Abstract
Cell proliferation, a main target in cancer therapy, is influenced by the surrounding three-dimensional (3D) extracellular matrix (ECM). In vitro drug screening is, thus, optimally performed under conditions in which cells are grown (embedded or trapped) in dense 3D matrices, as these most closely mimic the adhesive and mechanical properties of natural ECM. Measuring cell proliferation under these conditions is, however, technically more challenging compared with two-dimensional (2D) culture and other “3D culture conditions,” such as growth on top of a matrix (pseudo-3D) or in spongy scaffolds with large pore sizes. Consequently, such measurements are only slowly applied on a wider scale. To advance this, we report on the equal quality (dynamic range, background, linearity) of measuring the proliferation of cell layers embedded in dense 3D matrices (collagen, Matrigel) compared with cells in 2D culture using the easy (one-step) and in 2D well-validated, 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT)-assay. The comparison stresses the differences in proliferation kinetics and drug sensitivity of matrix-embedded cells versus 2D culture. Using the specific cell-layer-embedded 3D matrix setup, quantitative measurements of cell proliferation and cell invasion are shown to be possible in similar assay conditions, and cytostatic, cytotoxic, and anti-invasive drug effects can thus be reliably determined and compared in physiologically relevant settings. This approach in the 3D matrix holds promise for improving early-stage, high-throughput drug screening, targeting either highly invasive or highly proliferative subpopulations of cancers or both.
Introduction
Cell proliferation, survival, angiogenesis, and invasion form major targets in anti-cancer therapy. Potentially bioactive molecules are continuously being isolated from living matter or synthesized de novo in the search for new therapeutic cancer agents. Drugs or drug combinations that target multiple aspects of this malignant process are an ultimate goal in both first-line therapy and relapse prevention. The objective of gaining reliable insights into the multifaceted potential of drugs should preferably be incorporated in the design of early-stage screening steps. Cell proliferation, survival, and invasion should consequently be measured using similar conditions. Traditionally, in vitro screening for antiproliferative or toxic effects is performed on cells cultured on two-dimensional (2D) substrates.1–3 The predictability of clinical efficacy for hits identified in in vitro screening using 2D cultures is, however, generally low,4 and it is suggested that this could be increased by introducing a higher complexity in the early screening steps, that is, by using more physiological “in vitro three-dimensional (3D) culture” conditions.5 Indeed, cell-cell contacts and cell-extracellular matrix (ECM) interactions, and their associated signaling, have been implicated not only in the invasive process,6 but also in the cell proliferation.7–10 “In vitro 3D culture” is an increasingly fashionable term, but it is—confusingly—used for very diverse cell culture conditions displaying different levels of complexity. First, it is used for cells grown in a pseudo-3D culture, that is, on top of a natural matrix composed of ECM proteins.11–13 Second, the term 3D culture may indicate the cells grown on the curved 2D surface presented by porous scaffolds, usually characterized by a large surface area, high porosity, and a pore size (∼100 μm) that is typically much larger than the cell diameter and that enables effective fluid transport.14,15 Last, the 3D culture is used for cells grown fully encapsulated (embedded, trapped) within dense 3D matrices or hydrogels that have small pore sizes (as in natural ECM) and that can either be synthetic or biological in composition.11,16,17 In this article, we specifically focus on the measurements of cell proliferation of cancer cell lines embedded in dense 3D matrices, in particular those composed of collagen or Matrigel. These present a physiologically relevant topology as well as mechanical and adhesive properties reminiscent of a native ECM to cells.16,18 Moreover, given their density, collagen gels and Matrigel display a matrix fiber cross-linking degree and a pore size that is also found in tumor micro-environments and are still most frequently used for in vitro assaying of drug effects on cancer cell invasion.11,18,19 In addition, these dense 3D matrices composed of natural ECM proteins can be remodeled by the inherent ECM degradation properties of cells, a process essential for cell differentiation, growth, and invasion.
For cells grown on a 2D surface, diverse cell proliferation assays are available.20–25 These include the method using 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT), which is a second-generation water-soluble variant of the tetrazolium salt 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). It measures proliferation based on the metabolic activity of the cells and is frequently used in 2D culture conditions.26–31 Given the similar accessibility of cells grown in a pseudo-3D culture or in porous or spongy 3D scaffolds compared with that in 2D culture, the XTT-assay (and MTT-assay) has also already been successfully applied for cells grown on top of ECM gels31,32 or on the surface of porous ECM scaffolds.14,33–35 The use of XTT or other methods that measure the proliferation of cells embedded in dense 3D matrices is, however, still very limited and, in general, this approach is only slowly gaining application on a wider scale, especially for higher throughput analysis.36,37
The aim of this study was to provide a comprehensive report on the high-quality applicability of the XTT-assay as an easy, one-step procedure for measuring the proliferation of cells embedded as a layer in dense 3D matrices. In this way, we aim at facilitating the advance of cell proliferation measurements in physiologically more relevant environments. The results show that the XTT-method26 can be used with an equally high quality in dense 3D matrices (with different densities) as in a 2D culture. We further demonstrate that in vitro cell proliferation and antiproliferative effects induced by drugs are indeed different in a dense 3D matrix compared with a 2D culture. Moreover, in relation to the need of measuring the features of cancer cells in a multifaceted manner, we validate the application of the XTT-assay under a condition (including a relevant dense 3D matrix) that is also compatible with measuring the invasive capacity. Using selected drugs, we demonstrate that this may allow a more in-depth in vitro characterization of potential anti-cancer drugs.
Materials and Methods
Cell Lines
Human breast adenocarcinoma fibroblast-like MDA-MB-231 (ATCC: HTB-26) and human fibrosarcoma HT-1080 (ATCC: CCL-121) were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (FCS; Gibco), 20 mM L-GlutaMaxI (Gibco), and 1% penicillin-streptomycin (Gibco). Human mammary adenocarcinoma epithelial-like MCF-7 (ATCC: HTB-22) were cultivated in a 1:1 ratio of DMEM/F12 nutritional component HAM medium (HamF12; Gibco) supplemented with 10% heat-inactivated FCS (Gibco), 20 mM L-GlutaMaxI (Gibco), and 1% penicillin-streptomycin (Gibco). All the cells were maintained at 37°C in a humidified 5% CO2 atmosphere.
ECM Protein Handling: 3D Surface Coating and Preparation of 3D Matrices
Rat-tail collagen type I (acid-extracted, nonpepsin treated) and Matrigel were obtained from BD Biosciences. For obtaining a 2D coating of monomeric collagen, the wells of a 96-well plate (NUNC; Thermo Fisher Scientific) were incubated for 1 h at room temperature with 100 μL of 40 μg/mL monomeric collagen in calcium and magnesium containing Dulbecco's Phosphate-Buffered Saline (D-PBS; Gibco). The collagen solution for polymerization and 3D matrix preparation consisted of 2 mg/mL type I collagen, 1× MEM (Gibco), and 8.3 mM NaHCO3 in Hank's Balanced Salt Solution (Gibco), and its pH was adjusted to pH 7.4–8 with 1 M NaOH. Matrigel was diluted at 1/2 or 1/5 with normal growth medium to a final concentration of 5 mg/mL or 2 mg/mL (based on an approximate concentration provided by the manufacturer). The collagen solution and Matrigel dilution were kept on ice until further use.
