059 | −0.0946 | −0.0091 | −0.0027 | 0.325 | ATP6AP2 |
| ilmn_1747227 | 3.2E-03 | 0.0009 | 0.1169 | 0.0004 | 0.0015 | 0.332 | ADORA1 |
| ilmn_1737398 | 3.3E-03 | −0.0012 | −0.0938 | −0.0018 | −0.0005 | 0.332 | PTPLAD1 |
| ilmn_1785439 | 4.1E-03 | 0.0046 | 0.1024 | 0.0020 | 0.0072 | 0.349 | CD79B |
| ilmn_1682312 | 6.5E-03 | −0.0025 | −0.0726 | −0.0040 | −0.0010 | 0.377 | CYBB |
| ilmn_1811049 | 6.8E-03 | 0.0012 | 0.0999 | 0.0005 | 0.0019 | 0.379 | POU2AF1 |
| ilmn_1771333 | 6.9E-03 | −0.0035 | −0.0877 | −0.0057 | −0.0014 | 0.380 | CD47 |
Models adjusted for: age, gender, highest education level, smoking-status, site, hybridization-batch, amplification-batch, leukocyte proportions (and MMSE-baseline score, in ‘change’ models).
MMSE, Mini-Mental State Examination; CI, confidence interval.
FIG. 2.
Box plot of CCR2 expression by Mini-Mental State Examination (MMSE) score at RNA collection. The box plot shows CCR2 transcript expression levels (relative units) as plotted on the y axis by MMSE score RNA collection on the x axis.
Modeling of relationship between CCR2 transcript expression and MMSE/delta MMSE using spline analysis
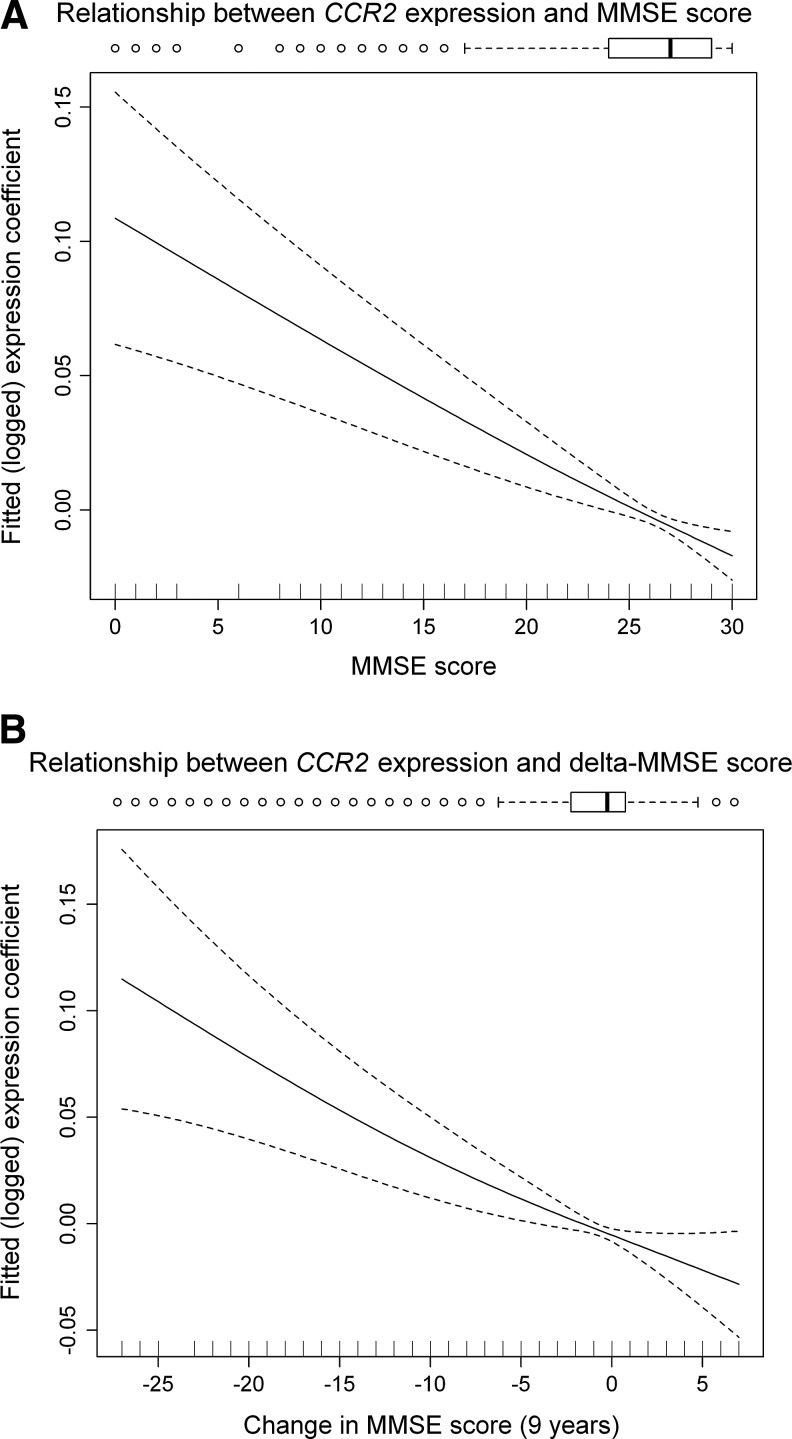
To understand any nonlinearity in the relationship between CCR2 expression and MMSE (and change in MMSE over 9 years, delta-MMSE), we fitted a penalized cubic spline regression model for: (A) CCR2 expression and MMSE at RNA collection, and (B) CCR2 expression and delta-MMSE over the preceding 9 years (see Fig. 3 for spline plots). We found the relationships to be approximately linear throughout the MMSE or delta-MMSE ranges.
FIG. 3.
