Abstract
An exo-β-(1→3)-D-galactanase (SGalase1) that specifically cleaves the β-(1→3)-D-galactan backbone of arabinogalactan-proteins (AGPs) was isolated from culture filtrates of a soil Streptomyces sp. Internal peptide sequence information was used to clone and recombinantly express the gene in E. coli. The molecular mass of the isolated enzyme was ~45 kDa, similar to the 48.2 kDa mass predicted from the amino acid sequence. The pI, pH and temperature optima for the enzyme were ~7.45, 3.8 and 48 °C, respectively. The native and recombinant enzymes specifically hydrolysed β-(1→3)-D-galacto-oligo- or poly-saccharides from the upstream (non-reducing) end, typical of an exo-acting enzyme. A second homologous Streptomyces gene (SGalase2) was also cloned and expressed. SGalase2 was similar in size (47.9 kDa) and enzyme activity to SGalase1 but differed in its pH optimum (pH 5). Both SGalase1 and SGalase2 are predicted to belong to the CAZy glycosyl hydrolase family GH 43 based on activity, sequence homology and phylogenetic analysis. The Km and Vmax of the native exo-β-(1→3)-D-galactanase for de-arabinosylated gum arabic (dGA) were 19 mg/ml and 9.7 μmol D-Gal/min/mg protein, respectively. The activity of these enzymes is well suited for the study of type II galactan structures and provides an important tool for the investigation of the biological role of AGPs in plants. De-arabinosylated gum arabic (dGA) was used as a model to investigate the use of these enzymes in defining type II galactan structure. Exhaustive hydrolysis of dGA resulted in a limited number of oligosaccharide products with a trisaccharide of Gal2GlcA1 predominating.
Keywords: β-D-Galactanases, Arabinogalactan-protein, Streptomyces sp, De-arabinosylate gum arabic, CAZy family GH 43
1. Introduction
Arabinogalactan-proteins (AGPs) are a class of plant proteoglycans containing 90–99% (w/w) carbohydrate and 1–10% (w/w) protein. The predominant carbohydrate components of AGPs are type II arabinogalactans (AGs)1 together with some short oligoarabinosides. 2,3 Type II AGs consist of a highly branched and complex framework of a β-(1→3)-D-galactan backbone and β-(1→6)-D-galactopyranosyl (Galp) side chains, which are further substituted with other sugars such as α-L-arabinofuranose/pyranose (Araf/p), glucopyranosyluronic acid (GlcpA), 4-O-methylglucopyranosyluronic acid (4-O-MeGlcpA), L-rhamnopyranose (L-Rhap) or L-fucopyranose (L-Fucp).4–7 Numerous studies have investigated the structures of the carbohydrate component of AGPs from various plants, for example Arabidopsis,8 gum arabic (GA),9–11 gum tragacanth, 12 larch,13 apple,14 grape,15 radish16,17 and ryegrass.18 Recently, the Hyp-contiguity hypothesis has also contributed to the better understanding of the glycosylation pattern of the protein backbones of AGPs. The Hyp-contiguity hypothesis predicts that non-contiguous Hyp residues would be exclusively substituted with AG polysaccharide chains, while contiguous Hyp residues would be glycosylated with oligoarabinosides.19–22 This hypothesis was generated by studying heterologously-expressed synthetic AGP protein backbones in tobacco and Arabidopsis.23 Hyp glycosylation may be tissue-specific and vary between species.22–24
Defining the precise structure of the carbohydrate moiety of AGPs is essential in furthering our understanding of their biosynthesis, as well as their biological and industrial functions. AGPs are proposed to be responsible for a range of functions in plants.7 For example, AGPs are proposed to play a role in signalling,25,26 embryogenesis, 27,28 xylem differentiation,26,29 as well as cell expansion and extension of apical tip growth in the moss Physcomitrella patens.30 The general structure of type II AG is well known, but its glycosyl sequences are yet to be elucidated, particularly the distribution and degree of polymerisation (DP) of (1→3)- and (1→6)-linked Galp chains.31 Chemical methods such as partial acid hydrolysis, alkaline degradation, acetolysis and Smith degradation have been invaluable in the analysis of the type II glycan chains of AGPs.15,18,32 However, due to the complexity of the carbohydrate moiety, chemical approaches alone are not sufficient.33 Therefore, specific enzymes, such as α-L-arabinofuranosidases (EC 3.2.1.55),14,15,34 β-glucuronidases (EC 3.2.1.31),35 and β-D-galactosidases (EC 3.2.1.23),34,36 exo-β-1,3-D-galactanases (EC 3.2.1.145)32,37 and endo-β-(1→6)-D-galactanases (EC 3.2.1.164)31,38,39 have been used to complement the chemical methods. Most of these enzymes are not commercially available and as a result, have only been used in a limited number of studies. This is especially true of the galactanases that cleave β-(1→3)- or (1→6)-D-Galp linkages in either an exo- or endo-manner. Furthermore, there is only one report to date of the isolation of an endo-β-(1→3)-D-galactanase.40 Enzymes may have different modes of action and/or catalytic sites. It is therefore important to isolate a wide spectrum of hydrolytic enzymes that cleave specific glycosyl linkages of the type II AGs. In the current study, an exo-β-(1→3)-Dgalactanase was isolated from culture filtrates of a Streptomyces sp. isolated from soil and the kinetic properties of the enzyme determined. This enzyme, and a second similar enzyme from the same source, were cloned and expressed in E. coli and their activity characterised. We also demonstrate how the activity of these enzymes is well suited for the study of type II galactan structure.
2. Results
2.1. Native enzyme purification
Isolate 19 was selected from 26 cultures for its ability to grow on medium supplemented with dGA and secrete protein with β-galactanase activity. The 16S rRNA gene sequence revealed isolate 19 to be a Streptomyces sp. related to a clade of species including S. achromogenes, S. acidiscabies, S. antibioticus, S. bobili, S. coelicolor, S. cyaneus, S. flavovariabilis, S. galilaeus, S. griseochromogenes, S. griseoruber, S. longisporus, S. mirabilis, and S. phaeofaciens (data not shown).
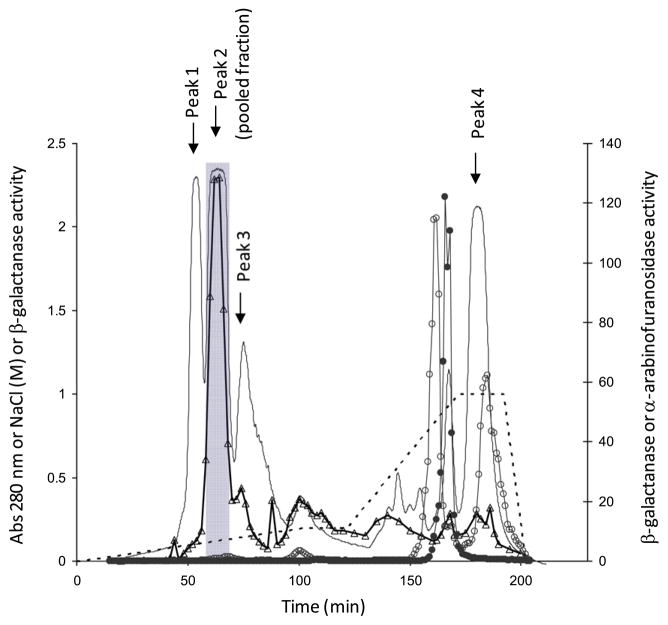
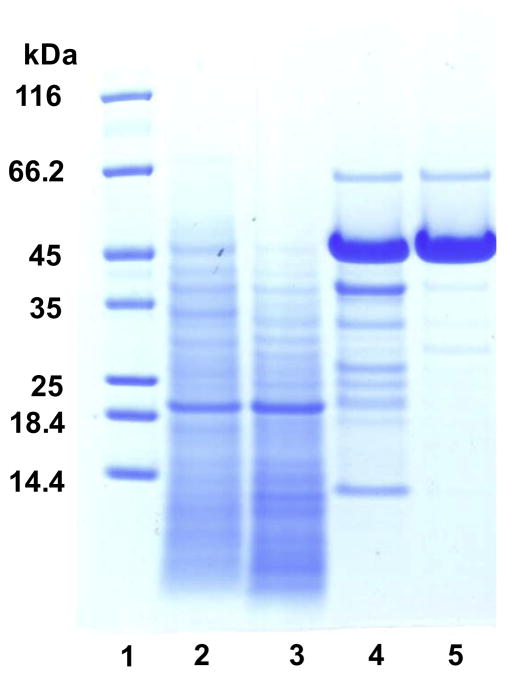
The culture filtrate (day 3) of Streptomyces sp. isolate 19 was used to obtain β-galactanase by precipitating the proteins using ammonium sulfate (crude enzyme preparation) followed by ion exchange and gel filtration chromatography (Table 1). Most of the unwanted β-D-galactosidase activity (~90%) in the crude enzyme preparation was removed by binding to DEAE Trisacryl (M) at pH 8 (data not shown). The remaining DEAE unbound fraction separated into four major protein peaks by cation exchange chromatography (Fig. 1). Most (89%) of the β-galactanase activity eluted as a single peak (EMD Peak 2, Fig. 1) in the range of 0.1–0.15 M NaCl, while most of the α-L-arabinofuranosidase and remaining β-D-galactosidase activities eluted at or above 0.75 M NaCl, well separated from the region associated with β-galactanase activity. EMD Peak 2 was further fractionated by size on a TSK HW 55 column and the β-galactanase activity eluted as a major peak, corresponding to a molecular mass of ~43 kDa (data not shown). The yield and the overall purification factor of the β-galactanase achieved in the current study were 3.3% and ~2-fold, respectively (Table 1). The enriched β-galactanase fraction, HW 55 (major peak), consisted of one major band at ~45 kDa, one minor band at ~66 kDa and three other protein bands between 25 and 40 kDa in very low abundance (Fig. 2, Lane 5).