Experimental Setups: 2D Culture and 3D Matrix-Embedded Cell Layer Culture
Cells were seeded in 96-well plates at a specified density. The cells in “2D culture” were seeded in normal growth medium on top of a monomeric collagen coating and maintained in the same medium. In the 3D setup, cells are trapped as a cell layer in the ECM. To obtain this “3D matrix-embedded cell layer” (Fig. 1A, middle), in the first step, the cells were seeded in normal growth medium on top of a thin layer of either polymerized collagen or Matrigel (polymerization ∼15 min at 37°C and 5% CO2) in a 96-well plate. After 2 h at 37°C and 5% CO2 allowing cell adhesion and spreading, the growth medium was removed, and the cells were overlaid with a second layer (40 μL) of the collagen solution or Matrigel dilution. After polymerization (30 min at 37°C and 5% CO2), 100 μL of the growth medium was added on top of the 3D collagen I gel or Matrigel.
Fig. 1.
XTT-based proliferation measurement in 2D and 3D matrix-embedded culture: optimization of cell number. (A) Schematic representation of experimental setups: 2D culture (left), 3D matrix-embedded cell layer (middle), and 3D matrix-embedded mixed culture (right). (B, C) XTT-based absorbance increases in function of cell number. The net absorbance (A450–620) is plotted; error bars represent SEM. (B) MDA-MB-231 and HT-1080 were seeded at cell densities from 5,000 cells to 50,000 cells in 2D culture (solid lines) or in a 3D environment, that is, as a cell layer in between two layers of collagen (dashed lines); n=2 (MDA-MB-231) or n=4 (HT-1080). Linear fitting on the data for cell densities ranging from 5,000 to 25,000 (dotted line). (C) MDA-MB-231 and HT-1080 cells seeded as a layer in Matrigel (at 5 mg/mL [solid line] or 2 mg/mL [dashed line]) at cell densities from 5,000 cells up to 50,000 cells/well; n=4. Linear fitting on the data corresponding to cell numbers ranging from 5,000 to 20,000 (dotted line). (D) Phase-contrast images of MDA-MB-231 cells seeded at 10,000 cells/well or 40,000 cells/well in a 2D culture (top) or a 3D matrix-embedded cell layer (bottom). Scale bar=200 μm. (E) MDA-MB-231 cells were seeded in a 2D culture at cell densities ranging from 5,000 to 100,000 cells and stained with DAPI. The cell nuclei were counted and plotted as cells/cm2 versus the number of seeded cells. 2D, two-dimensional; 3D, three-dimensional; XTT, 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide; SEM, standard error of the mean.
Cell Proliferation Assay in 2D and 3D
Cell Proliferation Kit II (XTT) (Roche) was used according to the manufacturer's protocol that was described for 2D cultures. In the 2D cultures, 50 μL of the XTT labeling mixture was added to the 100 μL of growth medium, whereas for the 3D matrix-embedded cells, 50 μL of the XTT labeling mixture was added to the 100 μL of growth medium present on top of the 40 μL matrix. The XTT labeling mixture was added in parallel samples at 0 h (t0) (subsequent to cell adhesion [2D] or polymerization of the matrix [3D]) and after a specified number of hours of cell growth (tx). A set of blanks were included for each condition. These do not contain cells and are treated identically as the normal samples. Absorbance for both 2D and 3D matrix-embedded cultures was measured at 3 h (or as specified) after XTT addition (see section on optimizing XTT incubation time in Results). Net absorbances (A450–A620) were calculated. All conditions were performed in multiple replicates as specified. Per condition, the mean net absorbance of all replicates at t0 and tx was background corrected (by subtracting the mean of the blanks) and designated as At0 and Atx, respectively. If not otherwise specified, the mean cell proliferation at tx was expressed as a fold increase versus t0 (Atx/At0).
Table 1 describes the step-wise protocol that measures cell proliferation in the 3D matrix-embedded cell layer culture.
Table 1.
Step-by-Step Protocol for XTT Assay on Cell Layers Embedded in 3D Matrix in 96-Well Plates
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | ECM solution | 40 μL/well | desired ECM concentration |
| 2 | Thin (bottom) layer of ECM | 100 μL | add to well and remove immediately |
| 3 | Incubation | 15 min | 37°C, 5% CO2 |
| 4 | Seed cells | 100 μL | 10,000 cells/100 μL |
| 5 | Incubation time (adhesion) | 2 h | 37°C, 5% CO2 |
| 6 | Second (top) layer of ECM | 40 μL/well | add ECM on top of cells |
| 7 | Incubation | 30 min | 37°C, 5% CO2 |
| 8 | Growth medium | 100 μL | add growth medium on top of ECM |
| 9 | Cell growth; drug incubation | 24–48 h | proliferation time |
| 10 | XTT labeling solution | 50 μL/well | detection of metabolic active cells |
| 11 | Incubation time | 3–4 h | 37°C, 5% CO2 |
| 12 | Assay readout | 450 and 620 nm | absorbance measurement using ELISA reader |
Step Notes
1. Prepare 1/3 extra of final volume needed, keep on ice.
2. This generates bottom ECM layer; recuperate ECM solution for use in step 6.
3. Polymerization of bottom ECM layer.
4. Cell density needs to be within determined linear range for each cell line used; useful test range 5,000–40,000 with 10,000 as recommended starting density; 2 or more replicates per condition; Include blank samples (no cells or cells killed using 0.2% triton-X-100) for backgrond correction.
5. Adhesion of cells; adhesion time dependent on cell type.
7. Polymerization of ECM.
8. Add drugs of interest to the growth medium.
9. Cell growth time and drug incubation time should exceed cell type- and experimental setup-dependent doubling time. Doubling time is determined in a separate experiment and is not identical in the 3D matrix compared with the 2D culture. As indication: in 3D matrix-embedded cell layer: 36 h for MDA-MB-231 and HT-1080.
10. XTT is reduced into a colored soluble formazan by metabolic active cells.
11. Incubation time may be varied depending on the cell number or cell type (as indication for adherent cells) (10,000–20,000 cells/well), preferably < 6 h to prevent saturation
12. Absorbance=A450–620nm−mean(Ablank).
10–12. Performed at t0 and at endpoint time in parallel samples.
2D, two-dimensional; 3D, three-dimensional; ECM, extracellular matrix.
Drug Treatment and Data Analysis
Cells, seeded at 10,000 (MDA-MB-231) or 5,000 (HT-1080) cells/well, were treated for 48 h (MDA-MB-231) or 44 h (HT-1080) (time points chosen to be larger than the doubling times of the respective cells, based on Figure 2) with y-27632 (Rho-associated protein kinase [ROCK] inhibitor) (Calbiochem, Merck KGaA), IPA 3 (group I p21-activated kinase [PAK] inhibitor) (Tocris Bioscience), or 5-fluoro-2′-deoxyuridine (dFUR) (Sigma-Aldrich). Overall, 1.33-fold (100.125-fold) serial dilutions were prepared for each drug to obtain dose-response curves leading to EC50 values. The starting concentration varied for each drug, that is, 400 μM y-27632, 120 μM IPA 3, and 90 μM 5-fluorouracil (5-FU). The inhibitors were administered at time t0 to the “2D cultures” by replacing the growth medium after cell adhesion with drug-containing medium. In 3D matrix-embedded cultures, the drugs were added at time t0 to the growth medium overlaying the matrix while taking into account the volume of the matrix to obtain the desired final drug concentration.