Cubic Spline plots for linearity of association between CCR2 expression and Mini-Mental State Examination (MMSE). (A) CCR2 expression and MMSE at RNA collection. (B) CCR2 expression and delta-MMSE over the preceding 9 years. These penalized cubic regression splines visualize the fitted (adjusted) relationship between CCR2 gene expression and MMSE score, and separately with change in MMSE over 9 years. The relationships are approximately linear after normalization (log-transformed expression data) and adjustment for multiple confounders.
Quantitative real-time PCR validation of CCR2 levels
To validate our microarray results, we also quantified CCR2 expression in a subset of our cohort using quantitative real-time PCR (qRT-PCR). We found that the expression level of CCR2 transcripts as measured by qRT-PCR correlated well with the expression levels measured by microarray analysis (r2=0.5, p=2×10−4).
Gene set enrichment analysis
No gene sets showed evidence of deregulation in association with MMSE score at RNA collection or change in score using GSEA, at a FDR q value of <0.1 (Table 4).
Table 4.
Biological and Molecular Functional Pathway Enrichments for Negative Correlation with MMSE Are Presented
| Pathway | Size | ES | p value | q value |
|---|---|---|---|---|
| Aromatic compound metabolic process | 17 | −1.82 | 0 | 0.85 |
| Regulation of DNA metabolic process | 29 | −1.74 | 0.008 | 0.95 |
| T cell activation | 27 | −1.56 | 0.031 | 1 |
| Transferase activity transferring glycosyl groups | 68 | −1.42 | 0.034 | 1 |
| Cellular defense response | 38 | −1.52 | 0.035 | 1 |
| Anatomical structure formation | 27 | −1.55 | 0.036 | 1 |
| Positive regulation of lymphocyte activation | 15 | −1.58 | 0.039 | 1 |
| Coenzyme metabolic process | 30 | −1.44 | 0.045 | 1 |
| RNA export from nucleus | 17 | −1.52 | 0.045 | 1 |
| DNA recombination | 32 | −1.5 | 0.047 | 1 |
ES refers to the nonspecific enrichment score.
MMSE, Mini-Mental State Examination; ES, enrichment score.