Table 1.
Protein content and enzyme activity of different fractions obtained in the purification of β-galactanase from filtrate of 3 day-old culture of Streptomyces sp.
| Fraction | Total protein (mg) | Protein yield (%) | Total activitya (Units) | Specific activity (Units/mg protein) | Yield factor (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| (NH4)2SO4 ppt (Crude enzyme) | 323 | 100 | 6975 | 22 | 100 | 1 |
| DEAE Trisacryl (Unbound) | 111 | 34 | 3720 | 34 | 53 | 1.5 |
| Fractogel EMD SO3− (EMD Peak 2) | 7 | 2.2 | 279 | 40 | 4 | 1.8 |
| Fractogel TSK HW 55 (Major peak) | 5.86 | 1.8 | 233 | 40 | 3.3 | 1.8 |
Units = One unit (U) of β-galactanase activity is defined as the amount of enzyme that liberates 1 nmol of low molecular weight substrate fragments bound with RB5 dye per minute under the assay conditions described.
Figure 1.
Elution profile of protein and enzyme activities on Fractogel EMD SO3~ resin. β-Galactanase activity was determined as described in the methods, except a different amount of sample (50 μL of fraction collected) and a longer incubation (1.5 h) were used. Absorbance at 280 nm (—), NaCl concentration (.....), β-galactanase activity (△; Ux3), β-galactosidase activity (○; U/10) and α-arabinofuranosidase activity (●; U/200). The pooled fractions are shaded (58–68 min).
Figure 2.
SDS–PAGE of various protein fractions obtained during the purification process. Lanes: (1) Molecular weight markers, (2) crude enzyme, (3) DEAE Trisacryl unbound fraction, (4) Fractogel EMD SO3− bound fraction (EMD Peak 2; Figure 1), and (5) Fractogel TSK HW 55 (major peak; Figure 2).
2.2. Galactanase cloning and heterologous expression
The ~45 kDa protein band was excised from the gel for Nterminal sequencing and internal sequencing post in-gel trypsin digestion. The N-terminal peptide sequence (A S ASFTLGATYTDQN) (Fig. 2 Lane 5) was compared to the NCBI non-redundant protein databases using the BLASTp algorithm. Thorough searches for short, nearly exact matches found no significant matches. However, the first 10 amino acids of the N-terminal peptide sequence were the same as one of five trypsin generated peptides (highlighted in Fig. 3). The de novo amino acid sequences of the remaining four trypsin-generated peptides (also in Fig. 3) from the same protein band had a relatively high similarity to a number of proteins across different regions of the amino acid sequences, with the top hit in the NCBI BLAST search being exo-β-(1→3)-D-galactanase sequences from the CAZy database (http://www.cazy.org/CAZY). These amino acid sequences were from Phanerochaete chrysosporium galactan (1→3)-β-galactosidase (Pc1,3Gal43A, gi 63108312), Coprinopsis cinerea hypothetical protein (CC1G_03773, gi 116510091), Streptomyces avermitilis exo-β-(1→3)-D-galactanase (Sal1,3Gal43A, gi 29828651) and Clostridium thermocellum exoβ-(1→3)-D-galactanase (Ct1,3Gal43A, gi 67916108). Commonly conserved features of these sequences include regions showing sequence similarity to CAZy glycosyl hydrolase family 43 (GH 43) and ricin carbohydrate-binding domains.
Figure 3.

Alignment of SGalase1 and SGalase2 amino acid sequences. Conserved amino acids are highlighted in black, predicted SGalase2 signal peptide is boxed, catalytic domain is underlined and carbohydrate-binding domain is double-underlined. SGalase1 peptides used to design degenerate primers are indicated by grey lines above the sequences. Numbers on the right indicate amino acid position.
Using degenerate primers based on the N-terminal and internal peptide sequences of the isolated protein, the SGalase1 gene sequence was amplified and sequenced, along with another putative Streptomyces sp. galactanase gene, SGalase2. The predicted proteins encoded by the two genes share 85% amino acid sequence similarity (using mature protein sequence only) and a predicted size of ~48 kDa for the mature enzymes (Fig. 3). SGalase2 has a predicted N-terminal signal peptide and conserved domain searches41 found that both of the predicted proteins contain a GH 43 catalytic domain at the N-terminal end, and a ricin-type β-trefoil carbohydrate- binding domain at the C-terminal end (Fig. 3). Despite repeated attempts, the very 5′ end of SGalase 1 proved difficult to obtain, however based on protein sequence analysis, the majority of the enzyme was cloned. Sequences for SGalase 1 and SGalase 2 were deposited in Genbank with accession numbers JQ683399 and JQ683400 respectively.
The amino acid sequences for the catalytic domains of the characterised GH43 enzymes were aligned with those of SGalase1 and SGalase2, and the program MEGA 5 was used to generate a Neighbor-Joining tree (Fig. 4). The tree shows, with high bootstrap support, that the two SGalase enzymes are more similar to each other than to any of the other enzymes, and that they are nested within the other exo-β-(1→3)-galactanases.
Figure 4.
Phylogenetic tree of the catalytic domains of the SGalase proteins and characterised CAZy family 43 glycosyl hydrolases. The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 8.35 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method and are in the units of the number of amino acid differences per site.
SGalase 1 and SGalase 2 were cloned into E. coli and expressed. Although both proteins formed protein inclusion bodies, their renatured forms (after urea solubilisation) retained galactanase activity, appeared as single protein bands by SDS–PAGE and were therefore used for further characterisation.
2.3. Enzyme characterisation
The pI of the isolated Streptomyces sp. β-galactanase was ~7.45 as determined by IEF gel electrophoresis and had a pH optimum of 3.8 at 37 °C, and temperature optimum of 47.5 °C at pH 3. The pH optima of recombinant SGalase1 and SGalase2 were 3.8 and 5.0 at 37 °C, respectively. The temperature optimum of both recombinant enzymes was 48 °C at their appropriate pH optima. All enzymes were most stable up to 37 °C.
Each of the enzymes was tested against a broad panel of oligoand poly-saccharides (Supplementary Table 1) and were found to be active only against oligo- and poly-saccharides containing β-(1→3)-D-Gal linkages. The specific activities against these substrates, as well as the percentage (%) relative to the specific activity towards a ‘β-(1→3)-D-galactan’ (Acacia) preparation, are presented in Table 2. The enzymes had no, or negligible, activity on pnitrophenyl (PNP) β-D-galactopyranoside, PNP β-D-fucopyranoside, PNP α-L-arabinofuranosidase PNP α-D-galactopyranoside, PNP β/α-D-glucopyranoside, PNP β/α-L-fucopyranoside, PNP β-Dcellobioside, PNP α-D-mannopyranoside or PNP β-D-xylopyranoside after 24 h incubation at 37 °C.
Table 2.
Activity of the native, and recombinant SGalase 1 and SGalase 2 on β-(1→3)/(1→6)-D-linked polysaccharides and galacto-oligosaccharides
| Substrate | Native enzyme
|
SGalase 1
|
SGalase 2
|
|||
|---|---|---|---|---|---|---|
| Specific activity ± SD (μmol D-Gal/min/mg protein) | Relative specific activity (%) | Specific activity ± SD (μmol D-Gal/min/mg protein) | Relative specific activity (%) | Specific activity ± SD (μmol D-Gal/min/mg protein) | Relative specific activity (%) | |
| Polysaccharides | ||||||
| β-(1→3)-D-Galactan (Acacia) | 10.8 ± 0.3 | 100 | 12.4 ± 0.4f,c | 100 | 3.8 ± 0.1f,c | 100 |
| dGA (Acacia) | 4.4 ± 0.1 | 20 | 3.8 ± 0.2e,g | 31 | 1.4 ± 0.1e,g | 37 |
| Gum arabic (Acacia) | 0.03 ± 0.0 | 30 | 0.0 ± 0.0e,h | 0 | 0.0 ± 0.0e,h | 0 |
| Arabinogalactan (Larch) | 0.05 ± 0.0 | 30 | 0.0 ± 0.0e,h | 0 | 0.0 ± 0.0e,h | 0 |
| β-(1→3;1→6)-D-Galactan (P. zopfii) | 0.1 ± 0.0 | 1 | 0.0 ± 0.0e,h | 0 | 0.0 ± 0.0e,h | 0 |
| Oligosaccharides | ||||||
| Methyl β-glycosides of β-(1→3)-D- | ||||||
| Galactobiopyranoside | 63.2 ± 1.6 | 293 | 92.0 ± 5.8a,b | 740 | 37.10 ± 1.9a,c | 972 |
| Galactotriopyranoside | nd | nd | 28.5 ± 3.9d,b | 229 | 5.2 ± 0.2e,c | 137 |
| Galactotetrapyranoside | 49.8 ± 0.4 | 230 | 27.9 ± 1.1d,b | 225 | 4.2 ± 0.1e,c | 111 |
| Galactopentaopyranoside | 44.2 | 205 | 32.6 ± 6.0d,b | 262 | 6.7 ± 0.1e,c | 175 |
| Galactoheptaopyranoside | nd | nd | 21.7 ± 0.6d,b | 175 | 3.4 ± 0.3e,c | 89 |
| Methyl β-glycosides of β-(1→6)-D- | ||||||
| Galactotriopyranoside | 0.0 ± 0.0 | 0 | 0.0 ± 0.0e,h | 0 | 0.0 ± 0.0e,h | 0 |
| Galactotetraopyranoside | nd | nd | 0.0 ± 0.0e,h | 0 | 0.0 ± 0.0e,h | 0 |
| Galactopentaopyranoside | 0.2 ± 0.0 | 2 | 0.0 ± 0.0e,h | 0 | 0.0 ± 0.0e,h | 0 |
| Galactohexaopyranoside | 0.2 ± 0.0 | 2 | 0.0 ± 0.0e,h | 0 | 0.0 ± 0.0e,h | 0 |
nd, not determined; Enzyme at a 0.1, d0.5, e0.7, f0.4 μg protein and incubated for b5, c15, g30 min, h24 h at 37 °C.
