Abstract
Background:
Response to radiotherapy varies between individuals both in terms of efficacy and adverse reactions. Finding genetic determinants of radiation response would allow the tailoring of the treatment, either by altering the radiation dose or by surgery. Despite a growing number of studies in radiogenomics, there are no well-replicated genetic association results.
Methods:
We carried out a candidate gene association study and replicated the result using three additional large cohorts, a total of 2036 women scored for adverse reactions to radiotherapy for breast cancer.
Results:
Genetic variation near the tumour necrosis factor alpha gene is shown to affect several clinical endpoints including breast induration, telangiectasia and overall toxicity. In the combined analysis homozygosity for the rare allele increases overall toxicity (P=0.001) and chance of being in the upper quartile of risk with odds ratio of 2.46 (95% confidence interval 1.52–3.98).
Conclusion:
We have identified that alleles of the class III major histocompatibility complex region associate with overall radiotherapy toxicity in breast cancer patients by using internal replication through a staged design. This is the first well-replicated report of a genetic predictor for radiotherapy reactions.
Keywords: breast cancer, normal tissue radiation injury, genetics, tumour necrosis factor alpha (TNFα).
Radiotherapy is an important treatment modality for local tumour control and has contributed to declining mortality rates, for example, in breast cancer. Most patients show either no reaction or only transient skin reactions to radiation treatment, but a minority show life-long adverse reactions including vascular damage and fibrosis (Bentzen et al, 2003). Depending upon the tissue irradiated these reactions can lead to severe and sometimes life-threatening complications such as pulmonary fibrosis or oesphagitis (O’Sullivan and Levin, 2003). In low survival rate cancers there would be a benefit to identifying patients resistant to radiotherapy reactions to target for higher radiation doses, whereas in high survival rate tumour types such as breast cancer, the drive is to identify patients sensitive to adverse reactions, with the aim of offering alternative treatment. In breast cancer patients could be offered mastectomy instead of wide local excision followed by radiotherapy.
The variation in clinical response to radiotherapy is partly explained by treatment factors such as radiation dose, medical history or body type, and with an unknown contribution from genetic factors. Direct estimation of the heritability of clinical radiosensitivity is difficult to achieve, but is likely to be somewhat lower than for chromosomal and cellular radiosensitivity, which have been calculated as 58–78% (Finnon et al, 2008; Curwen et al, 2010).
Acute reactions to radiotherapy are either inflammation-related or through target cell depletion, and predict late reactions especially fibrosis (Barnett et al, 2009a). Broadly, late adverse reactions fall into two different pathophysiological categories namely vascular damage and fibroblast proliferation. Genes that affect processes early in DNA repair or inflammation pathways may be expected to lead to a wide range of late reactions, with other gene effects being more specific for damage to particular tissues or manifestations.
Identified predictors of radiosensitivity are aimed to ultimately contribute to an algorithm for guiding oncology treatment decisions, but there is no unanimity in the field about the most useful output measure of clinical radiosensitivity. Possibilities would include estimating the risk that an individual would develop late reactions with a particular score on one of the commonly used clinical scales (e.g., Common Terminology Criteria for Adverse Events or Late Effects of Normal Tissue-Subjective Objective Management Analytical (LENT-SOMA)), a score of combined toxicity (e.g., Standardized Total Average Toxicity (STAT) score (Barnett et al, 2011b)) or for being in a high risk group (e.g., being in the upper quartile of overall toxic reactions).
Previous studies in radiogenomics have focussed on a wide range of candidate genes, but no genetic association results have yet proved replicable (Andreassen and Alsner, 2009, Barnett et al, 2009a, 2011a). More recently researchers have moved towards genome-wide association studies (GWAS) to overcome the well-documented problems with the candidate gene approach, and these are coming to fruition (Kerns et al, 2010). GWAS are very effective at detecting association with common causative alleles, with their power determined mainly by cohort size. Whether association results derive from candidate or genome-wide approaches, there is a pressing need for within-experiment replication through either staged design or combined analyses.
In this paper we describe a genetic association study used a three-staged design to ensure internal replication.