Post hoc sensitivity analyses
To determine if the observed association is dependent on the inclusion of some younger people in the predominantly older sample, or dependent on the oldest old, we carried out a sensitivity analysis in subjects aged ≥70 and <90 years old at RNA extraction; MMSE score and (logged) CCR2 expression were still strongly associated (beta=−0.15, p=2e−4) within this age range. The apolipoprotein E (ApoE) genotype was available on n=480 (excluding e2e4 n=4) in our sample: CCR2 expression was positively associated with the ApoE risk haplotype (trend test across e2e2, e2e3, e3e3, e3e4, e4e4 groups: coef=0.02; confidence interval (CI) 0.007–0.03, p=0.001). The association between CCR2 expression and MMSE at RNA collection was attenuated but remained significant after additional adjustment for ApoE status (beta=−0.1, p=0.018). There was no statistical interaction between ApoE status and CCR2 expression.
Co-morbidity is very common with cognitive impairment in later life in the general older population, and therefore our approach has been to avoid disease specific exclusions. We did examine the effect of removing those diagnosed as having had a stroke (n=76 removed), and CCR2 remained associated with MMSE score (beta=−0.14, p=0.001) in the stroke-free group.
Discussion
In this study, we have investigated associations between blood leukocyte-derived mRNA expression and MMSE scores in an aging population. CCR2 expression reached inflammation-specific significance (and only narrowly missed genome-wide significance after accounting for multiple testing), providing the first human population evidence of likely consistency with the Ccr2 mouse models. Our finding that ApoE haplotype status, the major inherited genetic risk factor associated with Alzheimer pathology, is associated with CCR2 expression is also consistent with CCR2 signaling having a direct role in cognitive decline in later life in human populations.
The MMSE score is a widely used measure of cognitive impairment in later life, although it has a ceiling effect that limits its ability to detect early subtle cognitive changes, particularly in younger highly educated participants11,22 and is not considered an appropriate measure to diagnose mild cognitive impairment or related prodromal diagnoses without supplementing it with other cognitive measures, although the population relevance of these clinically derived constructs remains disputed.23 Over several years follow-up, the MMSE does however provide a useful measure of cognitive impairment and cognitive decline in participants with and without dementia12 and is the most widely used measure to monitor cognitive function and decline in elderly adults in clinical settings. Therefore, we consider that it is an appropriate tool to assess age-related differences in cognitive function in our cross-sectional population.
Our functional, rather than pathological, classification of participants was important for several reasons. Cognitive decline leads to impairments in activities of daily living and is a major public health threat. It is also rarely determined purely by one pathophysiological mechanism—mixed dementia is now considered to be the most common form of dementia24 and functional decline has only a moderate association with the pathological changes of Alzheimer disease.25 Finally, our use of a large, population-based cohort, rather than from specialist clinics, gave us power to detect moderate biomarker effects that could be of clinical application in population testing and risk reduction. Biomarkers of functional cognitive change may hold more clinical application in the population than markers of single pathological processes.