All three enzyme preparations had similar substrate specificity, but the native enzyme and its recombinant form, SGalase1, had higher specific activities than SGalase2. Among the methyl βglycosides of β-(1→3)-linked D-galacto-oligosaccharides tested, the specific activity of the enzyme was highest towards the galactobioside. The specific activities of the enzymes decreased dramatically at DP 3 and remained relatively similar across DP 3–5 and 7. The enzymes hydrolysed the ‘β-(1→3)-D-galactan’ and dGA at lower rates compared to the methyl β-glycosides of β-(1→3)-linked Dgalacto- oligosaccharides. The specific activity of the SGalase1 and 2 towards dGA were only 30% and 37%, respectively, of that towards the ‘β-(1→3)-D-galactan’. The kinetic parameters of the enzyme towards dGA were determined by a Lineweaver–Burk plot from which Km = 19 mg/ml and Vmax = 9.7 μmol D-Gal/min/mg protein were obtained. Native AGPs from Acacia, the AG from larch (Larix), β-(1→3;1→6)-galactan from P. zopfii and the methyl β-glycoside of β-(1→6)-linked D-galacto-oligosaccharides were resistant to hydrolysis by the enzymes after extended incubation of 24 h (Table 1).
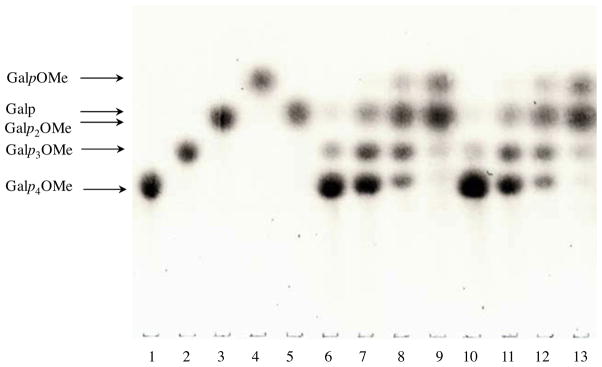
The modes of action of SGalase1 and SGalase2 were studied by TLC (Fig. 5). Hydrolysates of the β-(1→3)-linked D-galactotetrapyranoside showed the release of Gal/methyl β-glycosides of β-(1→3)- linked D-galacto-oligopyranosides with DP lower than DP 4. Notably, methyl β-D-galactopyranoside (GalpOMe) was not observed initially. As the incubation progressed, Gal and/or Galp2OMe (Gal and Galp2OMe were not well resolved on the TLC) and GalpOMe continued to accumulate with the concomitant disappearance of the higher oligomers. The final hydrolysis products showed two major spots on the TLC plate, corresponding to GalpOMe, and Gal and/or Galp2OMe. The Gal and Galp2OMe may be better resolved by modification of solvent conditions or alternatively, identified by mass spectrometry. Nevertheless, the absence of GalpOMe at the initial stages of hydrolysis, demonstrated that SGalase1 and SGalase2 released Gal from the upstream end of β-(1→3)-linked D-galactotetraopyranosides (the non-reducing end in the non-methylated equivalent), thus catalysing the hydrolysis in an exo-manner.
Figure 5.
Mode of action of the SGalase1 and 2 on methyl β-glycosides of β-(1→3)-linked D-galactotetrapyranoside analysed by TLC. The reaction mixture (4.2 μL final volume) contained 0.1 μg enzyme and 1.4 μL of methyl β-glycosides of β-(1→3)-linked D-galactotetraopyranoside (Galp4OMe, 50 mM) and was incubated for 0 min (lane 6 and 10 for SGalase 1 and 2, respectively), 5 min (lane 7 and 11 for SGalase 1 and 2, respectively), 30 min (lane 8 and 12 for SGalase 1 and 2, respectively) or 3 h (lane 9 and 13 for SGalase 1 and 2, respectively) at 37 °C. Standards on lanes: (1) Galp4OMe, (2) methyl β-glycosides of β-(1→3)-linked D-galactotriopyranoside (Galp3OMe), (3) methyl βglycosides of β-(1→3)-linked D-galactobiopyranoside (Galp2OMe), (4) methyl β-D-galactopyranoside (GalpOMe), (5) galactose (Galp). *Galp and Galp2OMe virtually comigrated.
dGA was treated with enzyme and then ethanol precipitated to separate the high molecular weight (ppt fraction) from the low molecular weight (supt fraction) components, to study the structures of the products of enzyme digestion. The monosaccharide composition of the dGA (starting material, Table 3) determined by methanolysis showed that the most abundant sugar was Gal (73%), followed by a significant amount of GlcA (25%), and low levels of Rha (2%) and Ara (<0.5%). The monosaccharide compositions of enzyme hydrolysed dGA ppt and supt fractions were similar in Gal (61% and 65%, respectively) and Rha (4% and 2%, respectively), and both had 34% GlcA, and a minimal amount of Ara (1%). A low level of Glc was detected in all the samples in linkage analysis, with a higher amount in the enzyme hydrolysed dGA ppt fraction (Table 3). However, no Glcp was detected in the monosaccharide composition analysis (by methanolysis) in any of the samples, suggesting Glcp is likely to be a contaminant sugar introduced during the methylation analysis.
Table 3.
Composition of the dGA and the Streptomyces sp. exo-β-(1→3)-D-galactanase hydrolysed dGA product
| Sample | GA1 | Starting material2 | Enzyme hydrolysed dGA3
|
Control dGA (no enzyme)4
|
|||
|---|---|---|---|---|---|---|---|
| dGA | ppt | supt | ppt | supt | |||
| Total sugar content5 (% ±SD) | ND | ND | 23 ± 5 | 77 ± 8 | 100 ± 10 | 0 ± 0 | |
| Methanolysis6 | Monosaccharide composition (weight %) | ||||||
| Rha | ND | 2 | 4 | 2 | ND | ND | |
| Ara | ND | tr | 1 | 1 | ND | ND | |
| Gal | ND | 73 | 61 | 65 | ND | ND | |
| GlcA | ND | 25 | 34 | 34 | ND | ND | |
| Linkage analysis7 | Deduced glycosidic linkage | Linkage composition (mol%) | |||||
| Rhap | Terminal | 10 | 2 | 4 | 5 | 4 | — |
| Arap | Terminal | 1 | — | 2 | 1 | tr | — |
| Araf | Terminal | 14 | — | 2 | 2 | tr | — |
| 1,3- | 13 | tr | 1 | 2 | 1 | — | |
| GlcpA | Terminal | 2 | 28 | ND | ND | ND | ND |
| 1,4- | 16 | 2 | ND | ND | ND | ND | |
| Galp | Terminal | 14 | 5 | 16 | 33 | 9 | — |
| 1,3- | 1 | 3 | 4 | — | 6 | — | |
| 1,6- | tr | 34 | 40 | 54 | 38 | — | |
| 1,4,6- | — | — | 1 | 1 | 4 | — | |
| 1,3,4- | 1 | — | — | — | — | — | |
| 1,3,6- | 18 | 26 | 18 | 1 | 35 | — | |
| 1,2,6- | — | — | 1 | 1 | tr | — | |
| 1,3,4,6- | 10 | — | tr | tr | 1 | — | |
| Glcp | 1,4- | — | — | 9 | — | tr | — |
| 1,2,3,4,6- | — | — | 2 | tr | tr | — | |
ND, not determined; —, not detected; tr, trace <1 mol %; ppt, precipitate fraction; supt, supernatant fraction.
The linkage analysis for neutral sugars of the dGA showed that it was composed mainly of Galp (~68 mol %), with terminal (5 mol %), 1,3-linked (3 mol %), 1,6-linked (34 mol %) and 1,3,6- linked (26 mol %) (Table 3). Compared to the dGA, the enzyme hydrolysed dGA had increases in terminal Galp (from 5 to 16 and 33 mol % for ppt and supt fractions, respectively), and 1,6-linked Galp (from 34 to 40 and 54 mol % for supt fraction, respectively). Notably, a significant decrease in 1,3,6-linked Galp for both ppt and supt fractions (from 26 to 18 and 1 mol %, for ppt and supt fractions, respectively) was observed. Moreover, 1,3-linked Galp was absent in the enzyme hydrolysed dGA supt fraction, while it remained relatively unchanged in the ppt fraction (Table 3). No sugar was detected in the control dGA supt fraction, while the linkage composition of the control dGA ppt fraction was essentially the same as that of the untreated dGA (Table 3).