Fig. 2.
Cell proliferation kinetics differ in 2D culture and 3D matrix-embedded cell layer culture in a cell type-dependent manner; optimization of XTT reaction time. (A–D) Cell proliferation kinetics. Cell proliferation expressed as a fold increase versus t0 based on the net absorbance ratios Atx/At0 in a function of time. Error bars represent the SEM of two independent experiments; per experiment n=8 for all conditions; pair-wise comparison (post-hoc test) between different setups indicated by ***P<0.001; **P<0.01; *P<0.05. MDA-MB-231 at 10,000 cells/well (A, solid and dashed line) and HT-1080 at 5,000 cells/well (B, solid and dashed line) were seeded as a 2D culture (solid line) or a 3D matrix-embedded cell layer (2 mg/mL collagen I; dashed line) or both were seeded as a 3D matrix-embedded cell layer (2 mg/mL collagen I) at 40,000 cells/well (A and B, dotted line). (C, D) MDA-MB-231 cells (10,000/well) (C) and HT-1080 (5,000/well) (D) seeded in between two layers of Matrigel at 2 or 5 mg/mL. (E, F) Influence of XTT incubation time on mean absorbance (A450–620) in 2D culture (E) and in a 3D collagen-embedded cell layer setup (F) for MDA-MB-231 at a low (10,000 cells/well) and high (∼20,000 cells/well, i.e., after 40 h of growth) cell density. Absorbances were measured after 2, 3, 4, 6, and 24 h of XTT incubation. Background signals (based on blank samples) (dashed lines) and signals from the wells containing cells (solid lines) at a low (gray) and high (black) cell density are plotted.
The proliferation fold at tx versus t0 was first calculated as the ratio of the background subtracted net absorbances at time x versus time zero (Atx/At0). Consequently, Atx/At0=1 is indicative of a cytostatic effect, and Atx/At0<1 indicates cytotoxicity (data points in red in dose-response curves). Atx/At0 values were normalized by setting untreated samples to 100% viability and blank samples (no cells) to 0% viability (maximal toxicity). We verified for the two cell lines and for the 2D and 3D condition that these “maximal toxicity” samples gave identical absorbance as alternative “0% viability” samples in which the same number of cells are present as in the drug-treated samples, but in which the cells were killed by treatment with 0.2% Triton-X-100 (Sigma) before XTT-addition and measurement (100% dead cells). Indeed, the maximal deviation in absorbance A between these two “maximal toxicity” controls in all conditions was ΔAtx=0.021, whereas ΔAtx between the controls and the 100% viability samples was on average 0.885. Using normalized Atx/At0 data, four-parameter logistic or biphasic semi-log dose-response curves were generated using nonlinear regression to calculate EC50 values (GraphPad Prism). The EC50 values represent the concentration of the drug at which 50% of the effect is obtained.
Invasion Assay
MDA-MB-231 cells were seeded as a 3D matrix-embedded cell layer (collagen, 2 mg/mL) as just described. A cell-free area was, however, generated in the middle of the well, according to the ORIS cell invasion protocol (ORIS™; Platypus Technologies), and the cells were allowed to invade for 36 h in this cell-free central zone. Phase-contrast time-lapse movies were recorded for 36 h with an interval of 20 min.
Assay Quality
Signal-to-background (S/B), signal-to-noise (S/N), and Z-factors are determined as described earlier.38 The dynamic range of the assay in 2D culture or using the 3D matrix-embedded cell layer setup was derived from the comparison of the control and drug-treated cells. Untreated MDA-MB-231 cells were used as a positive (maximal) signal, and MDA-MB-231 treated with 90 μM 5-FU were used as a negative signal (minimal response for maximal inhibition). Overall, 20 replicates of each condition were included per experiment, and the experiment was performed on three different days. The mean positive and mean negative signal±SD were used to calculate the Z-factor.
Microscopy
Phase-contrast and fluorescent images were recorded using a 10× UPlanFL objective (N.A. 0.30) on a CellM system with a IX81 microscope (Olympus). For nuclei counting, MDA-MB-231 cells in the 96-well plate were fixed in 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 in D-PBS, blocked in 2% bovine serum albumin (BSA; Sigma-Aldrich)/1% Glycine in D-PBS, and stained with DAPI (Molecular Probes). The nuclei were manually counted using Cell Counter (ImageJ, http://rsbweb.nih.gov/ij).
Statistical Analysis
Statistical analysis of the time courses showing proliferation kinetics was carried out using two-way analysis of variance followed by a post-hoc test using Bonferroni adjustments (Graphpad Prism). The EC50 values were statistically compared using a Student's t-test according to the Graphpad Prism guidelines. P values less than 0.05 were considered as indicating statistically significant differences. Data are generally expressed as mean±standard error of the mean. In each experiment, multiple replicates per condition are performed (2 ≤ n ≤ 8), and one (of two) representative experiment is shown.
Results and Discussion
Assay Conditions and Optimization for Measuring Cell Proliferation in 3D Versus 2D Cell Layers; Performance Comparison
We applied the one-step XTT-assay for quantifying cell numbers, cell viability, and cell proliferation in cells grown in a 3D setup, that is, those embedded in a dense matrix (collagen type I or Matrigel). We detail the performance of the XTT-assay in these dense 3D matrices compared with its classical use in cells grown on a 2D surface (“2D culture” on monomeric collagen coating) (Fig. 1A, left). In the 3D setup used, the cells are embedded as a cell layer in the plane between two collagen layers (3D matrix-embedded cell layer) (Fig. 1A, middle; for details see Materials and Methods), which is distinct from “3D matrix-embedded mixed cultures” in which the cells are dispersed as single cells within the matrix (Fig. 1A, right). We specifically address proliferation in the “3D matrix-embedded cell layer” format, because this setup is highly compatible with measuring the invasive capacity of cells using a 3D woundhealing-like assay in which the cells are allowed to invade a central cell-free area (ORIS; Platypus Technologies). This assay is gaining interest for high-throughput screening of 2D migration39–41 and 3D invasion.42,43 We ultimately show that measuring the effects of compounds on invasion and proliferation under similar 3D conditions forms an asset in the in vitro characterization and evaluation of cancer targeting drugs.
To demonstrate the quality of employing the XTT-assay in dense 3D matrices, we addressed different issues that are expected to vary with cell type and assay format (3D vs. 2D). These include (i) cell starting numbers (ensuring a linear response as well as an appropriate signal over background), (ii) optimal XTT incubation time (preventing signal saturation), and (iii) proliferation rates and/or doubling times (required before screening antiproliferative compounds in a specific format).
To determine the linear range for the different 2D and 3D setups, the cancer cell lines MDA-MB-231 or HT-1080 were seeded and allowed to settle for 2 h before the addition of XTT at a wide range of densities (5,000 to 50,000 cells/well) in 96 wells (Fig. 1B, C). For the 2D culture and for the cell layer embedded in 3D collagen type I matrix, the net absorbance signal initially increases linearly with cell number until ∼20,000–25,000 cells/well, and, subsequently, levels off toward a maximum (Fig. 1B). We also extended the assay to a different matrix, that is, Matrigel, representing a reconstituted laminin-rich basement membrane. The cell number analysis of MDA-MB-231 and HT-1080, embedded as a layer in Matrigel at a concentration of 2 (Fig. 1C, dashed line) and 5 mg/mL (solid line), is similar as in the 3D collagen matrix with a linear increase in absorbance for the range of 5,000–20,000 cells/well. Figure 1D and E documents the basis of the deviation from linearity. At cell densities higher than 30,000 cells/well, confluency is reached in the 2D- and 3D layer-based setups (Fig. 1D). For the tested adherent cell types, the limited surface available for cell adhesion determines the linear range, as supported by the fact that the DAPI-based cell count was only linearly correlated to the seeded cell numbers below ∼40,000 cells/well (Fig. 1E).
