Abstract
Background:
Colon cancer predisposition is associated with mutations in BRCA1. BRCA1 protein stability depends on binding to BARD1. In different cancers, expression of differentially spliced BARD1 isoforms is correlated with poor prognosis and decreased patient survival. We therefore suspected a role of BARD1 isoforms in colon cancer.
Methods:
We performed immunohistochemistry in 168 colorectal cancers, using four antibodies directed against differentially expressed regions of BARD1. We determined structure and relative expression of BARD1 mRNA isoforms in 40 tumour and paired normal peri-tumour tissues. BARD1 expression was correlated with clinical outcome.
Results:
BARD1 isoforms were expressed in 98% of cases and not correlated with BRCA1. BARD1 mRNA isoforms were upregulated in all tumours as compared with paired normal peri-tumour tissues. Non-correlated expression and localisation of different epitopes suggested insignificant expression of full-length (FL) BARD1. Expression of N- and C-terminal epitopes correlated with increased survival, but expression of epitopes mapping to the middle of BARD1 correlated with decreased survival. Middle epitopes are present in oncogenic BARD1 isoforms, which have pro-proliferative functions. Correlated upregulation of only N- and C-terminal epitopes reflects the expression of isoforms BARD1δ and BARD1φ.
Conclusion:
Our results suggest that BARD1 isoforms, but not FL BARD1, are expressed in colon cancer and affect its progression and clinical outcome.
Keywords: colorectal cancer, BARD1 isoforms, BRCA1, differential splicing, predictive biomarker
Survival and prognosis for colorectal cancer, the third leading cause of cancer-related death (WHO), depend on the stage of the disease at the time of diagnosis (Hewitson et al, 2007; Figueredo et al, 2008). Therefore, a better understanding of the molecular events involved in colorectal cancer onset and metastatic progression is needed for early detection and treatment (Rudmik and Magliocco, 2005).
MLH1, MSH2, β-Catenin, and p53 are important markers for the prediction of outcome and response to chemotherapy for colorectal cancer (Markowitz and Bertagnolli, 2009). A number of studies suggest a role for BRCA1 in colon cancer development. Loss of heterozygosity at chromosomal region 17q, comprising BRCA1, was found in 49% of colonic adenocarcinomas (Garcia-Patiño et al, 1998). Three-fold increase of colon cancer risk was reported for breast or ovarian cancer patients with BRCA1 mutations when compared with non-carrier patients (Ford et al, 1994; Brose et al, 2002; Kadouri et al, 2007). However, increased colon cancer risk for BRCA1 and BRCA2 mutation carriers, was found to be age dependent (Lin et al, 1999; Suchy et al, 2010), and was not confirmed in studies that did not take the patients’ age into account (Kirchhoff et al, 2004; Niell et al, 2004). BRCA1 and BARD1 also interact with hMSH2, a gene commonly associated with hereditary nonpolyposis colorectal cancer (HNPCC) (Lynch et al, 1997; Wang et al, 2001), and defects in the BRCA1-hMSH2 signalling pathway lead to the increased risk of cancer (Wang et al, 2001). These interactions might partially explain the high incidence of gynaecological tumours in HNPCC kindred, as well as the increased colon cancer susceptibility in BRCA1 kindred (Easton et al, 1995; Lynch et al, 1997).
BRCA1 acts in pathways of DNA repair and maintenance of genetic stability, and its deficiency might provide ground for carcinogenesis. The BRCA1-associated protein, BARD1 is required for most tumour suppressor functions of BRCA1 (Wu et al, 1996; Fabbro et al, 2002). The BRCA1–BARD1 heterodimer has ubiquitin ligase functions (Hashizume et al, 2001; Baer and Ludwig, 2002; Oyake et al, 2002; Morris and Solomon, 2004), specifically important for G2/M checkpoint control and genetic stability (Ouchi et al, 2004; Starita et al, 2004). Individually, BRCA1 and BARD1 have low ubiquitin ligase activities in vitro (Meza et al, 1999; Joukov et al, 2001), which implies that BRCA1 activity can be compromised not only by BRCA1 gene mutations but also by aberrant expression of BARD1 (Irminger-Finger and Jefford, 2006).