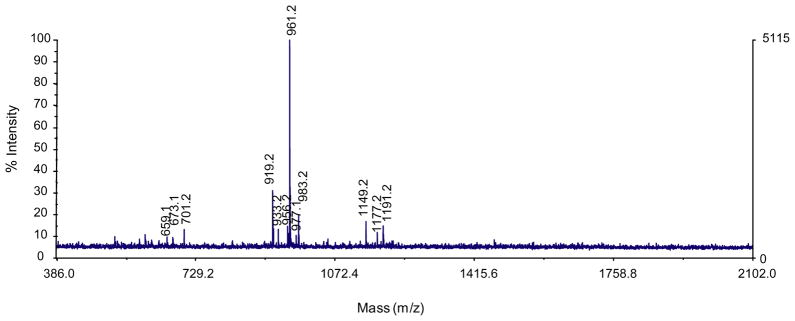
The MALDI-TOF MS spectrum of the final enzyme product after exhaustive hydrolysis displayed 6 major molecular ion species of m/z 673.2, 701.2, 919.2, 961.2, 1149.2 and 1191.2 (Fig. 6 and Table 4). These ions were present in replicate experiments and in each case the m/z 961 ion dominated. The deduced structure of each ion was as follows: (1) peracetylated Hex1HexA1 (m/z 673.2), (2) peracetylated Hex2 (m/z 701.2), (3) peracetylated Hex1HexA2 (m/z 933.2), (4) peracetylated Hex2HexA1 (m/z 961.2), (5) peracetylated Hex2HexA1- Pent (m/z 1177.2), and (6) peracetylated Hex2HexA1DeoxyHex1 (m/z 1191.2), as the sodium adducts. The peaks at m/z 919.2 and 1149.2 represent underacetylated [minus an acetyl (m/z 42) group] species of m/z 961.2 and 1191.2, respectively. The peak at m/z 977.1 represents the K+ adduct of the major ion, peracetylated Hex2HexA1, and it should be noted that the 1149.2 ionmayalso represent the Na+ adduct of Hex1HexA2Pent. Peaks at 605, 956 and 983 (m/z) were unidentified.
Figure 6.
MALDI TOF Mass Spectrometry of oligosaccharide products (peracetylated) released from dGA galactanase hydrolysis. Deduced oligosaccharide structures are presented in Table 4.
Table 4.
Oligosaccharide structures deduced from MALDI-TOF MS analysis of peracetylated supernatant (supt) fraction of the Streptomyces sp. exo-β-(13)-D-galactanase hydrolysed dGA product
| Peaks (m/z) | % Intensity (relative intensity) | Deduced and possible predominant structures |
|---|---|---|
| 673.2 | 10 (4%) | Hex1HexA1 |
| GlcpA-(1→6)-Galp | ||
| 659.1 (701—acetyl group) | 10 (10%) | Hex2 |
| 701.2 | 12 | Galp-(1→6)-Galp |
| 933.2 | 12 (5%) | Hex1HexA2 |
| GlcpA-(1→4)-GlcpA-(1→6)-Galp | ||
| 919.2 (961—acetyl group) | 30 (62%) | Hex2HexA1 |
| 961.2 | 100 | GlcpA-(1→6)-Galp-(1→6)-Galp |
| 977.1 (961.2 K + adduct) | 10 | |
| 1177.2 | 10 (4%) | Hex2HexA1Pent1 |
| GlcpA-(1→3)-Araf-(1→6)-Galp-(1→6)-Galp | ||
| GlcpA-(1→6)-Galp-(1→3)-Araf-(1→6)-Galp | ||
| 1149.2 (1191.2—acetyl group) | 18 (15%) | Hex2HexA1DeoxyHex1 |
| 1191.2 | 15 | Rhap-(1→6)-Galp-(1→4)-GlcpA-(1→6)-Galp |
| Rhap-(1→4)-GlcpA-(1→6)-Galp-(1→6)-Galp |
Unless otherwise stated, ions appear as sodium adducts.
3. Discussion
We have successfully purified, cloned and heterologously expressed an exo-β-(1→3)-D-galactanase from Streptomyces sp. The overall purification factor of the Streptomyces sp. exo-β-(1→3)-Dgalactanase (Table 1) was low compared to those reported for βgalactanases in other studies (>12-fold).33,37,42 The reason for a low purification factor is not fully understood, but could be attributed to the presence of several enzymes that all possess βgalactanase activity within the crude enzyme as well as the nature of the assay used. The total β-galactanase activity of the crude enzyme preparation may actually reflect the combined activities of multiple enzymes, β-(1→3)- and/or β-(1→6)-D-galactanases and their isozymes, rather than the activity of a single dominant enzyme. Faint clearing zones were seen in zymograms with gels containing the chromogenic substrate and run under native conditions (data not shown).43 The clearing patterns of the crude and final isolated enzyme fractions were different, suggesting that the final preparation was not representative of all β-galactanase activities in the crude preparation. Besides, the chromogenic substrate (RB5-dGA) contains both β-(1→3)- and (1→6)-galactosyl linkages, therefore precise measurement of a single β-galactanase in the presence of multiple β-galactanases is challenging. The purification fold could only be determined more accurately if linkage-specific substrates were available.
The molecular mass of the Streptomyces sp. exo-β-(1→3)-Dgalactanase is within the range (38–66 kDa as estimated by SDS– PAGE) of other exo-β-(1→3)-D-β-galactanases,33,37,42,44,45 while its pI is lower than those from Irpex lacteus.37 The pH optima of most other exo-β-(1→3)-D-galactanases were reported within the range of 4.5–6.33,37,44,42,45,46 Most of the β-(1→3)-D-galactanases appear to be stable in acidic to neutral, but not alkaline conditions,33,37,42,46 except that from Clostridium thermocellum, which is stable in both acidic and alkaline conditions, spanning a wide pH range of 3–10.44 The temperature optimum of the isolated and recombinant enzymes is similar to that of previously identified exo-β-(1→3)-Dgalactanases (40–50 °C).33,37,42,44
The Vmax/Km value of the native isolated enzyme for dGA (0.5 μmol D-Gal/min/mg protein/mg dGA/mL) is relatively low, suggesting dGA is unlikely to be the best substrate for the enzyme. This is not surprising, since dGA is a highly branched glycan consisting of 26 mol % 1,3,6-linked Galp and only a low level of 1,3-linked Galp (3 mol %).43,44 Based on the relative activity measured against different substrates (see Table 2), it is likely that the Vmax/Km values of this enzyme would be higher for methyl β-glycosides of β-(1→3)- linked D-galacto-oligosaccharides. However, due to limited availability of these synthetic substrates, the kinetic parameters could not be further explored. Some exo-β-(1→3)-D-galactanases were reported to have Vmax/Km values of 43.6 μmol galactobiose/min/mg protein/mg galactan III/mL33 and 64.3 μmol D-Gal/min/mg protein/ mM ‘β-(1→3)-D-galactan’,37 and Kcat/Km values of 263,45 30842 and 175846 min−1 mM−1 ‘β-(1→3)-D-galactan’.
The activity profile observed for the Streptomyces sp. exo-β-(1→3)-D-galactanases, SGalase 1 and SGalase 2, towards different substrates is distinct from the previously reported exo-(1→3)-Dgalactanases, which exhibit higher activities towards the ‘β-(1→3)-D-galactan’ than β-(1→3)-D-galacto-oligosaccharides or methyl β-glycosides of β-(1→3)-linked D-galacto-oligosaccharides. 37,42,46 The exo-β-(1→3)-D-galactanases from A. niger37 and S. avermitilis42 showed increasing activity towards β-(1→3)-galactooligosaccharides and their methyl β-galactosides with increasing DP from 2 to 5, while the exo-β-(1→3)-D-galactanase from P. chrysosporium46 had a higher activity towards β-(1→3)-D-galactobiose than towards β-(1→3)-D-galactotriose. The exo-β-(1→3)-Dgalactanase from C. thermocellum exhibited a slightly higher activity towards β-(1→3)-D-galactotriose than towards the ‘β-(1→3)-D-galactan’.45 All the β-(1→3)-D-galactanases reported thus far have high substrate specificity, except that from R. niveus,47 which hydrolysed coffee bean AG to liberate Ara, Gal and β-(1→6)-D-galactobiose. However, questions have been raised about the purity of the β-(1→3)-D-galactanase from R. niveus.31,33,37 The results on mode of action of the isolated enzyme are consistent with those reported for the exo-β-(1→3)-D-galactanases from I. lacteus,37 P. chrysosporium,46 C. thermocellum45 and S. avermitilis,60 where only Gal was reported as the hydrolysis product released from the nonreducing end of the ‘β-(1→3)-D-galactan’.42,45,46
After exhaustive hydrolysis of the dGA with the Streptomyces sp. exo-β-(1→3)-D-galactanase, the extent of hydrolysis was determined to be ~20%. This is defined as the reducing sugars released after extensive hydrolysis (equivalent to 1.2 mg Gal) relative to the total sugar content (equivalent to 6 mg Gal) of the substrate used. After ethanol precipitation of the extensively hydrolysed dGA, the precipitate (ppt) and supernatant (supt) fractions accounted for 23% and 77% of the total sugar content, respectively (Table 3). The extent of hydrolysis of dGA by Streptomyces sp. exo-β-(1→3)- D-galactanase is similar to that reported for exo-β-(1→3)-Dgalactanases from A. niger on galactan III (~18%),33 as well as that achieved by the exo-β (1→3)-D-galactanase from I. lacteus on the α-L-arabinofuranosidase-treated radish leaf AGP (21%).37 In addition, the β-galactosidase from radish48,49 and spinach34 were reported to achieve 17–31% extent of hydrolysis of an α-L-arabinofuranosidase- treated radish leaf AGP. Although the extent of hydrolysis was modest, a substantial carbohydrate component of the dGA was released as low DP glycosyl moieties after Streptomyces sp. exo-β-(1→3)-D-galactanase hydrolysis, as indicated by the high proportion (77%) of the total sugar content in the enzyme hydrolysed dGA supt fraction (Table 3). By comparison, the exo-β-(1→3)-D-galactanase from I. lacteus was able to achieve 26% extent of hydrolysis of an α-L-arabinofuranosidase-treated radish root AGP, leading to removal of 91% carbohydrate from the substrate.37
The limited extent of hydrolysis of various substrates containing β-(1→3)-linked Gal residues is presumably due to the presence of structure(s) that would interfere with the galactanases in gaining access to the β-(1→3)-Gal linkages,37,49 such as the presence of uronic acids at the non-reducing ends of β-(1→6)-linked Gal side chains of most of the AGPs.6,7,49 Supporting this conclusion is the reported increase in the extent of hydrolysis from ~14% to ~80% when α-L-arabinofuranosidase-treated radish leaf AGP was hydrolysed by the combined action of radish β-galactosidase and a microbial β-glucuronidase.49 The sugar composition data of dGA used in the present study also showed the presence of high levels of terminal uronic acids (Table 3), suggesting that future studies of the extent of hydrolysis of dGA when treated simultaneously with β-galactosidase and β-glucuronidase would be advisable to generate the preferred substrate for the Streptomyces sp. exo-β-(1→3)-Dgalactanase.