Together, this indicates that the useful range of cell numbers for adherent cells is similar for the 3D matrix cell layer assay and in the 2D culture and is limited by confluent growth and well surface area. Consequently, this range is lower than that for suspension cells (Figure 3 in Company Application Note, www.roche-applied-science.com).
Fig. 3.
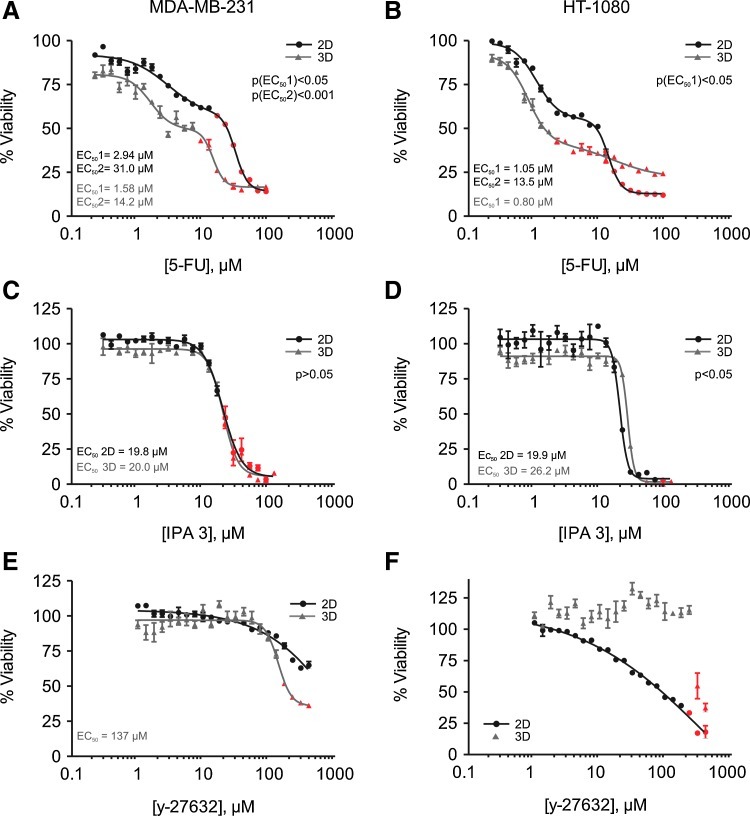
Dose-dependent inhibition of cell proliferation on drug treatments in 3D versus 2D conditions. (A–F) MDA-MB-231 and HT-1080 cells were seeded at a low cell density (10,000 and 5,000 cells/well, respectively) in 2D cultures (black lines, circles) and 3D matrix-embedded cell layers (gray lines, triangles) and were left untreated (control) or drug treated. Cell proliferation was measured after 48 h (MDA-MB-231) or 44 h (HT-1080), and the fold increase versus t0 (Atx/At0) of the control (untreated) samples was set to 100% viability. Symbols in red indicate the % viability corresponding with Atx/At0<1, that is, indicating toxicity (see also Materials and Methods). The error bars represent SEM; n=4 for each dilution; experiments were repeated twice, and one representative experiment is shown. (A, B) Biphasic semilog dose-response curves of MDA-MB-231 (A) and HT-1080 (B) cells treated with a 1.33 serial dilution of 5-FU using a starting concentration of 90 μM. (C, D) Four-parameter semi-log dose-response curves of MDA-MB-231 (C) or HT-1080 (D) cells treated with a 1.33 serial dilution of IPA 3 using a starting concentration of 120 μM. (E, F) Four-parameter semi-log dose-response curves of MDA-MB-231 (E) and HT-1080 (F) cells treated with a 1.33 serial dilution of y-27632 using a starting concentration of 400 μM. 5-FU, 5-Fluorouracil.
Based on the experiments with MDA-MB-231 shown in Figure 1B and C, we evaluated whether higher background values—as might be expected in a dense 3D matrix—influence the quality of the measurements. Since background effects can only be judged correctly by considering the variation in response,38 we also calculated, in addition to S/B, the parameters that take into account the variation on background (S/N) or on both background and signal (as in a Z-factor). Note that these calculations only provide details on signal to background levels; see next for an evaluation of the true dynamic range using minimal and maximal signals after background correction. Despite the fact that the S/B-values in 3D conditions are generally smaller than in 2D conditions (Table 2), the low variation in the mean absorbances results in sufficiently high S/N values (and Z-values ≥0.5) for nearly all the 3D conditions. This indicates that, using XTT, the background in the 3D matrix-embedded setup is not lowering the assay quality and that this quality in the 3D matrix-embedded setup is comparable to that in the 2D culture. If needed, cell numbers can be increased to optimize S/N (compare values for 10,000 and 20,000 cells in Matrigel; Table 2).
Table 2.
Evaluation of Signal to Background Ratio's in 2D and 3D Setups
| Setup | No. of cells (×103) | S/Ba | S/Nb | Zc |
|---|---|---|---|---|
| 2D culture | 10 | 2.72 | 25.48 | 0.68 |
| 20 | 4.40 | 50.45 | 0.86 | |
| 3D (cell layer embedded in collagen matrix) | 10 | 1.87 | 76.47 | 0.63 |
| 20 | 3.50 | 220.97 | 0.48 | |
| 3D (cell layer embedded in Matrigel [5 mg/mL]) | 10 | 1.75 | 34.29 | 0.68 |
| 20 | 2.60 | 72.90 | 0.78 | |
| 3D (cell layer embedded in Matrigel [2 mg/mL]) | 10 | 1.87 | 24.45 | 0.49 |
| 20 | 2.66 | 46.71 | 0.78 |
S/B=μs/μb, where μs is mean signal (450–620 nm) and μb is mean background (450–620 nm).
S/N=(μs−μb)/SDb, where SDb is standard deviation of μb.
Z=1−(3×[SDs+SDb])/|μs-μb|, where SDb and SDs are standard deviation of μb and μs.
S/B, signal-to-background; S/N, signal-to-noise.