Materials and methods
Description of cohorts
LeND cohort
This study had been undertaken with the written consent of patients attending the oncology departments or breast units from three hospitals in the East Midlands region of England; the University Hospitals of Leicester–Glenfield Hospital (n=566; 89.4%), the Nottingham University Hospitals–City Hospital Ca mpus (n=35; 5.5%) and the Derby Hospitals–Royal Derby Hospital (n=32; 5.0%). In total, 633 women with a unilateral or bilateral, histologically confirmed early breast cancer (T1-3, N0-1, M0 at presentation) or ductal carcinoma in situ (DCIS) receiving adjuvant breast or chest wall irradiation after complete macroscopic tumour excision by breast-conserving surgery (n=493) or mastectomy (no reconstruction; n=140) were recruited from the follow-up clinics. Radiotherapy was given with 6–10 MV photons using tangential opposed fields at a variety of dose-fractionation schedules, but for most patients 50 Gy in 25 fractions over 5 weeks prescribed to the ICRU reference point. Boost irradiation with 9–15 Gy in 3–5 fractions was given to women with poor prognostic features. The sample collection and study were carried out with local and national ethics approval. Adverse effects of radiotherapy were scored on the LENT-SOMA scale with a median follow-up time of 62 months. Further details of the LeND cohort are as described previously (Giotopoulos et al, 2007, 2008; Tanteles et al, 2009; Murray et al, 2011).
German cohorts: ISE and MARIERAD
For both the ISE and the MARIERAD study populations, histologically confirmed early breast cancer or in situ patients were recruited from the Rhine-Neckar-Karlsruhe region in Germany. Date of breast cancer diagnosis was between 1998 and 2001 for the ISE study (Lilla et al, 2007), and between 2002 and 2005 for the MARIE study, from which patients for the MARIERAD study were drawn (Flesch-Janys et al, 2008). Briefly, breast cancer patients were eligible if they were treated unilaterally with radiotherapy(but not with chemotherapy) after breast-conserving surgery. For the ISE study at three sites the radiotherapy was given to the whole breast, either 50 Gy in 25 fractions or 50.4 Gy in 28 fractions, followed by a photon or electron boost with doses ranging from 5 to 20 Gy. At the fourth site, patients received 56 Gy in 28 fractions without boost. In the MARIERAD study, radiotherapy was applied as whole-breast irradiation, followed by additional boost irradiation of the tumour bed for three quarters of patients. Whole-breast irradiation was either applied in daily doses of 1.8 Gy (28 fractions) or 2.0 Gy (25 fractions), summing up to a total whole-breast irradiation dose predominantly between 50.0 and 56.0 Gy. Median total irradiation dose applied including boost irradiation was 60.4 Gy. Two endpoints were assessed: skin alterations/telangiectasia and fibrosis at the irradiated breast. Late adverse effects were documented by a study physician according to a standard protocol using the RTOG/EORTC scoring, ranging from 0=no late adverse effects to 4=severe adverse effects. Late adverse effects were defined as ⩾grade 2. Median follow-up time was 51 months for the ISE study and 68 months for the MARIERAD study. For the ISE study, genotype data were available for 390 of the 418 patients with follow-up data who were treated with conventional radiotherapy (three patients treated with interstitial boost were omitted from the analysis). After exclusion of 27 patients who received intraoperative or interstitial boost irradiation in the MARIERAD study, 363 of 387 patients treated with conventional radiotherapy were included in the genotype analysis. Written informed consent was obtained and approval from the Local Ethics Committee.
RAPPER
The RAPPER study (UKCRN1471) is a large UK sample collection study, opened in 2005, which recruits patients from clinical trials and other well-designed studies. All patients in the Cambridge Intensity Modulated Radiotherapy (IMRT) and Manchester prospective trials were offered recruitment to RAPPER when they enroled in the component study; blood was taken for RAPPER before radiotherapy. Toxicity data for all patients were collected prospectively within the component clinical trial. RAPPER is approved by the Cambridgeshire 2 Research Ethics Committee (05/Q0108/365). All patients gave written informed consent that their samples could be used for genetic research.