The enzymes isolated and cloned in this study show substrate specificities similar to those of the exo-β-(1→3)-D-galactanases that are also classified in CAZy GH 43.42,45,46 The CAZy database (http://www.cazy.org/)50 classifies glycoyl hydrolases into 130 families based on amino acid sequence similarities. Recently, Kotake et al.40 reported on the isolation of an endo-β-(1→3)-Dgalactanase from the mushroom, Flammulina velutipes. Sequence information and phylogenetic analysis determined that this enzyme belongs to CAZy GH 16, a family containing other endo-acting enzymes, including endo-β-(1→3)- and endo-β-(1→3, 1→4)- glucanases. GH 43 contains several hundred enzymes, including xylosidases, arabinofuranosidases and galactanases. The galactanases in GH 43, as well as the enzymes from the current study, show only the galactan-β-(1→3)-galactosidase activity but not the activities of other enzymes reported for the GH 43 family, including βxylosidase (EC 3.2.1.37), α-L-arabinofuranosidase (EC 3.2.1.55), endo-α-(1→5)-L-arabinanase (EC 3.2.1.99) and β-(1→4)-xylanase (EC 3.2.1.8) activities (http://www.cazy.org/fam/GH43.html). Phylogenetic analysis of the characterised GH 43 family members showed that the galactanases and arabinanases each formed a monophyletic group with strong bootstrap support. The xylosidases generally formed two clades that are well separated from one another, with one being closer to the endo-arabinanases and the other closer to the galactanases. The dual-function xylosidase/ arabinofuranosidases and remaining arabinofuranosidases scatter throughout the phylogenetic tree and do not appear to form functional groups based on enzyme activity. The absence of clear phylogenetic grouping based on protein sequence alone demonstrates that the characterisation of the activities of these enzymes remains an important aspect in the study and use of glycosyl hydrolases. It enables further organisation of the members of each family into functional groups dependent on substrate specificity and potentially highlights sequence information that will aid in identifying particular enzyme activities at the transcript level.
Data of the hydrolysed dGA products from the current project are similar to those reported for other exo-β-(1→3)-D-galactanases acting on AGPs. For example, the exo-β-(1→3)-D-galactanases from A. niger degraded galactans II and III (prepared by acid hydrolysis and two successive Smith degradations of the gum arabic from A. senegal), releasing Gal, β-(1→6)-D-galacto-oligosaccharides with or without Ara attached, and other unidentified reaction products of higher DP.33 The exo-β-(1→3)-D-galactanases from I. lacteus,37 P. chrysosporium,46 C. thermocellum45 and S. avermitilis42 hydrolysed α-L-arabinofuranosidase-treated radish root AGPs releasing Gal and β-(1→6)-D-galacto-oligosaccharides with or without 4-OMeGlcpA. In the current study, the released oligosaccharides were analysed using MALDI-TOF MS. The assigned structures showed the presence of a mixture of oligosaccharides in the enzyme hydrolysed dGA 80% ethanol supt fraction. Several sequences are possible, but the proposed oligosaccharide structures have been deduced based on the following observations: (1) The reducing end sugar of the Streptomyces sp. exo-β-(1→3)-D-galactanase hydrolysed dGA must be Galp since the enzyme specifically cleaves only 1,3-Galp. (2) Most of the GlcpA in the dGA were terminal residues (28 mol %), with only 2 mol % 1,4-linked (Table 3). Therefore, the HexA-containing oligosaccharides predominantly consist of terminal GlcpA rather than 1,4-linked GlcpA. (3) The linkage analysis data also showed terminal Rhap being the only deoxyHexose residue present in the enzyme hydrolysed dGA supt fraction (Table 3). Therefore, the deoxyHexose containing oligosaccharide must contain Rhap as the terminal residue.
No 4-O-MeGlcpA or any other monosaccharide residue was evident from MALDI-TOF MS analyses (Fig. 6). This is not surprising because there is only a very low level of 4-O-MeGlcpA in gum arabic from A. senegal.51 As shown in Table 3, the enzyme hydrolysed dGA supt fraction contained mainly terminal Galp (33 mol %) and 1,6-linked Galp (54 mol %), with depletion of both 1,3- and 1,3,6- linked Galp; while the ppt fraction had a lower proportion of 1,3,6-linked Galp compared to the dGA. This suggested that Streptomyces sp. exo-β-(1→3)-D-galactanase was able to cleave the β-1,3-linkages of both the 1,3- and 1,3,6-linked Galp residues, releasing oligosaccharides of DP 2–4, made up of GlcpA-(1→6)-Galp and Galp-(1→6)-Galp with or without GlcpA, Araf or Rhap attached (MALDI-TOF MS data, Table 4). The rather small set of oligosaccharide structures enzymatically released from de-arabinosylated gum arabic AGPs in this study is consistent with the findings of Tan et al.52 that proposed small repetitive subunits combining to form the much larger AGs on AGPs.
4. Experimental
4.1. Source of enzyme and culturing
Dilutions of soil collected from a rotationally-grazed pasture at the Dairy Research Institute, Ellinbank, Australia, were plated onto plates of medium VL55 solidified with agar53 and containing 0.05% (w/v) gum arabic. Twenty-six rapidly-developing colonies were subcultured to purity, and then transferred to plates containing 0.05% (w/v) de-arabinosylated gum arabic (dGA). Isolate 19, which grew well on dGA and secreted β-galactanases, was selected for further investigation.
The isolate, identified as a Streptomyces species, was maintained at 28–30 °C in the dark on a synthetic agar medium containing MgSO4·7H2O (20 mM), CaCl2·2H2O (3 mM), (NH4)2HPO4 (20 mM), KCl (5 mM), NaCl (10 mM), selenite/tungstate solution (1 mL/L),54 trace element SL10 solution (2 mL/L),55 MES (9.75 g/L), dGA (5 g/ L) and granulated agar (15 g/L), adjusted to pH 5.5. A hyphal plug (0.5 cm diameter) was diced and added to medium (2 mL), prepared as above (without agar) in a 12 mL sterile polypropylene tube, and incubated at 30 °C with shaking (150 rpm). The culture was bulked up by a series of 3 day subcultures until it was inoculated (17.5 mL) into the liquid medium (350 mL) in a 1000 mL flask. After 3 days, the culture was harvested by centrifugation (3373×g, 10 min), and the supernatant filtered through Whatman No. 541 filter paper to give the culture filtrate.