Subsequently, we measured cell proliferation after 12, 24, 36, and 48 h for MDA-MB-231 (Fig. 2A, C) and HT-1080 (Fig. 2B, D) to compare cell-specific doubling times in the different setups (2D and 3D) and matrices (collagen and Matrigel). This is important to select assay times for drug screening. Proliferation kinetics of both MDA-MB-231 and HT-1080 (10,000 and 5,000 cells/well seeded at t0, respectively) are significantly slower in the 3D collagen-embedded layer setup (dashed line) versus the 2D culture (solid line) (Fig. 2A, B). Doubling (i.e., a proliferation fold vs. t0 equal to two) is reached at least 12 h later in the 3D matrix-embedded layers than in the 2D culture. As expected, based on the confluency shown in Figure 1D, MDA-MB-231 and HT-1080 that are seeded at 40,000 cells/well in the 3D collagen-embedded cell layer no longer showed proliferation due to contact inhibition (Fig. 2A, B, dotted lined). Proliferation kinetics of MDA-MB-231 and HT-1080 in 5 mg/mL Matrigel are comparable with those in 3D collagen embedded cultures, yielding a fold change of 2 after 36 h (Fig. 2C, D, solid lines). Similar proliferation kinetics for MDA-MB-231 are observed in 2 mg/mL Matrigel (Fig. 2C, dashed line), whereas HT-1080 cells show faster proliferation kinetics in less dense Matrigel (Fig. 2D, dashed line). Similar to reports using cells in 3D pseudo-culture, reports on the expansion of stem cells, and in the field of regenerative tissue research,44–47 these data agree with previous works that cell-ECM contact affects cell proliferation and does so in a cell type-dependent manner.
In addition to the assay time, we compared the effect of varying the time of the XTT reaction (XTT incubation times: 2–24 h) in 3D versus 2D conditions. Figure 2E and F shows for MDA-MB-231 the net absorbance (A450–620) in function of XTT incubation time for background samples and for samples with low and high cell numbers. Background values (dashed lines) are only slightly affected by longer XTT incubation time, and these are similar in 2D and 3D conditions. At high cell numbers (Fig. 2E, F, black), saturation of the signal is already reached at ∼6 h XTT incubation. For low cell numbers (Fig. 2E, F, gray), the increase is at least linear up to 6 h of the XTT reaction, but after 24 h of XTT incubation, the signal has no longer increased in a linear fashion. These analyses indicate that an XTT incubation time of 3 to 4 h is optimal to yield a linear response with cell number, and differences are hardly observed for 2D and 3D conditions. A longer (6–24 h) XTT incubation time may only be used for low cell numbers (or for cell types with apparent low metabolic activity; see Figure 3 in Company Application Note, www.roche-applied-science.com).
In summary, we demonstrate the equal applicability of the XTT-assay for measuring cell growth in a 3D matrix-embedded cell layer, as compared with 2D cultured cells. Measurements are possible in the two most commonly used dense 3D matrices (collagen gel [2 mg/mL] and Matrigel [2 and 5 mg/mL]). The background of 3D samples does not affect the quality of the measurements. However, a different optimization is required for different culture conditions (2D, 3D, matrix type, and cell type), especially with regard to signal linearity versus cell number and cell doubling time. Significantly, the differences observed in cell proliferation kinetics in 2D cultures versus 3D matrix-embedded cultures indicate the potential benefit of measuring cell growth in 3D matrices.
Measuring Compound Effects in 3D Versus 2D Conditions
We further validated proliferation measurements in the 3D setup by evaluating the effect of compound treatment on cell growth and viability and by comparing them with the effects on cells grown in 2D. Proliferation folds (Atx/At0) are plotted in semi-log dose response curves as “% cell viability” versus drug concentration (Fig. 3;, see Materials and Methods for details). This approach allows, in principle, the determination of EC50 values (defined as the concentration of the drug at which 50% of the effect is obtained), indicative of an antiproliferative (cytostatic) or cytotoxic effect or, where applicable, for both effects individually.
We first tested the effect of dFUR, which is metabolized into its active form, 5-FU, on MDA-MB-231 and HT-1080 (10,000 and 5,000 cells seeded per well, respectively). This compound is widely used in anti-cancer chemotherapy.48,49 We observe biphasic dose-dependent inhibition of cell growth for MDA-MB-231 (Fig. 3A) and HT-1080 (Fig. 3B), in both 2D culture (black line, circles) and 3D matrix (gray line, triangles). The first phase of the curve shows a dose-dependent antiproliferative effect. In the second phase, the ratios of measured cell number versus cell number at the start (∼Atx/At0) are smaller than 1 (presented in figure by red symbols), indicating dose-dependent toxicity. This second phase is more gradual for HT-1080 in 3D. We derive two EC50 values characterizing each dose-response curve (except for the second phase for HT-1080 in 3D). For MDA-MB-231, both EC501 and EC502 are significantly smaller for cells in the 3D matrix than those in the 2D culture (P<0.05, factor 2, Fig. 3A); in addition, for HT-1080, the EC501 value is lower in the 3D matrix than in the 2D culture (P<0.05, Fig. 3B). In addition, the EC501 values indicative of a cytostatic effect are in the high nM-low μM concentration range, whereas obvious cytotoxic effects (∼EC502) require a drug concentration that is approximately 10-fold higher.
These results demonstrate a different (i.e., two-fold higher) sensitivity to the cytostatic activity of 5-FU in the 3D collagen-embedded cell layer setup versus the culture on a 2D monomeric collagen coating in the tested cell lines. This underlines the potential benefit of scoring drug effects on cell proliferation measurements within a 3D matrix.
We used the effect of 5-FU to further investigate the dynamic range, reproducibility, and overall quality of the assay in 3D versus 2D conditions. We performed three measurements on different days to compare the effect on the 2D culture and the 3D matrix-embedded-layer setup on untreated MDA-MB-231 (maximal signal, no inhibition) and on MDA-MB-231 treated with 90 μM 5-FU (used as minimal signal, maximal inhibition). The Z-factors, derived as described earlier,38 for both the 2D culture and the 3D matrix-embedded cell layer setup (0.928 and 0.819, respectively, Table 3) are well above the critical Z-value of 0.5, demonstrating that, in the 3D matrix-embedded cell layer approach, the assay quality is high and the dynamic range is comparable to that in the 2D culture.
Table 3.
Dynamic Range of Proliferation Assay in 3D Matrix Versus 2D Culture
| Untreated (maximal signal) | 5-FU treated (minimal signal) | |
|---|---|---|
| 2D | ||
| Meana | 1.350 | 0.199 |
| SD | 0.023 | 0.005 |
| Zb | 0.928 | |
| 3D | ||
| Meana | 0.593 | 0.133 |
| SD | 0.021 | 0.007 |
| Zb | 0.819 | |
Mean of three experiments performed on three different days (n=20 per condition in each experiment)
Z=1−(3×[SDmax+SDmin])/|μmax−μmin|, where μmax is mean maximal signal (450–620 nm), μmin is mean minimal signal (450–620 nm), μmax, μmin: background corrected, SDmax is SD maximal signal, and SDmin is SD minimal signal.
5-FU, 5-Fluorouracil.
As mentioned previously, the 3D matrix-embedded cell layer setup is highly suitable for testing the invasive capacity of cells. Anti-invasive drugs may or may not affect cell proliferation, or may display identical or different dose dependency for anti-invasion or antiproliferation. The application of the XTT-assay in this experimental 3D setup described here that is also suited for invasion analysis allows the testing of these multiple aspects. As an illustration, we evaluated the effect on the proliferation of two compounds known to inhibit key molecules in the signals driving amoeboid or mesenchymal cell migration and invasion,50 namely inhibitors against ROCK or against PAK.