Samples were obtained from 942 of the 1145 women recruited into the Cambridge Breast IMRT Trial (ISRCTN21474421) who underwent conservative surgery followed by adjuvant radiotherapy (Barnett et al, 2009b, 2011). All patients were treated to a dose of 40 grey (Gy) in 15 fractions, 5 days a week over 3 weeks with 6 MV photons prescribed to the ICRU 50 reference point. In this study, patients with significant dose inhomogeneities, defined by a volume of 2 cm3 or more exceeding 107% of the prescribed dose, were randomised to either standard breast RT (control arm) or to a simple method of forward-planned IMRT (interventional arm). A total of 34 samples were from patients enroled in a prospective study of breast toxicity in women who received conservative surgery and adjuvant radiotherapy at the Christie Hospital, Manchester. Samples were obtained from 63 patients recruited to the intensity modulated and partial organ radiotherapy low trial of partial breast radiotherapy (Coles et al, 2006). Samples were obtained from 179 patients from the Radiation Complications and Epidemiology (RACE) study (Martin et al, 2010). The RACE study recruited 82 cases from the Royal Marsden Hospital/Gloucestershire Oncology Centre (RMH/GOC) Breast Fractionation trial (Yarnold et al, 2005) and the RMH Breast Radiotherapy Dosimetry trial (Donovan et al, 2007) with marked changes in photographic appearance and 108 controls with no evidence of radiation-induced change in breast appearance.
The Cambridge Breast IMRT trial had as primary end point photographic assessment of late cosmetic effects. Breast shrinkage changes were recorded on a three-point scale (none/minimal=1, mild=2 and marked=3) by three observers comparing baseline and 2-year photographs and generating a consensus score. Clinical assessment was made by a trained specialist radiographer with a 2-year follow-up time after radiotherapy. The breast was examined after treatment for telangiectasia, oedema, change in pigmentation and palpable induration. Induration of the breast was defined as hardening of the tissue and was used to assess fibrosis. Each of the secondary end points was scored 0–3 (none, a little, quite a bit and very much) on the scale used in the START trials. Pigmentation change was scored from 0 to 2 according to the LENT-SOMA scale.
Genotyping
The LeND cohort was genotyped by using SNPlex technology (Applied Biosystems, Foster City, CA, USA). In all, 5 SNPs failed genotyping (TGF-β1 rs2241718, RAD9A rs2286620, exp ACVR2A rs286385, CDC25A rs3731487 and RAD9A rs91757), and 43 SNPs were successfully genotyped (Table 1). Tests for deviation from Hardy–Weinberg equilibrium showed that none of the 43 assays had a P-value <0.01, and only 5 had a P-value <0.1 (close to the average expected number of 4.3).
Table 1. Genotyped SNPs and associated genes.
| Gene | Chr | Position | Pathway | SNP |
|---|---|---|---|---|
| APEX | 14 | 19 994 994 | DNA repair | rs1130409 |
| ATM | 11 | 107 730 871 | Cell cycle | rs664143 |
| ATR | 3 | 143 764 302 | Cell cycle | rs2227928 |
| AXIN2 | 17 | 60 979 143 | Wnt signalling | rs11079571 |
| CCND1 | 11 | 69 170 912 | Cell cycle | rs647451 |
| CDKN2A (p16) | 9 | 21 963 412 | Cell cycle | rs7036656 |
| CHK1 | 11 | 125 025 731 | Cell cycle | rs567889 |
| CHK2 | 22 | 29 132 990 | Cell cycle | rs5762764 |
| CTGF | 6 | 132 273 257 | Growth factor | rs6918698 |
| ERCC2 (XPD) | 19 | 45 854 919 | DNA repair (NER) | rs13181 |
| ERCC4 | 16 | 13 922 582 | DNA repair (NER) | rs744154 |
| ERCC5 | 13 | 102 326 003 | DNA repair (NER) | rs17655 |
| exp ACVR2A (AXIN1) | 16 | 316 781 | TGF-β | rs7195617 |
| exp E2F5 | 12 | 364 160 | TGF-β | rs1860360 |
| exp ID2 | 7 | 108 617 064 | TGF-β | rs10953597 |
| exp ID3 | 2 | 168 930 735 | TGF-β | rs6717927 |
| exp SMAD1 | 18 | 62 060 696 | TGF-β | rs1873481 |
| FGFR2 | 10 | 123 352 317 | Growth factor | rs2981582 |
| GSTP1 | 11 | 67 109 265 | Detoxification | rs1695 |
| IER2 | 19 | 12 836 792 | Growth factor | rs1042164 |
| IL12RB2 | 1 | 67 836 060 | Inflammation | rs3790568 |
| KLF4 | 9 | 109 289 326 | Cell cycle | rs2236599 |
| LIG3 | 17 | 30 355 688 | DNA repair (BER) | rs1052536 |