4.2. Substrates and sugar standards
Various p-nitrophenyl (PNP) glycosides and oligo-/poly-saccharides, including methyl β-(1→3)-D-galactobioside, and α-(1→3)-Dgalactobiose were purchased from Sigma–Aldrich (St. Louis, Missouri, USA), Megazyme (Bray, Co. Wicklow, Ireland), Applichem Inc. (Cheshire, CT, USA), or Biosupplies (Melbourne, Australia). ‘β-(1→3)-D-Galactan’ (Acacia senegal), a Smith degraded product of gum arabic (GA) with an estimated DP of 50,46 and β-(1→3;1→6)-galactan from Prototheaca zopfii with molecular mass of 85 kDa38 were generous gifts from Professor Y. Tsumuraya (Department of Biochemistry and Molecular Biology, Faculty of Science, Saitama University, 255 Shimo-okubo, Sakuraku, Saitama 338-8570, Japan). Methyl β-glycosides of β-(1→3)- and β-(1→6)-linked Dgalacto- oligosaccharides were synthetic materials described previously. 56,57 dGA (molecular mass of ~10 kDa and DP ~60) was prepared from a partial acid hydrolysis (0.25 M CF3CO2H for 2 h at 90–95 °C) of GA (A. senegal, Sigma–Aldrich, St. Louis, Missouri, USA).43,44 Reactive Black 5 dye (RB5, Sigma–Aldrich, St. Louis, Missouri, USA) coupled dGA (RB5-dGA) was prepared as described by Ling et al.43,44
4.3. Enzyme purification
Purification was conducted at 4 °C unless otherwise indicated. The column fractions were desalted and buffer exchanged between chromatography steps on pre-equilibrated EconoPac 10DG (BioRad, Hercules, USA) disposable columns unless otherwise stated. Protein in the culture filtrate of the Streptomyces isolate was concentrated by precipitation with 90% (w/v) ammonium sulfate, followed by three chromatography steps. The precipitate was dissolved in DEAE binding buffer (25 mM Tris–HCl, pH 8) and the solution (47 mL, 323 mg protein) loaded under gravitational flow onto a DEAE Trisacryl (M) column (1.5 cm × 12.3 cm; PALL BioSepra Life Sciences, New York, USA) pre-equilibrated in binding buffer. The unbound protein was collected and the column was washed with 4 bed vols of buffer. The DEAE unbound fraction was concentrated and the buffer was exchanged into EMD binding buffer (25 mM citric acid–Na2HPO4, pH 3.5) by ultrafiltration on a YM 10 membrane (MWCO 10,000; Millipore, Australia).
The DEAE unbound fraction (111 mg protein) was applied to a Fractogel EMD SO3− (M) column (1 cm × 29.3 cm, Merck KGaA, Germany) pre-equilibrated in EMD binding buffer (25 mM citric acid–Na2HPO4, pH 3.5) at 1 mL/min using a 50 mL Superloop (GE Healthcare Life Sciences, Australia) and a Beckman System Gold HPLC (Fullerton, USA) at room temperature. The column was washed with 3 bed vols of binding buffer, and the bound proteins eluted with a stepwise increase in NaCl to 0.2 M over 100 min, maintained at 100 mM for 20 min and increased to 1 M over the next 50 min and maintained at this concentration for 20 min and then reduced to 0 M over 10 min. Protein elution was monitored by absorbance at 280 nm. Fractions (2 mL) were collected and stored at 4 °C for enzyme assays. The fractions with β-galactanase activity were pooled (EMD Peak 2) and concentrated to 1.8 mL (7 mg protein) and then buffer exchanged into TSK HW 55 (S) running buffer (150 mM sodium acetate, pH 5 with 0.2 M NaCl) using Amicon ultra-15 units (MWCO 10,000; Millipore, Australia). EMD Peak 2 was chromatographed on a TSK HW 55 (S) column (1.5 cm × 83.2 cm, Merck KGaA, Germany) in TSK HW 55 (S) running buffer at gravitational flow (~0.06 mL/min). Fractions were collected every 30 min. The gel filtration column was calibrated using the following standards from Sigma–Aldrich (St. Louis, Missouri, USA): albumin from bovine serum (67 kDa), ovalbumin from chicken egg white (43 kDa), carbonic anhydrase from bovine erythrocytes (29 kDa) and aprotinin from bovine lung (6.5 kDa).
4.4. Assays
β-Galactanase activity was determined using a chromogenic assay.44 The assay mixture contained 0.05 mL sample (~10 μg protein) and 0.25 mL dyed dGA substrate (RB5-dGA, 0.04 g/mL) prepared in sodium acetate buffer (0.1 M, pH 5). The mixture was incubated at 37 °C for 20 min, terminated by adding 1.25 mL of the cold precipitating reagent [20% (v/v) acetic acid, 80% (v/v) 2-methoxyethanol and 0.4% (w/v) zinc acetate] and released products measured at 590 nm and activity determined as described in Ling et al. (2009).44
Glycosidase activities were determined using the respective synthetic p-nitrophenyl (PNP) glycosides as described by Tsumuraya et al.37 with modification. The assay mixture contained the substrate (25 μL of 3.3 mM) and enzyme, both in sodium acetate buffer (0.1 M, pH 5). The mixture (50 μL final volume) was incubated at 37 °C for 15–30 min (or 24 h for the specificity study), and the reaction terminated with sodium carbonate (200 mM, 0.2 mL) and absorbance was then measured in a 96-well microtitre plate at 405 nm. One unit (U) of glycosidase activity is defined as the amount of enzyme that liberates 1 nmol of p-nitrophenol per minute under the assay conditions described.
Reducing sugar content was measured with the copper arsenomolybdate reagent of Nelson58 as modified by Somogyi.59 The assay was modified to a micro-scale consisting of enzyme and substrate mixture (50 μL), and the copper reagent (50 μL). The mixture was incubated for 20 min in a boiling water bath and cooled on ice before arsenomolybdate reagent (50 μL) was added. The absorbance of the final assay mixture (140 μL each) was measured in a 96-well micro plate at 490 nm. Galactose (Gal) was used as the standard.
The soluble protein concentration was determined by BioRad protein assay according to the micro-assay procedure in the supplier’s instruction manual (BioRad, Hercules, USA). Bovine serum albumin (BSA, Sigma–Aldrich, St. Louis, Missouri, USA, A7030- 50G) was used as the standard.
4.5. Electrophoresis and protein sequencing
The purified enzyme (4 μg protein) was run on a Novex IEF gel (according to the instructions of the manufacture) and the pI estimated by comparison with the Novex IEF markers pI 3–10 (20 μL per lane; Invitrogen, Carlsbad, USA).
Aliquots of the fractions (6–8 μg protein) were analysed by SDS–PAGE on Nu PAGE 4–12% Bis-Tris pre-cast gels (1 mm × 10 well, Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. The gel was stained with GelCode blue stain reagent (Pierce, Rockford, Illinois, USA) and calibrated with molecular weight markers (Fermentas Life Sciences, Canada).
Proteins in the SDS–PAGE gel were electro-blotted onto a PVDFPlus- membrane (Osmonics Nitrobind 0.45 μm, Minnetonka, USA) in a BioRad Trans blot cell (Biorad, Hercules, USA). The protein band with Mr ~45 kDa was excised from the membrane and sequenced by Edman N-terminal amino acid sequencing (Biomolecular Research Facility, University of Newcastle, Newcastle, Australia). Internal peptide sequence was obtained from similar SDS–PAGE gels that were stained with Coomassie Brilliant Blue R-250, and de-stained with de-staining reagent (5% acetic acid, 50% methanol). The gel was rinsed in water before the protein bands (Mr ~45 kDa and ~66.2 kDa) were excised and trypsin ingel digested and analysed essentially as described in Natera et al.60 The amino acid sequences were manually cluster aligned to the amino acid sequences with the top hit of the BLAST search on the NCBI database, as well as to the exo-1,3-D-galactanase sequences made available from the Carbohydrate Active Enzymes (CAZy) database (http://www.cazy.org/CAZY).
4.6. Cloning of exo-β-(1→3)-galactanase genes
Total genomic DNA was purified by grinding freeze-dried Streptomyces sp. tissue, homogenising in extraction buffer (50 mM Tris–HCl, pH 7.6; 100 mM NaCl; 50 mM EDTA; 2% SDS; 0.2% PVP; 0.1% β-mercaptoethanol), and extracting with phenol/ chloroform/isoamylalcohol (25:24:1), phenol/chloroform (1:1) and chloroform. DNA was precipitated from the aqueous phase with isopropanol, and treated with RNase.
Degenerate primers (Supplementary Table 2) based on SGalase1 peptide sequences were used to amplify internal portions of SGalase1 and SGalase2. Primer pairs were DF1 and DR1 for SGalase1, and DF2 and DR1 for SGalase2. All PCR products were subcloned into the pGEM-T Easy vector (Promega) according to the manufacturer’s instructions, and sequenced by Macrogen on ABI 3730xl DNA Analyser machines.
Flanking 5′ and 3′ gene sequence was obtained using the adaptor- ligation PCR method of Zheng et al.,61 following digestion of genomic DNA with restriction enzymes NaeI, NruI, PvuII and SmaI. The following nested, gene-specific primers (Supplementary Table 2) were used: AF1, AF2 and AF3 for SGalase1, and AR1, AR2, AF4, AF5, AF6 and AF7 for SGalase2.
4.7. Phylogenetic analysis
The catalytic domain sequences of SGalase 1 and SGalase 2 were aligned with those of the biochemically characterised GH 43 family members for phylogenetic analysis using MEGA5.62 Analysis involved 34 amino acid sequences (positions containing gaps and missing data were eliminated) to construct a phylogenetic tree using the Neighbor-Joining method.
4.8. Expression and purification of recombinant exo-β-(1→3)- galactanases
The coding sequences of mature SGalase1 and SGalase2 proteins were amplified by PCR, using gene-specific primers GF1, GR1, GF2 and GF2 (Table 1). Products were ligated into the Gateway entry vector pENTR/D-TOPO (Invitrogen) according to the manufacturer’s instructions, then transferred into pTOOL7, a Gateway-enabled vector derived from pET-32a(+) (Novagen) and obtained from Natalie Kibble (ACPFG, Adelaide, Australia). Constructs were transformed into Origami DE3 E. coli cells (Novagen) by heat-shock. Selected clones were grown in LB medium containing ampicillin (50 μg/mL), induced with 0.5 mM IPTG, and harvested at 3 h post-induction. Inclusion bodies containing the recombinant enzymes were purified using BugBuster Master Mix (Novagen) according to the manufacturer’s instructions, then solubilised in 8 M urea. Enzymes were renatured by dialysis against 20 mM sodium acetate buffer, pH 5. A third construct containing YFP was designed and analysed alongside SGalase 1 and SGalase 2 to ensure that any activity obtained by the recombinantly expressed protein preparations was specific to the enzymes and not an endogenous E. coli protein. The YFP preparation displayed no enzyme activity in any of the assays used.