Compound IPA 3 targets the auto-regulatory mechanism of group I PAKs that are possible targets in cancer therapy as a consequence of their role in cell proliferation, morphogenic processes, and cell motility.51–53 A concentration range of IPA 3 (up to 120 μM) was administered to MDA-MB-231 and HT-1080 cells, in 2D culture, or to the cell layer embedded in 3D collagen. For MDA-MB-231, this results in an inhibition of cell viability with very similar dose responses for 2D and 3D (EC50 values ∼20 μM) (Fig. 3C). At EC50 or higher doses, the cell numbers after drug treatment are lower than those initially seeded (Atx/At0<1), indicating toxicity. Although the EC50 values for HT-1080 cells are in the same range as for MDA-MB-231, HT-1080 are slightly less sensitive to IPA 3 in a 3D matrix-embedded cell layer (EC50=26.2 μM) compared with 2D (EC50=19.9 μM) (P<0.05, Fig. 3D). In addition, compared with MDA-MB-231, the effect is cytostatic rather than cytotoxic in the tested concentration range (Fig. 3C, D). In comparison, for these cells and using the same experimental 3D setup, this compound typically already displays a significant inhibition (40%) of invasion of the matrix embedded cell layer in a central cell-free zone at lower concentrations (5 μM) (Huyck et al., data not shown).
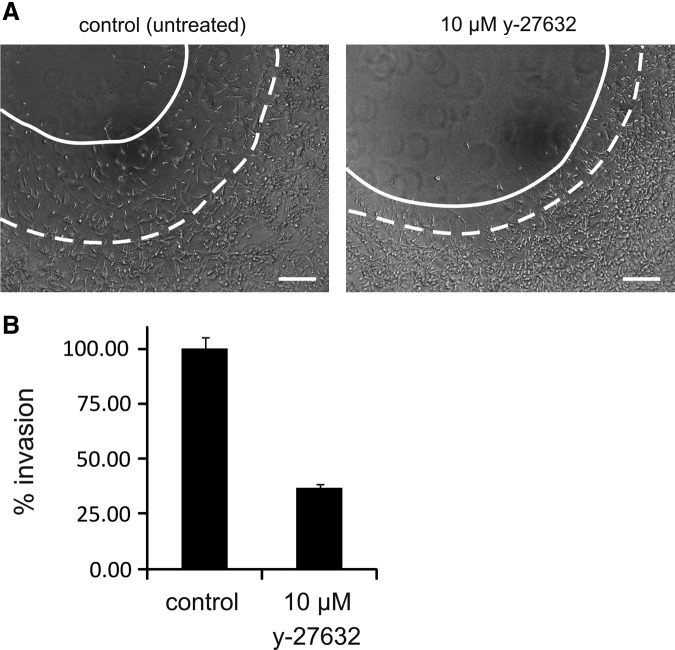
As shown in Figure 4, 10 μM of the ROCK inhibiting compound y-27632 yielded in the 3D matrix-embedded cell layer format more than 50% invasion inhibition; a similar inhibition is obtained for HT-1080 cells (not shown). However, at this drug concentration, cell growth—measured in the same experimental 3D setup—is completely unaffected (Fig. 3E, F). Tenfold higher concentrations of this drug are required to induce inhibition of MDA-MB-231 growth and toxicity in the 3D matrix as demonstrated by an EC50 of 137 μM (Fig. 3E). For HT-1080 cells in the 3D matrix-embedded setup, a drug effect (toxicity) is even not observed until a concentration of 300 μM or higher is achieved (Fig. 3F). In contrast to the low response in the 3D setting, proliferation in the 2D culture of MDA-MB-231 and, especially, of HT-1080 is more sensitive to ROCK inhibition: a gradual dose-dependent inhibition is observed, although even at 400 μM (the highest concentration tested), a maximal effect was not yet reached (Fig. 3E, F). Rho and ROCK have generally been linked to proliferation and cell survival in diverse cell lines, a.o. based on their role in integrin-based adhesion complex formation and in their control of cyclin and cyclin-dependent kinase expression.54–56 The ECM is known to provide cells with both chemical and mechanical cues a.o. via FAK-Rho-Erk signaling and Rho-ROCK-actomyosin contractility,57 and these cues are substantially different in 2D versus 3D matrix.58 Given the difference in sensitivity of HT-1080 proliferation to ROCK inhibition in 2D versus 3D, it will be of interest to determine whether the extent of adhesion and of ROCK-dependent activation of key proliferation factors differs in the 2D versus 3D conditions. This example of drug effect further documents the different responses when drug-treated cells are cultured in 2D or 3D conditions and, in addition, the importance of testing different cellular properties (in casu invasion and proliferation) in the same experimental setup. Based on the analyses in the 3D matrix, y-27632 can be viewed as a drug that acts purely as an anti-invasive agent in MDA-MB-231 and HT-1080 at low doses (lower μM range) and only becomes toxic to the cells at, at least, 10-fold higher concentrations.
Fig. 4.
3D matrix-embedded cell layer invasion is inhibited at low doses of y-27632. MDA-MB-231 cells were seeded as a 3D matrix-embedded cell layer (2 mg/mL collagen) with a central cell-free area (according to ORIS™ cell invasion assay; Platypus Technologies) and were allowed to invade for 36 h. (A) Phase-contrast images of MDA-MB-231 after 36 h invasion. MDA-MB-231 were left untreated (left) or treated with 10 μM y-27632 (right). The dashed line indicates the edge of the peripheral cell layer at start (time 0); the solid line indicates the edge of the cell layer after 36 h of invasion. Scale bar=200 μm. (B) Invasion efficiency for untreated and y-27632 treated cells. The change in the area of the cell layer was measured at different time points, and the velocity of the area increase was calculated. The increase over time in the area of the untreated cells was set to 100% invasion. The error bars represent SEM; n=6.
Conclusions
This article, in which we employed a 3D setup in which a cell layer is fully matrix embedded, establishes that proliferation measurements in dense 3D matrices using the one-step XTT proliferation assay are as easy as and qualitatively equal to current approaches in 2D culture (and, by extrapolation, in porous 3D scaffolds). As such, this article will accelerate wider application of this 3D approach, which is strongly suggested as yielding more physiologically relevant answers. Using a quality comparison of measurements conducted on cells embedded in a dense 3D matrix versus cells in 2D, we show a similar dynamic range and no negative effects of background. The assay allows the measuring of cell-type specific proliferation kinetics and antiproliferative, cytostatic, and cytotoxic effects of compounds in different dense 3D matrices (collagen, Matrigel). We show that cell growth and drug sensitivity in a 3D matrix are not necessarily identical as in a 2D culture, and these differences do not follow a predictable pattern but are cell type- and compound-dependent. Although the higher predictability of results obtained in 3D matrices still needs confirmation in vivo, the observed differences already suggest a possible benefit of (also) screening in more complex in vitro conditions. Moreover, by validating proliferation measurements in the 3D matrix-embedded cell layer setup that is compatible with invasion measurements, the drug effects on proliferation and invasion can be more reliably compared. This results in a more optimal in vitro drug characterization. This validated protocol in the 3D matrix, thus, holds promise for improving early-stage, high-throughput drug screening for targeting—by individual drugs or by combinations—both highly invasive and highly proliferative subpopulations of cancers. The validation of the assay is established here in collagen and Matrigel, but may also prove useful in microporous synthetic ECM mimics or nanofibrous scaffolds with bio-inspired cell-instructive and degradable properties that are being developed.16,59
Abbreviations
- 2D
two-dimensional
- 3D
three-dimensional
- 5-FU
5-Fluorouracil
- BSA
bovine serum albumin
- dFUR
5-Fluoro-2′-deoxyuridine
- DMEM
Dulbecco's modified Eagle's medium
- D-PBS
Dulbecco's Phosphate-Buffered Saline
- ECM
extracellular matrix
- FCS
fetal calf serum
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PAK
p21-activated kinase
- ROCK
Rho-associated protein kinase
- S/B
signal-to-background
- SEM
standard error of the mean
- S/N
signal-to-noise
- XTT
2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide
Acknowledgments
This work was supported by fund G.0441.10N from the FWO Vlaanderen and fund 01601207 from the GOA (Geconcerteerde Onderzoeksactie) granted to C.A. L.H. was funded by the BOF (Bijzonder Onderzoeksfonds) 01J04806.