| LIG3 | 17 | 30 313 159 | DNA repair (BER) | rs3744355 |
| LIG3 | 17 | 30 358 225 | DNA repair (BER) | rs3744357 |
| NEK11 | 3 | 130 947 435 | Cell cycle | rs3738000 |
| PTTG1 | 5 | 159 775 043 | Cell cycle | rs2910190 |
| PTTG1 | 5 | 159 786 856 | Cell cycle | rs2961951 |
| PTTG1 | 5 | 159 779 450 | Cell cycle | rs3811999 |
| RAD21 | 8 | 117 938 641 | DNA repair | rs16888927 |
| RAD21 | 8 | 117 945 589 | DNA repair | rs16888997 |
| SOD2 | 6 | 160 033 862 | Oxidative stress | rs4880 |
| TGFB1 | 19 | 41 865 643 | TGF-β | rs11083616 |
| TGFB1 | 19 | 41 845 801 | TGF-β | rs11466338 |
| TGFB1 | 19 | 41 851 509 | TGF-β | rs4803455 |
| TGFBR2 | 3 | 30 637 332 | TGF-β | rs1036095 |
| TGFBR2 | 3 | 30 643 688 | TGF-β | rs4522809 |
| TNFα | 6 | 31 651 010 | Cytokine | rs1800629 |
| VEGF | 6 | 43 846 328 | Growth factor | rs2010963 |
| XRCC1 | 19 | 48 747 566 | DNA repair (BER) | rs25487 |
| XRCC3 | 14 | 103 235 680 | DNA repair (HR) | rs1799796 |
| XRCC4 | 5 | 82 684 699 | DNA repair (NHEJ) | rs1805377 |
| XRCC5 (KU80) | 2 | 216 774 882 | DNA repair (NHEJ) | rs3835 |
Abbreviations: SNP=single nucleotide polymorphism; TGF=transforming growth factor.
Genes preceded by ‘exp’ indicate that the SNP was found to associate with expression of the gene in Smirnov et al, 2009, but is not physically in the gene.
For both German study populations, the tumour necrosis factor alpha (TNFα) SNP rs1800629 was genotyped by using iPlex application (Sequenom, SanDiego, CA, USA). There was no deviation from Hardy–Weinberg in both study populations.
In the RAPPER cohort the rs2857595 SNP was genotyped on an Illumina CytoSNP 12 array (Illumina Inc., San Diego, CA, USA). Genotype calling was performed with the GenCall software application to automatically cluster, call genotypes and assign confidence scores. The GenCall application incorporates a clustering algorithm (GenTrain) and a calling algorithm (GenCall software, Illumina Inc.).
Statistical analysis
Phenotypes used in this study were the residuals from linear regression incorporating known predictors on a study-specific basis into the model. For the LeND cohort the telangiectasia regression covariates were type of surgery, radiation boost and bra cup size. The STAT score incorporated induration, telangiectasia, oedema and atrophy. Standardized Total Average Toxicity regression covariates were surgery and bra cup size. For the German cohorts the telangiectasia regression covariates were as follows: BMI, radiation dose at the skin and clinic. The fibrosis regression covariates were as follows: BMI, radiation dose at the skin and bra cup size. The STAT score incorporated telangiectasia and fibrosis. The STAT regression covariates wereas follows: bra cup, smoking pack years, BMI and dose at the skin. For the RAPPER cohorts STAT incorporates telangiectasia, oedema, shrinkage, pigmentation and pain. The STAT regression covariates were as follows: breast volume, smoking, diabetes, post-op infection and acute score+boost.
Clinical endpoints were combined within and between cohorts by conversion to Z scores, i.e., number of s.d.s from the cohort mean and then averaging the Z scores to generate the STAT score (Barnett et al, 2011b). To enable bivariate analyses an arbitrary breakpoint was chosen by defining ‘cases’ as being in the upper quartile of risk (i.e., the 25% of patients with the highest unexplained adverse reactions).
Statistical analysis used SPSS v16.0 (IBM software, Armonk, NY, USA). Genetic analysis was carried out using the Plink v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) and SimHap v1.02 (http://www.genepi.meddent.uwa.edu.au/ software/simhap). For calculation of empirical P-values 20 000 permutations were performed.
Results
LeND cohort genotyping
In all, 43 candidate SNPs were genotyped in 35 genes, most of them as putatively affecting gene expression or protein function directly, or by trans effects on expression of TGFβ pathway genes. The latter SNPs were derived from unpublished data in Smirnov et al, 2009 provided by the authors. The direct candidate genes were drawn mainly from DNA repair, cell cycle and TGFβ pathways (Table 1).