We investigated blood leukocytes to identify clinically useful, minimally invasive markers of cognitive function because cortical tissue is inappropriate for the identification of cognitive decline risk or progression in the population. Furthermore, recent studies suggest that circulating systemic factors, such as chemokines and their receptors, have an important role in neurogenesis and cognitive function in animal models.8 Peripheral inflammation has also been shown to impact negatively on human cognitive function26 with chronic immune activation in the brain having an association with neurodegenerative disease and cognitive decline.27,28
There are both similarities and differences between our data and previous studies.9,10,29,30 It has been suggested that genes involved in cytoskeletal maintenance, cellular trafficking, cellular stress response, redox homeostasis, transcription, and DNA repair may be associated with Alzheimer disease,23 and proteomics studies have suggested that circulating lymphocytes may be promising biomarkers for this disorder.31 The differences could arise from different patient selection criteria; we used population data rather than data from clinical settings. Differences with prior proteomic findings in peripheral blood in cognitive decline32,33may also be due to small study sizes or the possibility that circulating cytokines previously found associated with cognitive decline may be expressed by activated central nervous system (CNS) inflammatory cells and released into the circulation, rather than being expressed by circulating leukocytes.34
The CCR2 receptor binds CCL2, a protein that is abundantly expressed in macrophage-rich areas of atherosclerotic plaques and in brain microglia,7 resulting in migration of macrophages and brain microglial cells to their site of action. In mouse models, vascular disruption of Ccr2 or Ccl2 leads to a reduction in atherosclerotic plaque formation in ApoE- or LDL-R null mice, even when fed a high-fat diet.6,31 This may be mediated by Ccr2 depletion, reducing macrophage infiltration. In mouse brain, Ccr2 knockout has been shown to result in accelerated disease progression, increased mortality, and an increase in soluble antibody (Ab) assemblies.7,35 Work on CCR2 gene knockout mice bred on a background of chimeric mouse/humanβ-amyloid precursor/presenilin overexpression (APPSwe/PS1/CCR2−/−) suggested that Alzheimer disease might be associated with a decreased expression of CCR2.35 Although our results at first appear to conflict with this, we suggest that the increased CCR2 levels we note in cognitively impaired individuals probably reflect a reactive increase in the need for chemoattractants in subjects with increased β-amyloid deposition. This is supported by the observation that the ligand for CCR2, CCL2, has previously been reported to be upregulated in the brain of patients suffering from Alzheimer disease.36 Recent studies have also suggested that CCR2 is with a key modulator of negative regulation of neurogenesis and cognitive function in mice.8 Our data support a role for CCR2 in the etiology of age-related cognitive decline, and indicates that the CCR2 mouse models7,35 may have particular relevance to the human population, although much more work is needed to confirm this mechanism in human populations.
Key limitations of our study include the possibility that that there are gene expression correlates of cognitive function in specific white cell subtypes that we have not measured separately. These are not, however, likely to be very marked or common, as we have found only limited overall expression changes. Similarly, as noted above, the MMSE score, a widely used clinical measure of cognitive function in older people, suffers from a relative insensitivity to frontal-executive dysfunction and visuo-spatial deficits37 and a ceiling effect inhibiting sensitive differentiation between medium and high cognitive performers.38 Our cohort may also be subject to informative loss to follow-up and reflect a higher functioning group, because people with very impaired cognitive function may not have reported for blood sampling in year 9.
We note also that several probes to CCR2 transcripts were present on the chip, but only one showed associations with MMSE score. Differences in the relationship between phenotype and alternative specific probes for the same gene are not atypical in microarray studies. These discrepancies can arise from several factors, including differences in binding dynamic of the individual probes, differences in the interindividual intensities between probe signal that can add noise to the data and reduce power, and the presence of alternatively processed isoforms that may bind probes differentially. In the case of CCR2, three alternatively expressed isoforms exist, with only a single probe, ilmn_1774761, capable of binding to all isoforms. If the effect we note is driven by a specific isoform, only those probes that identify the transcript in question will show a significant association. Because up to 90% of all genes are alternatively spliced, and the full transcriptomic output from any gene is not fully known at present, this can lead to apparent differences in the association of particular probes with disease phenotype in association studies.
Future work should first seek to replicate our CCR2 finding and explore associations with the full range of isoform-specific probes for the main target genes. Work should also seek to gather more evidence on which of the near-significant probes may in fact be associated with cognitive function. Future prospective studies will also provide valuable information as to the role of chemoattractant proteins in age-related cognitive decline. Functional work in humans is also needed to clarify the mechanisms involved in the raised expression of CCR2 we observed in those with lower or declining MMSE scores. Studies of the role of CCR2 in specific forms of dementia (Alzheimer, vascular, Lewy body, etc., or mixed) would also help characterize the role of CCR2. If raised expression of CCR2 is a helpful but insufficient response to the accumulation of β-amyloid, then attempts to increase this response further may potentially be effective. Studies of the role of CCR2 in vascular disease and proinflammatory response would also be informative. Our findings raise the possibility that CCR2 expression levels may also be associated with early, presymptomatic cognitive changes, although a more sensitive approach than change in MMSE score would be required to detect this.