Disclosure Statement
The authors declare that no competing financial interests exist.
References
- 1.Shum D. Radu C. Kim E, et al. A high density assay format for the detection of novel cytotoxic agents in large chemical libraries. J Enzyme Inhib Med Chem. 2008;23:931–945. doi: 10.1080/14756360701810082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomelleri C. Dalla Pellegrina C. Chignola R. Microplate spectrophotometry for high-throughput screening of cytotoxic molecules. Cell Prolif. 2010;43:130–138. doi: 10.1111/j.1365-2184.2009.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slater K. Cytotoxicity tests for high-throughput drug discovery. Curr Opin Biotechnol. 2001;12:70–74. doi: 10.1016/s0958-1669(00)00177-4. [DOI] [PubMed] [Google Scholar]
- 4.Carnero A. High throughput screening in drug discovery. Clin Transl Oncol. 2006;8:482–490. doi: 10.1007/s12094-006-0048-2. [DOI] [PubMed] [Google Scholar]
- 5.Justice BA. Badr NA. Felder RA. 3D cell culture opens new dimensions in cell-based assays. Drug Discov Today. 2009;14:102–107. doi: 10.1016/j.drudis.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Pathak A. Kumar S. Biophysical regulation of tumor cell invasion: moving beyond matrix stiffness. Integr Biol (Camb) 2011;3:267–278. doi: 10.1039/c0ib00095g. [DOI] [PubMed] [Google Scholar]
- 7.Bissell MJ. Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbott A. Cell culture: biology's new dimension. Nature. 2003;424:870–872. doi: 10.1038/424870a. [DOI] [PubMed] [Google Scholar]
- 9.Hodkinson PS. Elliott T. Wong WS, et al. ECM overrides DNA damage-induced cell cycle arrest and apoptosis in small-cell lung cancer cells through beta1 integrin-dependent activation of PI3-kinase. Cell Death Differ. 2006;13:1776–1788. doi: 10.1038/sj.cdd.4401849. [DOI] [PubMed] [Google Scholar]
- 10.Yamada KM. Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Lee GY. Kenny PA. Lee EH. Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhiman HK. Ray AR. Panda AK. Three-dimensional chitosan scaffold-based MCF-7 cell culture for the determination of the cytotoxicity of tamoxifen. Biomaterials. 2005;26:979–986. doi: 10.1016/j.biomaterials.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Yoshii Y. Waki A. Yoshida K, et al. The use of nanoimprinted scaffolds as 3D culture models to facilitate spontaneous tumor cell migration and well-regulated spheroid formation. Biomaterials. 2011;32:6052–6058. doi: 10.1016/j.biomaterials.2011.04.076. [DOI] [PubMed] [Google Scholar]
- 14.Shen YH. Shoichet MS. Radisic M. Vascular endothelial growth factor immobilized in collagen scaffold promotes penetration and proliferation of endothelial cells. Acta Biomater. 2008;4:477–489. doi: 10.1016/j.actbio.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Maltman DJ. Przyborski SA. Developments in three-dimensional cell culture technology aimed at improving the accuracy of in vitro analyses. Biochem Soc Trans. 2010;38:1072–1075. doi: 10.1042/BST0381072. [DOI] [PubMed] [Google Scholar]
- 16.Tibbitt MW. Anseth KS. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng. 2009;103:655–663. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prestwich GD. Evaluating drug efficacy and toxicology in three dimensions: using synthetic extracellular matrices in drug discovery. Acc Chem Res. 2008;41:139–148. doi: 10.1021/ar7000827. [DOI] [PubMed] [Google Scholar]
- 18.Cukierman E. Bassi DE. Physico-mechanical aspects of extracellular matrix influences on tumorigenic behaviors. Semin Cancer Biol. 2010;20:139–145. doi: 10.1016/j.semcancer.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolf K. Alexander S. Schacht V, et al. Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 21.Blaheta RA. Kronenberger B. Woitaschek D, et al. Development of an ultrasensitive in vitro assay to monitor growth of primary cell cultures with reduced mitotic activity. J Immunol Methods. 1998;211:159–169. doi: 10.1016/s0022-1759(97)00202-0. [DOI] [PubMed] [Google Scholar]
- 22.Jones LJ. Gray M. Yue ST. Haugland RP. Singer VL. Sensitive determination of cell number using the CyQUANT cell proliferation assay. J Immunol Methods. 2001;254:85–98. doi: 10.1016/s0022-1759(01)00404-5. [DOI] [PubMed] [Google Scholar]
- 23.Sekhon BK. Roubin RH. Tan A. Chan WK. Sze DM. High-throughput screening platform for anticancer therapeutic drug cytotoxicity. Assay Drug Dev Technol. 2008;6:711–721. doi: 10.1089/adt.2008.148. [DOI] [PubMed] [Google Scholar]
- 24.Quent VM. Loessner D. Friis T. Reichert JC. Hutmacher DW. Discrepancies between metabolic activity and DNA content as tool to assess cell proliferation in cancer research. J Cell Mol Med. 2010;14:1003–1013. doi: 10.1111/j.1582-4934.2010.01013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nociari MM. Shalev A. Benias P. Russo C. A novel one-step, highly sensitive fluorometric assay to evaluate cell-mediated cytotoxicity. J Immunol Methods. 1998;213:157–167. doi: 10.1016/s0022-1759(98)00028-3. [DOI] [PubMed] [Google Scholar]
- 26.Scudiero DA. Shoemaker RH. Paull KD, et al. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 1988;48:4827–4833. [PubMed] [Google Scholar]
- 27.Huertas D. Soler M. Moreto J, et al. Antitumor activity of a small-molecule inhibitor of the histone kinase Haspin. Oncogene. 2012;31:1408–1418. doi: 10.1038/onc.2011.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jurado R. Lopez-Flores A. Alvarez A. Garcia-Lopez P. Cisplatin cytotoxicity is increased by mifepristone in cervical carcinoma: an in vitro and in vivo study. Oncol Rep. 2009;22:1237–1245. doi: 10.3892/or_00000560. [DOI] [PubMed] [Google Scholar]
- 29.Mad'arova J. Lukesova M. Hlobilkova A, et al. Synthetic inhibitors of CDKs induce different responses in androgen sensitive and androgen insensitive prostatic cancer cell lines. Mol Pathol. 2002;55:227–234. doi: 10.1136/mp.55.4.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishimura M. Abiko Y. Mitamura J. Kaku T. Effect of cell plating density and extracellular matrix protein on cell growth of epithelial rests of Malassez in vitro. Med Electron Microsc. 1999;32:127–132. doi: 10.1007/s007950050019. [DOI] [PubMed] [Google Scholar]
- 31.Ohbayashi M. Yasuda M. Kawakami I. Kohyama N. Kobayashi Y. Yamamoto T. Effect of interleukins response to ECM-induced acquisition of drug resistance in MCF-7 cells. Exp Oncol. 2008;30:276–282. [PubMed] [Google Scholar]