Differentially spliced and highly upregulated BARD1 isoforms were identified in breast and ovarian cancer (Li et al, 2007; Zhang et al, 2012). Most BARD1 isoforms lack the RING finger, which is required for BRCA1 interaction, but retain the BRCT domains. Aberrant BARD1 isoforms are also expressed in non-small cell lung cancer (NSCLC) and significantly correlated with decreased patient survival (Zhang et al, 2012). Furthermore, BARD1 isoforms have been shown to encode functions essential for cancer cell viability antagonising the BRCA1-BARD1 ubiquitin ligase activity (Li et al, 2007; Ryser et al, 2009; Dizin and Irminger-Finger, 2010; Zhang et al, 2012).
BARD1 isoforms were recently reported in colorectal cancer (Gautier et al, 2000), and it was suggested, based on lack of expression of an N-terminal BARD1 epitope, that lack of full-length (FL) BARD1 is a prognostic marker for poor outcome (Sporn et al, 2011). In this study, we investigated BARD1 mRNA and protein expression, by RT–PCR and immunodetection of epitopes from different regions of BARD1 in 168 colorectal cancer samples, and tested their correlation with clinical characteristics and patient outcome.
Patients and methods
Patients’ characteristics
Pathological diagnoses were made by experienced pathologists based on WHO criteria and staged according to American Joint Committee on Cancer classification. All patients were informed and compliance was obtained as well as approval of the local ethical committees. A total of 168 cases with colorectal cancer containing 20 cases from Italy and 148 cases from Germany were examined (Table 1).
Table 1. Patient characteristics of colorectal cancer.
| Samples | Germany | Cagliari | Total |
|---|---|---|---|
| Cases | 148 | 20 | 168 |
| Gender | |||
| Male | 83 | 10 | 93 |
| Female | 65 | 10 | 75 |
| Age | |||
| Range | 41–97 | 33–73 | 33–97 |
| Median | 73 | 60.5 | 71 |
| Normal (peri-tumour) | 0 | 20 | 20 |
| Tumour | 148 | 20 | 168 |
| Histology | |||
| Adenocarcinoma | 148 | 20 | 168 |
| Grade | |||
| Well differentiated | 10 | 0 | 10 |
| Moderately differentiated | 105 | 12 | 117 |
| Poorly differentiated | 32 | 6 | 38 |
| Undifferentiated | 0 | 0 | 0 |
| Unspecified | 1 | 2 | 3 |
| Tumour | |||
| T1 | 3 | 1 | 4 |
| T2 | 32 | 4 | 36 |
| T3 | 69 | 13 | 82 |
| T4 | 42 | 2 | 44 |
| TX | 2 | 0 | 2 |
| Node | |||
| N0 | 74 | 7 | 81 |
| N1 | 34 | 7 | 41 |
| N2 | 36 | 6 | 42 |
| N3 | 1 | 0 | 1 |
| NX | 3 | 0 | 3 |
| Metastasis | |||
| M0 | 101 | 15 | 116 |
| M1 | 44 | 5 | 49 |
| MX | 3 | 0 | 3 |
| Stage | |||
| I | 26 | 4 | 30 |
| II | 38 | 3 | 41 |
| III | 35 | 9 | 44 |
| IV | 44 | 4 | 48 |
| Unknown | 5 | 0 | 5 |
The sections used for immunochemical staining were tissue microarrays with tetramerous for each case. Of 148 cases, 75 had follow-up records and 73 patients had no survival data. Follow-up was from 1 to 72 months. Of the 75 patients with follow-up records, 22 were dead, 48 were lost, and 5 were still alive during last follow-up period.
Immunohistochemistry (IHC)
Formalin-fixed and paraffin-embedded 5-μm tissue sections were immunostained as described previously (Wu et al, 2006). The primary antibodies used for BARD1 detection were N19 (sc-7373, Santa Cruz Biotechnology, Santa Cruz, CA, USA) (1 : 25), PVC (1 : 100), WFS (1 : 100), and C20 (sc-7372, Santa Cruz Biotechnology) (1 : 20), which recognise epitopes in exons 1, 3, 4, and 11, respectively; the BRCA1 antibody was C20 (sc-642, Santa Cruz Biotechnology) (1 : 100). P8 antibody specific for exon 11 was raised against oligopeptide ILSRKPKPDSDVTQC at GenScript (www.genscript.com). All BARD1 antibodies and the BRCA1 antibody have been used previously (Irminger-Finger et al, 1998; Wu et al, 2006; Li et al, 2007; Zhang et al, 2012).
A selection of cases (N=8) was stained with a commercial antibody BL (A300-263, Bethyl Laboratories, Montgomery, TX, USA), mapping to exon 4, and a newly generated antibody (Zhang et al, 2012), directed against exon 11.