To conclude, we have carried out the largest assessment of in vivo leukocyte human gene expression alterations in conjunction with MMSE-measured cognitive function in a predominantly older population to date. We identified associations between MMSE score and CCR2 transcript levels, which may reflect the key role CCR2 plays in the removal of β-amyloid and in regulation of neurogenesis. Studies are needed to confirm our findings, establish whether these reflect the proposed mechanisms, and relate the circulating transcriptome changes to specific forms of brain pathology.
Supplementary Material
Acknowledgments
This study was supported in part by the Intramural Research Program, National Institute on Aging, and the U.S. National Institutes of Health. We thank the many people who contributed to the InCHIANTI study, including all of the anonymous participants. The U.S. National Institute on Aging intramural research program contributed the array data and supported this analysis. William Henley completed this work while seconded to PenCLARHC, which is funded by the National Institute of Health Research (NIHR). The views expressed in this publication are those of the author(s) and not necessarily those of the National Health Service (NHS), the NIHR, or the Department of Health.
Author Disclosure Statement
No competing financial interests exist.
L.W.H. oversaw the validation experiments, interpreted the data, and co-wrote the manuscript; R.B.S. aided in study design interpreted the data, and contributed to the manuscript; D.L. added clinical input, aided in study design, and contributed to the manuscript; L.P. carried out the multivariable regression analysis and contributed to the manuscript; A.F. carried out the real-time PCR validation of CCR2 levels; W.H. oversaw the statistical analysis and contributed to the manuscript; D.H. carried out the microarray experiments; J.M.G. aided in design of the InCHIANTI study and contributed to the manuscript; S.B. oversees the InCHIANTI study and contributed to the manuscript; A.S. oversaw the microarray experiments; L.F. organized the sample cohort and contributed to the manuscript; D.M. managed the project, interpreted the data, co-wrote the manuscript, and contributed funding.
References
- 1.Deary I. Wright A. Harris S. Whalley L. Starr J. Searching for genetic influences on normal cognitive ageing. Trends Cogn Sci. 2004;8:178–184. doi: 10.1016/j.tics.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Bishop N. Lu T. Yankner B. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464:529–535. doi: 10.1038/nature08983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes TF. Ganguli M. Modifiable midlife risk factors for late-life cognitive impairment and dementia. Curr Psychiatry Rev. 2009;5:73–92. doi: 10.2174/157340009788167347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorelick PB. Role of inflammation in cognitive impairment: Results of observational epidemiological studies and clinical trials. Ann NY Acad Sci. 2010;1207:155–162. doi: 10.1111/j.1749-6632.2010.05726.x. [DOI] [PubMed] [Google Scholar]
- 5.Philipson O. Lord A. Gumucio A. O'Callaghan P. Lannfelt L. Nilsson LN. Animal models of amyloid-beta-related pathologies in Alzheimer's disease. FEBS J. 2010;277:1389–1409. doi: 10.1111/j.1742-4658.2010.07564.x. [DOI] [PubMed] [Google Scholar]
- 6.Gu L. Okada Y. Clinton SK. Gerard C. Sukhova GK. Libby P. Rollins BJ. Absence of monocyte chemoattractant protein-1 reduces atherosclerosis in low density lipoprotein receptor-deficient mice. Mol Cell. 1998;2:275–281. doi: 10.1016/s1097-2765(00)80139-2. [DOI] [PubMed] [Google Scholar]
- 7.El Khoury J. Toft M. Hickman SE. Means TK. Terada K. Geula C. Luster AD. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat Med. 2007;13:432–438. doi: 10.1038/nm1555. [DOI] [PubMed] [Google Scholar]