Polymorphisms were assessed for association with two specific endpoints, fibrosis and telangiectasia, and for overall toxicity as measured by STAT score. All three phenotypes were the residuals from linear regression with known predictors included in the model (see Methods). Multiple testing was corrected for by permutation of the data. The most significant pointwise P-value was for telangiectasia with the minor A allele of rs1800629 in the TNFα gene (P=0.0028), but which was not significant experiment wide as calculated by permutation (P=0.13). The mode of inheritance observed from these data is intermediate between additive and recessive, as evidenced by the heterozygote AG telangiectasia score being between the GG homozygote score and the mid-point between the homozygotes (mid-point=+0.27; Table 2). The only other SNP with pointwise significant association with overall toxicity was rs3738000, a coding polymorphism in NEK11 (p.E488V; pointwise P=0.04, empirical P=0.83).
Table 2. s1800629 in LeND cohort.
| Genotype | GG | AG | AA |
|---|---|---|---|
| Count | 197 | 128 | 15 |
| Telangiectasia score Mean (25%, 75%) | −0.07 (−0.43, −0.11) | +0.07 (−0.27, +0.05) | +0.29 (−0.19, +0.57) |
| STAT score Mean (25%, 75%) | 0.00 (−0.43, +0.27) | −0.02 (−0.44, +0.26) | +0.26 (−0.03, +0.65) |
Abbreviation: STAT=Standardized Total Average Toxicity.
Telangiectasia and STAT scores are residuals calculated from regression of clinical endpoints on known predictive factors. Standardized Total Average Toxicity score is a measure of the overall toxicity calculated by combining clinical endpoints.
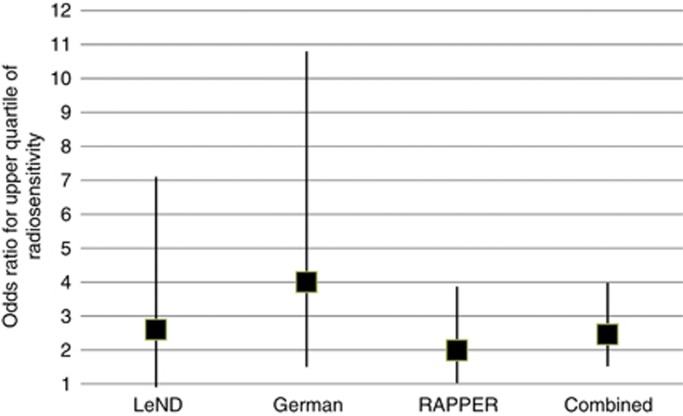
The rare allele of the rs1800629 TNFα SNP is only marginally associated with fibrosis in the LeND cohort, but is significantly associated with overall toxicity (Mann–Whitney P=0.02). For risk of being in the top quartile of adverse reactions, homozygosity for the rare allele shows marginal association (odds ratio 2.6, 95% confidence interval (CI) 0.9–7.1; Figure 1).
Figure 1.
Plot showing odds ratios under a recessive model (genotype AA vs AG, GG) for being in upper quartile of adverse reactions to radiotherapy by cohort. Square shows central estimate of odds ratio, whiskers shows 95% confidence intervals.
Replication in German cohorts
To follow-up the suggestive association of rs1800629, we genotyped two German cohorts for the same SNP. Residual telangiectasia was associated with the rs1800629 genotype under a recessive model in the two cohorts combined (P=0.01; Table 3). Furthermore fibrosis was also significantly raised in AA homozygotes (P=0.02), and therefore overall toxicity, as measured by STAT score, was also increased (P=0.02). Regression analysis shows that rs1800629 genotype accounts for 0.6% of the phenotypic variance of STAT score in the German cohorts, with all known predictive factors accounting for 9.4% of phenotypic variance. If we define the patients with serious adverse reactions as being the top quartile, then being homozygote for rs1800629 gives an increased risk for being in this group with an odds ratio of 4.0 (95% CI 1.5–10.8; Figure 1).