Expression levels of BARD1 and BRCA1 epitopes were measured semi-quantitatively. Staining was scored using intensity and percentage of the stained tumour cells as described before (Li et al, 2007; Zhang et al, 2012). The value of the staining intensity and positive cell percentage were multiplied to get the final staining score. The total staining score of each antibody is from 0 to 100. A score of 25 or less was defined as negative (‘−’), more than 25 was defined as positive (‘+’ for >25, ‘++’ for >50, and ‘+++’ for >75). For statistical analysis, only positive vs negative cases were considered, except the correlation of different antibodies staining using staining score. Four different regions were chosen for each tumour section and scored independently by three observers (YQ Zhang, L Li, and J Wu) without knowledge of clinical data.
Total RNA extraction, reverse transcription and PCR
Matched pairs of colorectal cancers and non-tumoural surrounding tissues were obtained from patients who underwent surgical resection of the tumour. Immediately after surgical specimen extraction, the colon was opened and both, tumoural tissue and normal mucosa, were collected. To preserve only the mucosal layer, a mucosectomy was performed after injecting saline solution to separate it from the submucosal layer.
RNA was isolated from frozen tissue sections using TRIzol (Invitrogen, Lucerne, Switzerland) according to manufacturer’s instruction. RT–PCR was performed to qualitatively show expression of different isoforms and to determine their structure.
Reverse transcription was performed using Promega (Madison, WI, USA) M-MLV reverse transcriptase according to manufacturer’s guidelines. A total of 2 μl of the reversed transcription reaction mixture was used for amplification of various fragments of BARD1 mRNAs with Taq polymerase (Qiagen) in a 50-μl reaction mix, according to manufacturer’s protocols.
PCR reactions were optimised semi-quantitatively for a cycle number that permits detection of all isoforms without reaching a plateau for the most abundant ones. BARD1 exon 1 to exon 11 amplification primers: forward (5′-GAGGAGCCTTTCATCCGAAG-3′), reverse (5′-CGAACCCTCTCTGGGTGATA-3′), 120 s elongation time, 56 °C annealing temperature, 35 cycles. BARD1 exon 1 to exon 4 amplification primers: forward (5′-GAGGAGCCTTTCATCCGAAG-3′), reverse (5′-ATTGCAGGCTGGGTTTGCACTGAAG-3′), 60 s elongation time, 56 °C annealing temperature, 35 cycles. PCR reactions were quantified as described (Zhang et al, 2012).
Glyceraldehyde 3 phosphate dehydrogenase (GAPDH) was amplified as internal reference – forward primer (5′-AGCCACATCGCTCAGACACC-3′), reversed primer (5′-GTACTCAGCGCCAGCATCG-3′), 57 °C annealing temperature, 30 s elongation time, 25 cycles. ER-alpha PCR forward primer (5′-ACAAGCGCCAGAGAGATGAT-3′), reverse primer (5′-GATGTGGGAGAGGATGAGGA-3′), 57 °C annealing temperature, 60 s elongation time, 30 cycles.
DNA purification and sequencing
The QIAEX II kit (Qiagen, Hombrechtikon, Switzerland) was used for DNA purification of RT–PCR products according to the manufacturer’s instructions, followed by sequencing.
Methylation-specific PCR (MSP)
The methylation status of BARD1 was evaluated by MSP (methylation-specific PCR) as described previously (Herman et al, 1996; Schneider-Stock et al, 2003). Primers were used as described in a previous study on BARD1 methylation (Li et al, 2007).
Statistical analysis
The Spearman’s correlation coefficient ρ was used to assess the correlation between expression levels of distinct epitopes of BARD1 and BRCA1. The χ2 test was used to compare the percentage of positive cases in tumour vs peri-tumour tissues and correlation of positive cases of BARD1 expression with clinical variables. Survival differences were estimated using Kaplan–Meier method compared by the log-rank test. For all calculations, the tests performed were two-sided, a value of P<0.05 was considered statistically significant. Analyses were performed using Statistical Package for the Social Sciences (SPSS) for Windows version 13 (SPSS Inc., Chicago, IL, USA).