- 8.Villeda SA. Luo J. Mosher KI. Zou B. Britschgi M. Bieri G. Stan TM. Fainberg N. Ding Z. Eggel A. Lucin KM. Czirr E. Park JS. Couillard-Despres S. Aigner L. Li G. Peskind ER. Kaye JA. Quinn JF. Galasko DR. Xie XS. Rando TA. Wyss-Coray T. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature. 2011;477:90–94. doi: 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maes O. Xu S. Yu B. Chertkow H. Wang E. Schipper H. Transcriptional profiling of Alzheimer blood mononuclear cells by microarray. Neurobiol Aging. 2007;28:1795–1809. doi: 10.1016/j.neurobiolaging.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Grunblatt E. Bartl J. Zehetmayer S. Ringel T. Bauer P. Riederer P. Jacob C. Gene expression as peripheral biomarkers for sporadic Alzheimer's disease. J Alzheimers Dis. 2009;16:627–634. doi: 10.3233/JAD-2009-0996. [DOI] [PubMed] [Google Scholar]
- 11.de Jager CA. Budge MM. Clarke R. Utility of TICS-M for the assessment of cognitive function in older adults. Int J Geriatr Psychiatry. 2003;18:318–324. doi: 10.1002/gps.830. [DOI] [PubMed] [Google Scholar]
- 12.Terrera GM. Matthews F. Brayne C. A comparison of parametric models for the investigation of the shape of cognitive change in the older population. BMC Neurol. 2008;8:16. doi: 10.1186/1471-2377-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrucci L. Bandinelli S. Benvenuti E. Di Iorio A. Macchi C. Harris TB. Guralnik JM. Subsystems contributing to the decline in ability to walk: Bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc. 2000;48:1618. doi: 10.1111/j.1532-5415.2000.tb03873.x. [DOI] [PubMed] [Google Scholar]
- 14.Zeller T. Wild P. Szymczak S. Rotival M. Schillert A. Castagne R. Maouche S. Germain M. Lackner K. Rossmann H. Eleftheriadis M. Sinning CR. Schnabel RB. Lubos E. Mennerich D. Rust W. Perret C. Proust C. Nicaud V. Loscalzo J. Hubner N. Tregouet D. Munzel T. Ziegler A. Tiret L. Blankenberg S. Cambien F. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brazma A. Hingamp P. Quackenbush J. Sherlock G. Spellman P. Stoeckert C. Aach J. Ansorge W. Ball C. Causton H. Gaasterland T. Glenisson P. Holstege F. Kim I. Markowitz V. Matese J. Parkinson H. Robinson A. Sarkans U. Schulze-Kremer S. Stewart J. Taylor R. Vilo J. Vingron M. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell A. A meta-analysis of the accuracy of the mini-mental state examination in the detection of dementia and mild cognitive impairment. J Psychiatr Res. 2009;43:411–431. doi: 10.1016/j.jpsychires.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 17.Folstein M. Folstein S. McHugh P. Mini-mental state. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:203–209. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 18.Storey JD. Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A. Tamayo P. Mootha VK. Mukherjee S. Ebert BL. Gillette MA. Paulovich A. Pomeroy SL. Golub TR. Lander ES. Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ. Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta]CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Wood S, editor. Generalized Additive Models: An Introduction with R. CRC Texts in Statistical Science. Chapman and Hall/CRC; 2006. [Google Scholar]
- 22.Ihl R. Frolich L. Dierks T. Martin EM. Maurer K. Differential validity of psychometric tests in dementia of the Alzheimer type. Psychiatry Res. 1992;44:93–106. doi: 10.1016/0165-1781(92)90044-4. [DOI] [PubMed] [Google Scholar]
- 23.Matthews FE. Stephan BC. McKeith IG. Bond J. Brayne C. Two-year progression from mild cognitive impairment to dementia: To what extent do different definitions agree? J Am Geriatr Soc. 2008;56:1424–1433. doi: 10.1111/j.1532-5415.2008.01820.x. [DOI] [PubMed] [Google Scholar]