Table 3. rs1800629 in German cohorts.
| Genotype | GG | AG | AA |
|---|---|---|---|
| Count | 522 | 210 | 16 |
| Telangiectasia score Mean (25%, 75%) | −0.024 (−0.728, +0.764) | +0.030 (−0.729, +0.910) | +0.490 (−0.244, +1.185) |
| Fibrosis score Mean (25%, 75%) | −0.054 (−1.006, +0.455) | +0.108 (−0.958,+0.486) | +0.421 (−0.370, +1.235) |
| STAT score Mean (25%, 75%) | −0.038 (−0.694, +0.580) | +0.061 (−0.640, +0.823) | +0.517 (+0.092, +1.214) |
Abbreviation: STAT=Standardized Total Average Toxicity.
Telangiectasia, fibrosis and STAT scores are residuals calculated from regression of clinical endpoints on known predictive factors. Standardized Total Average Toxicity score is a measure of overall toxicity calculated by combining clinical endpoints.
Replication in RAPPER cohort
For a second replication we used data from the RAPPER consortium. Data on rs1800629 were lacking and we therefore selected a SNP with the strongest linkage disequilibrium to rs1800629 as calculated from the HapMap data, which is rs2857595 (D’=0.95, R2=0.86). rs2857595 is an intergenic SNP that is 25.7 kb from rs1800629, lying between the NCR3 and AIF1 genes.
Under a recessive model rs2857595 is significantly associated with STAT score in the breast cancer RAPPER cohort (P=0.01; Table 4). For risk of being in upper STAT quartile for rs2857595 homozygotes odds ratio is 1.99 (95% CI 1.03–3.87; Figure 1).
Table 4. rs2857595 in RAPPER breast cohort.
| Genotype | GG | AG | AA |
|---|---|---|---|
| Count | 625 | 284 | 39 |
| STAT score Mean (25%, 75%) | −0.041 (−0.710, +0.410) | +0.038 (−0.640, +0.555) | +0.157 (−0.635, +0.795) |
Abbreviation: STAT=Standardized Total Average Toxicity.
Combined analysis
A combined analysis of association between STAT score and homozygosity for the rare allele of the typed SNP in 2036 women from all four cohorts combined gives a Mann–Whitney P-value of 0.001. For risk of being in the top quartile of radiotherapy toxicity, the P-value is 1.5 × 10−4 with an odds ratio of 2.46 (95% CI 1.52–3.98; Figure 1).
Discussion
The purpose of finding genetic loci in studies of complex diseases is normally to shed light on the underlying pathophysiology of the condition, with a long-term aim of developing therapies that target the causative genes. In therapeutic genetics, however, (e.g., pharmacogenetics and radiogenetics), the primary aim is to identify predictors to guide treatment, with any biological insights as secondary benefits. To be realistic components of a predictive algorithm for adverse reactions to radiotherapy, genetic effects will need to be robust in terms of replication and the central question will be their effect on an individual not a population, i.e., positive predictive value is more important than population attributable risk.
The candidate genes and SNPs tested in this study are drawn from a variety of sources, including SNPs that control radiation-dependent expression of genes in the TGFβ pathway, which is known to be central to fibrogenesis. Tumour necrosis factor alpha is a multi-functional cytokine produced by a variety of cells after injury or infection, which also increases TGFβ expression in fibroblasts (Sullivan et al, 2009). The TNFα SNP typed in the LeND and German cohorts, rs1800629, is in the promoter lying 308-bp upstream of the transcript start site, and >1 kb downstream of the lymphotoxin alpha (LTA) gene. A recent meta-analysis concluded that the G and A alleles do not significantly alter TNFα expression, although it cannot be discounted that they affect LTA expression (Mekinian et al, 2011). The rs2857595 SNP tested in the RAPPER cohort, lies in the same linkage disequilibrium block as rs1800629, being in the major histocompatibility complex (MHC) class III region.
Given that we find association with both SNPs, we are unable to determine which is the causative variation affecting radiosensitivity, or that neither are. In this study, the important question is whether we have found genetic variation that predicts radiosensitivity, and whether another polymorphism might provide higher predictive value. The MHC region is known to harbour long distance haplotypes, so it is conceivable that SNPs some considerable distance away may yield higher odds ratios. Major histocompatibility complex associations have been found for many diseases, but it remains problematic to identify the causative variation. Current radiogenomics GWAS will help in this as will developing methods for data analysis (Raychaudhuri et al, 2012).
In conclusion, we have identified that alleles of the class III MHC region associate with overall radiotherapy toxicity in breast cancer patients by using internal replication through a staged design. This is the first well-replicated report of a genetic predictor for radiotherapy reactions.