- 32.Ranta V. Mikkola T. Ylikorkala O. Viinikka L. Orpana A. Reduced viability of human vascular endothelial cells cultured on Matrigel. J Cell Physiol. 1998;176:92–98. doi: 10.1002/(SICI)1097-4652(199807)176:1<92::AID-JCP11>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 33.Miranda SC. Silva GA. Hell RC. Martins MD. Alves JB. Goes AM. Three-dimensional culture of rat BMMSCs in a porous chitosan-gelatin scaffold: A promising association for bone tissue engineering in oral reconstruction. Arch Oral Biol. 2011;56:1–15. doi: 10.1016/j.archoralbio.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 34.Tang ZP. Liu N. Li ZW, et al. In vitro evaluation of the compatibility of a novel collagen-heparan sulfate biological scaffold with olfactory ensheathing cells. Chin Med J (Engl) 2010;123:1299–1304. [PubMed] [Google Scholar]
- 35.Chiu LL. Weisel RD. Li RK. Radisic M. Defining conditions for covalent immobilization of angiogenic growth factors onto scaffolds for tissue engineering. J Tissue Eng Regen Med. 2011;5:69–84. doi: 10.1002/term.292. [DOI] [PubMed] [Google Scholar]
- 36.Zhang F. Xu R. Zhao MJ. QSG-7701 human hepatocytes form polarized acini in three-dimensional culture. J Cell Biochem. 2010;110:1175–1186. doi: 10.1002/jcb.22632. [DOI] [PubMed] [Google Scholar]
- 37.Oh SA. Lee HY. Lee JH, et al. Collagen three-dimensional hydrogel matrix carrying basic fibroblast growth factor for the cultivation of mesenchymal stem cells, osteogenic differentiation. Tissue Eng Part A. 2012;18:1087–1100. doi: 10.1089/ten.TEA.2011.0360. [DOI] [PubMed] [Google Scholar]
- 38.Zhang JH. Chung TD. Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 39.Gough W. Hulkower KI. Lynch R, et al. A quantitative, facile, and high-throughput image-based cell migration method is a robust alternative to the scratch assay. J Biomol Screen. 2011;16:155–163. doi: 10.1177/1087057110393340. [DOI] [PubMed] [Google Scholar]
- 40.Nizamutdinova IT. Kim YM. Chung JI, et al. Anthocyanins from black soybean seed coats stimulate wound healing in fibroblasts and keratinocytes and prevent inflammation in endothelial cells. Food Chem Toxicol. 2009;47:2806–2812. doi: 10.1016/j.fct.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 41.Hulkower RL. Casanova LM. Rutala WA. Weber DJ. Sobsey MD. Inactivation of surrogate coronaviruses on hard surfaces by health care germicides. Am J Infect Control. 2011;39:401–407. doi: 10.1016/j.ajic.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim SO. Kim H. Jung G. p53 inhibits tumor cell invasion via the degradation of snail protein in hepatocellular carcinoma. FEBS Lett. 2010;584:2231–2236. doi: 10.1016/j.febslet.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Attoub S. Hassan AH. Vanhoecke B, et al. Inhibition of cell survival, invasion, tumor growth and histone deacetylase activity by the dietary flavonoid luteolin in human epithelioid cancer cells. Eur J Pharmacol. 2011;651:18–25. doi: 10.1016/j.ejphar.2010.10.063. [DOI] [PubMed] [Google Scholar]
- 44.Weaver VM. Petersen OW. Wang F, et al. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexa A. Varga J. Remenyi A. Scaffolds are “active” regulators of signaling modules. FEBS J. 2010;277:4376–4382. doi: 10.1111/j.1742-4658.2010.07867.x. [DOI] [PubMed] [Google Scholar]
- 46.Chen XD. Extracellular matrix provides an optimal niche for the maintenance and propagation of mesenchymal stem cells. Birth Defects Res C Embryo Today. 2010;90:45–54. doi: 10.1002/bdrc.20171. [DOI] [PubMed] [Google Scholar]
- 47.Shih YR. Chen CN. Tsai SW. Wang YJ. Lee OK. Growth of mesenchymal stem cells on electrospun type I collagen nanofibers. Stem Cells. 2006;24:2391–2397. doi: 10.1634/stemcells.2006-0253. [DOI] [PubMed] [Google Scholar]
- 48.Santi DV. McHenry CS. Sommer H. Mechanism of interaction of thymidylate synthetase with 5-fluorodeoxyuridylate. Biochemistry. 1974;13:471–481. doi: 10.1021/bi00700a012. [DOI] [PubMed] [Google Scholar]
- 49.Ghoshal K. Jacob ST. An alternative molecular mechanism of action of 5-fluorouracil, a potent anticancer drug. Biochem Pharmacol. 1997;53:1569–1575. doi: 10.1016/s0006-2952(97)00040-3. [DOI] [PubMed] [Google Scholar]
- 50.Friedl P. Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2009;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deacon SW. Beeser A. Fukui JA, et al. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem Biol. 2008;15:322–331. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arias-Romero LE. Chernoff J. A tale of two Paks. Biol Cell. 2008;100:97–108. doi: 10.1042/BC20070109. [DOI] [PubMed] [Google Scholar]
- 53.Molli PR. Li DQ. Murray BW. Rayala SK. Kumar R. PAK signaling in oncogenesis. Oncogene. 2009;28:2545–2555. doi: 10.1038/onc.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim TY. Lee JW. Kim HP. Jong HS. Jung M. Bang YJ. DLC-1, a GTPase-activating protein for Rho, is associated with cell proliferation, morphology, and migration in human hepatocellular carcinoma. Biochem Biophys Res Commun. 2007;355:72–77. doi: 10.1016/j.bbrc.2007.01.121. [DOI] [PubMed] [Google Scholar]
- 55.Street CA. Bryan BA. Rho kinase proteins—pleiotropic modulators of cell survival and apoptosis. Anticancer Res. 2011;31:3645–3657. [PMC free article] [PubMed] [Google Scholar]
- 56.Zohrabian VM. Forzani B. Chau Z. Murali R. Jhanwar-Uniyal M. Rho/ROCK and MAPK signaling pathways are involved in glioblastoma cell migration and proliferation. Anticancer Res. 2009;29:119–123. [PubMed] [Google Scholar]
- 57.Provenzano PP. Keely PJ. Mechanical signaling through the cytoskeleton regulates cell proliferation by coordinated focal adhesion and Rho GTPase signaling. J Cell Sci. 2011;124(Pt 8):1195–1205. doi: 10.1242/jcs.067009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hakkinen KM. Harunaga JS. Doyle AD. Yamada KM. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Eng Part A. 2011;17:713–724. doi: 10.1089/ten.tea.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woolfson DN. Mahmoud ZN. More than just bare scaffolds: towards multi-component and decorated fibrous biomaterials. Chem Soc Rev. 2010;39:3464–3479. doi: 10.1039/c0cs00032a. [DOI] [PubMed] [Google Scholar]