4.9. pH and temperature optimum
All buffers were prepared with reference to Dawson et al.63 Glycine–HCl buffer (0.05 M) for pH 2.2, 2.6 and 3, sodium acetate buffer (0.1 M) for pH 3.8 and 5, MES–NaOH buffer (0.05 M) for pH 6, MOPS–KOH buffer (0.05 M) for pH 7 and Tris–HCl buffer (0.05 M) for pH 8. The activity of the enzyme in the different buffers was determined using the reducing sugar assay.
The temperature optimum of the purified enzyme was determined using dGA as the substrate at the following temperatures: 1 °C, 6 °C, 24 °C, 32 °C, 36.5 °C, 48 °C, 52.5 °C and 63.5 °C in glycine– HCl buffer (pH 3) as described above.
4.10. Determination of kinetic parameters
Michaelis constant (Km) and maximum velocity (Vmax) of the enzyme towards dGA at pH 3.8 and 37 °C were determined by a Lineweaver–Burk plot.64 The substrate concentration was varied over the range of 4.2–83.3 mg/mL, using 0.6 μg protein of the enzyme. The initial reaction velocity was determined by measuring the amount of reducing sugar increase per unit time using the reducing sugar assay.
4.11. Substrate specificity
The activity of the enzyme towards various substrates was measured using the reducing sugar assay. The reaction mixture contained the enzyme at a concentration of 0.51 mg/mL in TSK 55 (S) running buffer (0.05 μg or 1μg protein) and substrate (1.25 mM for methyl glycosides of β-(1→6)-linked D-galactooligosaccharides, 2.5 mM for methyl glycosides of β-(1→3)-linked D-galacto-oligosaccharides, 1.25 mg/mL for β-(1→3)-galactan and β-(1→3;1→6)-galactan or 5 mg/mL for gum arabic, larch arabinogalactan and dGA)) in sodium acetate buffer (0.1 M, pH 3.8). The total reaction mixture (50 μL for those that contain methyl β-glycosides of β-(1→3)-linked D-galacto-oligosaccharides and dGA or 25 μL for all other substrates) was incubated for 5 min, 10 min, 1 h or 24 h.
4.12. Mode of action
The mode of action of the purified enzyme on methyl βglycosides of β-(1→3)-linked D-galacto-tetra-/penta-/heptapyranosides was studied by analysing the products liberated from the substrates on a TLC plate of Silica Gel 60 (20 cm × 20 cm) with aluminium backing (Merck, Darmstadt, Germany). The reaction mixture (~33 μL final volume) consisted of the enzyme (0.6 μg protein) and substrate (5 mM, 15 μL) in sodium acetate buffer (25 mM, pH 3.8). The reaction mixture was incubated for 0, 5, 10 and 15 min at 37 °C and boiled for 5 min in a water bath to terminate the reaction. TLC was performed in a solvent mixture of 1-propanol/ethanol/water (7:1:2, v/v/v; ~75 mL). The TLC plate was air-dried and sprayed with 20% (v/v) H2SO4 in methanol, and the sugars were detected by charring the plate at 100 °C for ~5 min.
4.13. Monosaccharide analysis
Methanolysis was carried out by the method of Chaplin (1982),65 as modified by Ferguson (1992).66 Monosaccharide linkage composition was determined by GCMS of the partially methylated alditol acetates with and without prior carboxyl reduction, essentially as described by Sims and Bacic (1995)67 and Kim et al. (2006).68
4.14. MALDI-TOF MS analysis
The oligosaccharide products of enzyme hydrolysis were analysed by MALDI-TOF MS after derivatisation by acetylation to enhance sensitivity.69 Peracetylated sample (1 μL) was mixed with matrix (1 μL, 10 mg/mL dihydroxybenzoic acid in 50% acetonitrile) and 1 μL of the mixture was spotted onto a sample well of a MALDI plate and air dried. Spectra were obtained at 50 shots/spectrum on a MALDI-TOF MS (Applied Biosystems Voyager DE-STR System 4180, Foster City, CA, USA) in delayed (200 ns) reflector mode at positive polarity with an accelerating voltage of 20 kV and grid voltage of 69%.
Supplementary Material
Acknowledgments
Microbial isolation work for the source of enzyme was carried out by Kathryn Davis (Department of Microbiology and Immunology, University of Melbourne). All sample-derived tryptic peptide analyses were run by Dr. Siria Natera, and the MS and MS/MS data were interpreted and de novo sequenced by Ms. Kristina Ford and Dr. Siria Natera (School of Botany, The University of Melbourne). This project was supported by funds from the Co-operative Research Centre (CRC) for Bioproducts and Melbourne International Research Scholarship (MIRS).
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.carres.2012.02.033.
References
- 1.Aspinall GO. Carbohydrate Polymers of Plant Cell Walls. In: Loewus F, editor. Biogenesis of Plant Cell Wall Polysaccharides. Academic Press; New York: 1973. pp. 95–115. [Google Scholar]
- 2.Qi W, Fong C, Lamport DTA. Plant Physiol. 1991;96:848–855. doi: 10.1104/pp.96.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodrum LJ, Patel A, Leykam JF, Kieliszewski MJ. Phytochemistry. 2000;54:99–106. doi: 10.1016/s0031-9422(00)00043-1. [DOI] [PubMed] [Google Scholar]
- 4.Clarke AE, Anderson RL, Stone BA. Phytochemistry. 1979;18:521–540. [Google Scholar]
- 5.Churms SC, Merrifield EH, Stephen AM. Carbohydr Res. 1983;123:267–279. doi: 10.1016/s0008-6215(00)85676-1. [DOI] [PubMed] [Google Scholar]
- 6.Fincher GB, Stone BA, Clarke AE. Annu Rev Plant Physiol. 1983;34:47–70. [Google Scholar]
- 7.Nothnagel EA. Int Rev Cytol. 1997;174:195–291. doi: 10.1016/s0074-7696(08)62118-x. [DOI] [PubMed] [Google Scholar]
- 8.Sun W, Xu J, Yang J, Kieliszewski MJ, Showalter AM. Plant Cell Physiol. 2005;46:975–998. doi: 10.1093/pcp/pci106. [DOI] [PubMed] [Google Scholar]
- 9.Osman ME, Menzies AR, Williams PA, Phillips GO, Baldwin TC. Carbohydr Res. 1993;246:303–318. [Google Scholar]
- 10.Osman ME, Menzies AR, Martin BA, Williams PA, Phillips GO, Baldwin TC. Phytochemistry. 1995;38:409–417. [Google Scholar]
- 11.Tischer CA, Gorin PAJ, Iacomini M. Carbohydr Polym. 2002;47:151–158. [Google Scholar]
- 12.Tischer CA, Iacomini M, Gorin PAJ. Carbohydr Res. 2002;337:1647–1655. doi: 10.1016/s0008-6215(02)00023-x. [DOI] [PubMed] [Google Scholar]
- 13.Ponder GR, Richards GN. Carbohydr Polym. 1997;34:251–261. [Google Scholar]
- 14.Brillouet J-M, Williams P, Will F, Muller G, Pellerin P. Carbohydr Polym. 1996;29:271–275. [Google Scholar]
- 15.Saulnier L, Brillouet J-M, Moutounet M, du Penhoat CH, Michon V. Carbohydr Res. 1992;224:219–235. doi: 10.1016/0008-6215(92)84108-5. [DOI] [PubMed] [Google Scholar]
- 16.Tsumuraya Y, Hashimoto Y, Yamamoto S. Carbohydr Res. 1987;161:113– 126. [Google Scholar]
- 17.Tsumuraya Y, Ogura K, Hashimoto Y, Mukoyama H, Yamamoto S. Plant Physiol. 1988;86:155–160. doi: 10.1104/pp.86.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bacic A, Churms SC, Stephen AM, Cohen PB, Fincher GB. Carbohydr Res. 1987;162:85–93. [Google Scholar]
- 19.Kieliszewski MJ, Dezacks R, Leykam JF, Lamport DTA. Plant Physiol. 1992;98:919–926. doi: 10.1104/pp.98.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieliszewski MJ. Phytochemistry. 2001;57:319–323. doi: 10.1016/s0031-9422(01)00029-2. [DOI] [PubMed] [Google Scholar]