Acknowledgments
LeND cohort collection and genotyping were funded by the Breast Cancer Campaign (UK), MARIE study by the Deutsche Krebshilfe eV and the Dietmar Hopp Stiftung (Germany), ISE study by the Office for Radiation Protection in Germany, RAPPER are funded by the Cancer Research UK and acknowledge support from the Manchester Experimental Cancer Medicine Centre. The Royal Marsden NHS Foundation Trust and The Institute of Cancer Research acknowledge NHS funding to the NIHR Biomedical Research Centre. NGB and CEC are supported by the National Institute for Health Research Cambridge Biomedical Research Centre. The research collaboration leading to this paper was developed under the framework of the Radiogenomics Consortium.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
The authors declare no conflict of interest.
References
- Andreassen CN, Alsner J (2009) Genetic variants and normal tissue toxicity after radiotherapy: a systematic review. Radiother Oncol 92: 299–309 [DOI] [PubMed] [Google Scholar]
- Barnett GC, Coles CE, Elliott RM, Baynes C, Luccarini C, Conroy D, Wilkinson JS, Tyrer J, Misra V, Platte R, Gulliford SL, Sydes MR, Hall E, Bentzen SM, Dearnaley DP, Burnet NG, Pharoah PD, Dunning AM, West CM (2011a) Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: a prospective analysis study. Lancet Oncol 13(1): 65–77 [DOI] [PubMed] [Google Scholar]
- Barnett GC, West CM, Coles CE, Pharoah PD, Talbot CJ, Elliott RM, Tanteles GA, Symonds RP, Wilkinson JS, Dunning AM, Burnet NG, Bentzen SM (2011b) Standardized total average toxicity score: a scale- and grade-independent measure of late radiotherapy toxicity to Facilitate pooling of data from different studies. Int J Radiat Oncol Biol Phys 82(3): 1065–1074 [DOI] [PubMed] [Google Scholar]
- Barnett GC, West CM, Dunning AM, Elliott RM, Coles CE, Pharoah PD, Burnet NG (2009a) Normal tissue reactions to radiotherapy: towards tailoring treatment dose by genotype. Nat Rev Cancer 9: 134–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett GC, Wilkinson J, Moody AM, Wilson CB, Sharma R, Klager S, Hoole AC, Twyman N, Burnet NG, Coles CE (2009b) A randomised controlled trial of forward-planned radiotherapy (IMRT) for early breast cancer: baseline characteristics and dosimetry results. Radiother Oncol 92: 34–41 [DOI] [PubMed] [Google Scholar]
- Barnett GC, Wilkinson JS, Moody AM, Wilson CB, Twyman N, Wishart GC, Burnet NG, Coles CE (2011) Randomized controlled trial of forward-planned intensity-modulated radiotherapy for early breast cancer: interim results at 2 years. Int J Radiat Oncol Biol Phys 82(2): 715–723 [DOI] [PubMed] [Google Scholar]
- Bentzen SM, Dorr W, Anscher MS, Denham JW, Hauer-Jensen M, Marks LB, Williams J (2003) Normal tissue effects: reporting and analysis. Semin Radiat Oncol 13(3): 189–202 [DOI] [PubMed] [Google Scholar]
- Coles C, Yarnold J, Trials IMPORT, Management Group (2006) The IMPORT trials are launched (September 2006). Clin Oncol (R Coll Radiol) 18: 587–590 [DOI] [PubMed] [Google Scholar]
- Curwen GB, Cadwell KK, Winther JF, Tawn EJ, Rees GS, Olsen JH, Rechnitzer C, Schroeder H, Guldberg P, Cordell HJ, Boice JD (2010) The heritability of G2 chromosomal radiosensitivity and its association with cancer in Danish cancer survivors and their offspring. Int J Radiat Biol 86: 986–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan E, Bleakley N, Denholm E, Evans P, Gothard L, Hanson J, Peckitt C, Reise S, Ross G, Sharp G, Symonds-Tayler R, Tait D, Yarnold J, Breast Technology Group (2007) Randomised trial of standard 2D radiotherapy (RT) versus intensity modulated radiotherapy (IMRT) in patients prescribed breast radiotherapy. Radiother Oncol 82: 254–264 [DOI] [PubMed] [Google Scholar]
- Finnon P, Robertson N, Dziwura S, Raffy C, Zhang W, Ainsbury L, Kaprio J, Badie C, Bouffler S (2008) Evidence for significant heritability of apoptotic and cell cycle responses to ionising radiation. Hum Genet 123: 485–493 [DOI] [PubMed] [Google Scholar]