- 24.Wilson R. Leurgans S. Boyle P. Schneider J. Bennett D. Neurodegenerative basis of age-related cognitive decline. Neurology. 2010;75:1070–1078. doi: 10.1212/WNL.0b013e3181f39adc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramakers IH. Visser PJ. Aalten P. Bekers O. Sleegers K. van Broeckhoven CL. Jolles J. Verhey FR. The association between APOE genotype and memory dysfunction in subjects with mild cognitive impairment is related to age and Alzheimer pathology. Dement Geriatr Cogn Disord. 2008;26:101–108. doi: 10.1159/000144072. [DOI] [PubMed] [Google Scholar]
- 26.Terrando N. Monaco C. Ma D. Foxwell B. Feldmann M. Maze M. Tumor necrosis factor-a triggers a cytokine cascade yielding postoperative cognitive decline. Proc Natl Acad Sci USA. 2010;107:20518–20522. doi: 10.1073/pnas.1014557107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lue L. Kuo Y. Beach T. Walker D. Microglia activation and anti-inflammatory regulation in Alzheimer's disease. Mol Neurobiol. 2010;41:115–128. doi: 10.1007/s12035-010-8106-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenberg G. Inflammation and white matter damage in vascular cognitive impairment. Stroke. 2009;40:S20–S23. doi: 10.1161/STROKEAHA.108.533133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rezai-Zadeh K. Gate D. Szekely C. Town T. Can peripheral leukocytes by used as Alzheimer's disease biomarkers? Exp Rev Neurotherapeut. 2009;9:1623–1633. doi: 10.1586/ern.09.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cedazo-Minguez A. Winblad B. Biomarkers for Alzheimer's disease and other forms of dementia: Clinical needs, limitations and future aspects. Exp Gerontol. 2010;45:5–14. doi: 10.1016/j.exger.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Dawson TC. Kuziel WA. Osahar TA. Maeda N. Absence of CC chemokine receptor-2 reduces atherosclerosis in apolipoprotein E-deficient mice. Atherosclerosis. 1999;143:205–211. doi: 10.1016/s0021-9150(98)00318-9. [DOI] [PubMed] [Google Scholar]
- 32.Liao P. Yu L. Kuo C. Lin C. Kuo Y. Proteomics analysis of plasma for potential biomarkers in the diagnosis of Alzheimer's disease. Proteomics Clin Applic. 2007;1:506–512. doi: 10.1002/prca.200600684. [DOI] [PubMed] [Google Scholar]
- 33.Ray S. Britschgh M. Herbert C. Takeda-Uchimura Y. Boxer A. Blennow K. Friedman L. Galasko D. Jutel M. Karydas A. Kaye J. Leszek J. Miller B. Minthon L. Quinn J. Rabinovici G. Robinson W. Sabbagh M. So Y. Sparks D. Tabaton M. Tinklenberg J. Yesavage J. Tibshirani R. Wyss-Coray T. Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 34.Licastro F. Pedrini S. Caputo L. Annoni G. Davis L. Ferri C. Casadei V. Grimaldi L. Increased plasma levels of interleukin-1, interleukin-6 and a-1-antichymotrypsin in patients with Alzheimer's disease: Peripheral inflammation or signals from the brain? J Neuroimmunol. 2000;103:97–102. doi: 10.1016/s0165-5728(99)00226-x. [DOI] [PubMed] [Google Scholar]
- 35.Naert G. Rivest S. CC chemokine receptor 2 deficiency aggravates cognitive impairments and amyloid pathology in a transgenic mouse model of Alzheimer's disease. J Meurosci. 2011;31:6208–6220. doi: 10.1523/JNEUROSCI.0299-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishizuka K. Kimura T. Igata-yi R. Katsuragi S. Takamatsu J. Miyakawa T. Identification of monocyte chemoattractant protein-1 in senile plaques and reactive microglia of Alzheimer's disease. Psychiatry Clin Neurosci. 1997;51:135–138. doi: 10.1111/j.1440-1819.1997.tb02375.x. [DOI] [PubMed] [Google Scholar]
- 37.Bak T. A cognitive bedside assessment beyond the MMSE: The Addenbrooke's Cognitive Examination. Pract Neurol. 2007;7:245–249. [PubMed] [Google Scholar]
- 38.Franco-Marina F. Garcia-Gonzalez J. Wagner-Echeagaray F. Gallo J. Ugalde O. Sanchez-Garcia S. The Mini-mental State examination revisited: Ceiling and floor effects after score adjustment for educational level in an aging Mexican population. Int Psychogeriatr. 2010;22:72–81. doi: 10.1017/S1041610209990822. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.