- 21.Shpak E, Leykam JF, Kieliszewski MJ. Proc Natl Acad Sci USA [PNAS] 1999;96:14736–14741. doi: 10.1073/pnas.96.26.14736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shpak E, Barbar E, Leykam JF, Kieliszewski MJ. J Biol Chem. 2001;276:11272–11278. doi: 10.1074/jbc.M011323200. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Tan L, Lamport DTA, Showalter AM, Kieliszewski MJ. Phytochemistry. 2008;69:1631–1640. doi: 10.1016/j.phytochem.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Estévez JM, Kieliszewski MJ, Khitrov N, Somerville C. Plant Physiol. 2006;142:458–470. doi: 10.1104/pp.106.084244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svetek J, Yadav MP, Nothnagel EA. J Biol Chem. 1999;274:14724–14733. doi: 10.1074/jbc.274.21.14724. [DOI] [PubMed] [Google Scholar]
- 26.Johnson KL, Jones BJ, Schultz CJ, Bacic A. Non-Enzymic Cell Wall (Glycol) Proteins. In: Rose JCK, editor. The Plant Cell Wall: Annual Plant Reviews. Vol. 8. Blackwell Publishing; Australia: 2003. pp. 111–154. [Google Scholar]
- 27.van Hengel AJ, van Kammen Ab, de Vries SC. Physiol Plantarum. 2002;114:637–644. doi: 10.1034/j.1399-3054.2002.1140418.x. [DOI] [PubMed] [Google Scholar]
- 28.Letarte J, Simion E, Miner M, Kasha KJ. Plant Cell Rep. 2006;24:691–698. doi: 10.1007/s00299-005-0013-5. [DOI] [PubMed] [Google Scholar]
- 29.Putoczki TL, Pettolino F, Griffin MDW, Möller R, Gerrard JA, Bacic A, Jackson SL. Planta. 2007;226:1131–1142. doi: 10.1007/s00425-007-0559-2. [DOI] [PubMed] [Google Scholar]
- 30.Lee KJD, Sakata Y, Mau S-L, Pettolino F, Bacic A, Quatrano RS, Knight CD, Knox JP. Plant Cell. 2005;17:3109–3112. doi: 10.1105/tpc.105.034413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brillouet J-M, Williams P, Moutounet M. Agric Biol Chem. 1991;55:1565– 1571. [Google Scholar]
- 32.Menestrina JM, Iacomini M, Jones C, Gorin PAJ. Phytochemistry. 1998;47:715–721. doi: 10.1016/s0031-9422(97)00666-3. [DOI] [PubMed] [Google Scholar]
- 33.Pellerin P, Brillouet J-M. Carbohydr Res. 1994;264:281–291. doi: 10.1016/s0008-6215(05)80012-6. [DOI] [PubMed] [Google Scholar]
- 34.Hirano Y, Tsumuraya Y, Hashimoto Y. Physiol Plantarum. 1994;92:286–296. [Google Scholar]
- 35.Haque MA, Kotake T, Tsumuraya Y. Biosci Biotechnol Biochem. 2005;69:2170–2177. doi: 10.1271/bbb.69.2170. [DOI] [PubMed] [Google Scholar]
- 36.Li S-C, Han J-W, Chen K-C, Chen C-S. Phytochemistry. 2001;57:349–359. doi: 10.1016/s0031-9422(01)00022-x. [DOI] [PubMed] [Google Scholar]
- 37.Tsumuraya Y, Mochizuki N, Hashimoto Y, Kováč P. J Biol Chem. 1990;265:7207–7215. [PubMed] [Google Scholar]
- 38.Okemoto K, Uekita T, Tsumuraya Y, Hashimoto Y, Kasama T. Carbohydr Res. 2003;338:219–230. doi: 10.1016/s0008-6215(02)00405-6. [DOI] [PubMed] [Google Scholar]
- 39.Kotake T, Kaneko S, Kubomoto A, Haque MA, Kobayashi H, Tsumuraya Y. Biochem J. 2004;377:749–755. doi: 10.1042/BJ20031145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotake T, Hirata N, Degi Y, Ishiguro M, Kitazawa K, Takata R, Ichinose H, Kaneko S, Igarashi K, Samejima M, Tsumuraya Y. J Biol Chem. 2011;286:27848–27854. doi: 10.1074/jbc.M111.251736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchler-Bauer A, Anderson JB, Derbyshire MK, et al. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ichinose H, Kotake T, Tsumuraya Y, Kaneko S. Biosci, Biotechnol, Biochem. 2006;70:2745–2750. doi: 10.1271/bbb.60365. [DOI] [PubMed] [Google Scholar]
- 43.Ling NX-Y. PhD Thesis. The University of Melbourne; 2008. Isolation and characterization of an exo-β-(1→3)-D-galactanase from Streptomyces sp. with activity on plant arabinogalactan-proteins. [Google Scholar]
- 44.Ling NX-Y, Pettolino F, Liao M-L, Bacic A. Carbohydr Res. 2009;344:1941–1946. doi: 10.1016/j.carres.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 45.Ichinose H, Kuno A, Kotake T, Yoshida M, Sakka K, Hirabayashi J, Tsumuraya Y, Kaneko S. Appl Environ Microbiol. 2006;72:3515–3523. doi: 10.1128/AEM.72.5.3515-3523.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ichinose H, Yoshida M, Kotake T, Kuno A, Igarashi K, Tsumuraya Y, Samejima M, Hirabayashi J, Kobayashi H, Kaneko S. J Biol Chem. 2005;280:25820–25829. doi: 10.1074/jbc.M501024200. [DOI] [PubMed] [Google Scholar]
- 47.Hashimoto Y, Tsujisaka Y, Fukumoto J. Nogeikagaku Zasshi. 1969;43:831– 836. [Google Scholar]
- 48.Sekimata M, Ogura K, Tsumuray Y, Hashimoto Y, Yamamoto S. Plant Physiol. 1989;90:567–574. doi: 10.1104/pp.90.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kotake T, Dina S, Konishi T, Kaneko S, Igarashi K, Samejima M, Watanabe Y, Kimura K, Tsumuraya Y. Plant Physiol. 2005;138:1563–1576. doi: 10.1104/pp.105.062562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coutinho PM, Henrissat B. Carbohydrate-Active Enzymes. 1999 Server at URL: http://afmb.cnrs-mrs.fr/~pedro/CAZY/db/html.
- 51.Al-Assaf S, Katayama T, Phillips GO, Sasaki Y, Williams P. FFI J. 2003;208:771–780. [Google Scholar]
- 52.Tan L, Xu J, Lamport D, Qiu F, Cottrell C, Qian J, Kieliszewski M. Physiol Plantarum. 2007;130:Abstract 68. [Google Scholar]
- 53.Sait M, Hugenholz P, Janssen PH. Environ Microbiol. 2002;4:654–666. doi: 10.1046/j.1462-2920.2002.00352.x. [DOI] [PubMed] [Google Scholar]
- 54.Tschech A, Pfennig N. Arch Microbiol. 1984;137:163–167. [Google Scholar]
- 55.Widdel F, Kohring GW, Mayer F. Arch Microbiol. 1983;134:286–294. [Google Scholar]
- 56.Kováč P, Taylor RB, Glaudemans CPJ. J Org Chem. 1985;50:5323–5333. [Google Scholar]
- 57.Kováč P. Carbohydr Res. 1986;153:237–251. doi: 10.1016/s0008-6215(00)90266-0. [DOI] [PubMed] [Google Scholar]
- 58.Nelson N. J Biol Chem. 1944;153:375–380. [Google Scholar]
- 59.Somogyi M. J Biol Chem. 1952;195:19–23. [PubMed] [Google Scholar]
- 60.Natera SHA, Ford KL, Cassin AM, Patterson JH, Newbigin EJ, Bacic A. J Proteome Res. 2008;7:1159–1187. doi: 10.1021/pr070255c. [DOI] [PubMed] [Google Scholar]
- 61.Zheng S-J, Henken B, Sofiari E, Jacobsen E, Krens FA, Kik C. Transgenic Res. 2001;10:237–245. doi: 10.1023/a:1016633410041. [DOI] [PubMed] [Google Scholar]
- 62.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dawson RM, Elliott DC, Elliott WH, Jones KM. pH, Buffers, and Physiological Media. 3. Chapter 18 Clarendon Press; Oxford, USA: 1986. Data for Biochemical Research; pp. 417–448. [Google Scholar]
- 64.Segel IH. Enzymes. 2. Chapter 4 John Wiley & Sons; New York: 1976. Biochemical Calculations: How to Solve Mathematical Problems in General Biochemistry; pp. 208–323. [Google Scholar]
- 65.Chaplin MF. Anal Biochem. 1982;123:336–341. doi: 10.1016/0003-2697(82)90455-9. [DOI] [PubMed] [Google Scholar]
- 66.Ferguson MAJ. Chemical and Enzymic Analysis of Glycosyl- Phopatidylinositol Anchors. In: Hooper NM, Turner AJ, editors. Lipid Modification of Proteins: A Practical Approach. Oxford University Press; 1992. pp. 191–230. [Google Scholar]
- 67.Sims IM, Bacic A. Phytochemistry. 1995;38:1397–1405. [Google Scholar]
- 68.Kim JS, Reuhs BL, Michon F, Kaiser RE, Arumugham RG. Carbohydr Res. 2006;341:1061–1064. doi: 10.1016/j.carres.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 69.Dell A, Chalabi S, Hitchen PG, Jang-Lee J, Ledger V, North SJ, Pang P-C, Parry S, Sutton-Smith M, Tissot B, Morris HR, Panico M, Haslam SM. Mass Spectrometry of Glycoprotein Glycans: Glycomics and Glycoproteomics. In: Kamerling JP, Boons G-J, Lee YC, Suzuki A, Taniguchi N, Voragen AGJ, editors. Comprehensive Glycoscience: from Chemistry to System Biology. Vol. 2. Elsevier; Amsterdam: 2007. pp. 69–93. Analysis of Glycans Polysaccharide Functional Properties. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.