- Flesch-Janys D, Slanger T, Mutschelknauss E, Kropp S, Obi N, Vettorazzi E, Braendle W, Bastert G, Hentschel S, Berger J, Chang-Claude J (2008) Risk of different histological types of postmenopausal breast cancer by type and regimen of menopausal hormone therapy. Int J Cancer 123: 933–941 [DOI] [PubMed] [Google Scholar]
- Giotopoulos G, Armstrong C, Osman A, Peat I, Symonds RP, Talbot CJ (2008) Refining the evidence for GSTA1 and eNOS genetic effects on risk of radiotherapy-induced telangiectasia. Int J Cancer 123: 2973–2974 [DOI] [PubMed] [Google Scholar]
- Giotopoulos G, Symonds RP, Foweraker K, Griffin M, Peat I, Osman A, Plumb M (2007) The late radiotherapy normal tissue injury phenotypes of telangiectasia, fibrosis and atrophy in breast cancer patients have distinct genotype-dependent causes. Br J Cancer 96: 1001–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns SL, Ostrer H, Stock R, Li W, Moore J, Pearlman A, Campbell C, Shao Y, Stone N, Kusnetz L, Rosenstein BS (2010) Genome-wide association study to identify single nucleotide polymorphisms (SNPs) associated with the development of erectile dysfunction in African-American men after radiotherapy for prostate cancer. Int J Radiat Oncol Biol Phys 78: 1292–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilla C, Ambrosone CB, Kropp S, Helmbold I, Schmezer P, von Fournier D, Haase W, Sautter-Bihl ML, Wenz F, Chang-Claude J (2007) Predictive factors for late normal tissue complications following radiotherapy for breast cancer. Breast Cancer Res Treat 106: 143–150 [DOI] [PubMed] [Google Scholar]
- Martin S, Sydenham M, Haviland J, A'Hern R, Owen R, Bliss J, Yarnold J (2010) Test of association between variant tgbeta1 alleles and late adverse effects of breast radiotherapy. Radiother Oncol 97: 15–18 [DOI] [PubMed] [Google Scholar]
- Mekinian A, Tamouza R, Pavy S, Gestermann N, Ittah M, Mariette X, Miceli-Richard C (2011) Functional study of TNF-alpha promoter polymorphisms: literature review and meta-analysis. Eur Cytokine Netw 22: 88–102 [DOI] [PubMed] [Google Scholar]
- Murray RJ, Tanteles GA, Mills J, Perry A, Peat I, Osman A, Chan S, Cheung KL, Chakraborti PR, Woodings PL, Barwell JG, Symonds RP, Talbot CJ (2011) Association between single nucleotide polymorphisms in the DNA repair gene LIG3 and acute adverse skin reactions following radiotherapy. Radiother Oncol 99: 231–234 [DOI] [PubMed] [Google Scholar]
- O'Sullivan B, Levin W (2003) Late radiation-related fibrosis: pathogenesis, manifestations, and current management. Semin Radiat Oncol 13: 274–289 [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, Alfredsson L, Padyukov L, Klareskog L, Worthington J, Siminovitch KA, Bae SC, Plenge RM, Gregersen PK, de Bakker PI (2012) Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet 44: 291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG (2009) Genetic analysis of radiation-induced changes in human gene expression. Nature 459: 587–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DE, Ferris M, Nguyen H, Abboud E, Brody AR (2009) TNF-alpha induces TGF-beta1 expression in lung fibroblasts at the transcriptional level via AP-1 activation. J Cell Mol Med 13: 1866–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanteles GA, Whitworth J, Mills J, Peat I, Osman A, McCann GP, Chan S, Barwell JG, Talbot CJ, Symonds RP (2009) Can cutaneous telangiectasiae as late normal-tissue injury predict cardiovascular disease in women receiving radiotherapy for breast cancer? Br J Cancer 101: 403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnold J, Ashton A, Bliss J, Homewood J, Harper C, Hanson J, Haviland J, Bentzen S, Owen R (2005) Fractionation sensitivity and dose response of late adverse effects in the breast after radiotherapy for early breast cancer: long-term results of a randomised trial. Radiother Oncol 75: 9–17 [DOI] [PubMed] [Google Scholar]