Results
BARD1 mRNA expression pattern in colorectal cancer
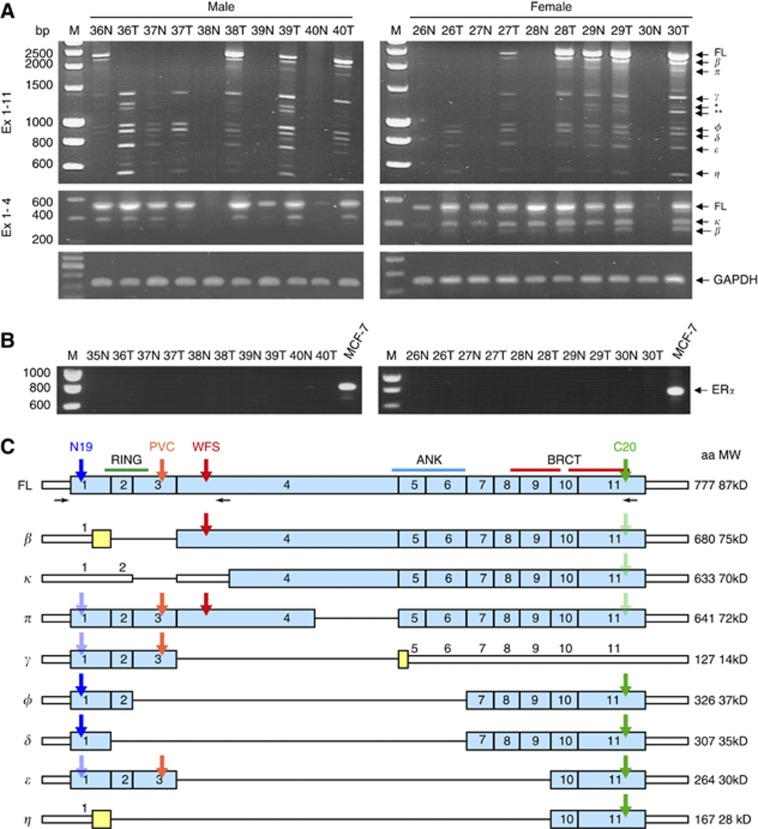
As in other cancers, we suspected that splice isoforms of BARD1 were expressed in colon cancer tissues. We assessed BARD1 mRNA expression in 20 tumour and peri-tumour tissues including 10 male and 10 female cases. We performed RT–PCR using forward primer specific to exon 1 and reverse primers in exons 4 or 11 to amplify the corresponding BARD1 coding regions (see Patients and Methods). Human GAPDH cDNA was amplified as internal control (Figure 1A).
Figure 1.
BARD1 mRNA isoform expression in colorectal cancer. (A) Amplification of FL BARD1 and/or isoforms using forward primer in exon 1, and reverse primer in exon 11 (Ex 1–11) or exon 4 (Ex 1–4). As examples, pairs of peri-tumour (N) and tumour (T) tissues of five male and five female patients are shown. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression is shown for the same samples as standard. Molecular marker is shown on the left (M). Presumed FL BARD1 and truncated isoforms are indicated on the right. The isoforms labelled * and ** in correspond to the deletion of exons 3 and 4 (gamma del-3), and deletion of exons 2–4 (gamma del 2–3), respectively, which were reported previously (Sporn et al, 2011). Patterns of isoforms were different in peri-tumour and tumour tissues expressed and expression was less frequent in peri-tumour tissues. (B) Amplification of oestrogen receptor α (ERα) in the same samples. MCF-7 was used as positive control (right). No ERα expression was observed in colorectal tissues, neither in peri-tumour nor in tumour samples of males and females. (C) Schematic presentation of exon structure of BARD1 and isoforms. Exons with ORF are shown as light blue bars, non-coding sequences as white narrow bars and alternatively translated sequences are shown as yellow bars. The positions of the primers used for RT–PCR are shown as horizontal black arrows below BARD1 FL scheme. BARD1 epitopes recognised by N19, C20, PVC, WFS antibodies are indicated with coloured arrows. The antibody epitopes presumably hidden owing to the protein conformation are shown as pale arrows. RING domain (RING), Ankyrin repeats (ANK) and BRCT domains (BRCT) are shown. The color reproduction of this figure is available on the British Journal of Cancer online.
We have sequenced all isoforms from at least one patient sample and determined the presence of FL BARD1, beta, kappa, and pi in colon cancer, but not alpha, which is expressed in lung and gynaecological cancers. The isoforms labelled * and ** in Figure 1A, correspond to deletion of exons 3 and 4 (gamma del-3), and deletion of exons 2–4 (gamma del 2–3), respectively, which were reported previously (Sporn et al, 2011) (Supplementary Figure 1).
We have also investigated the use of alternative transcript start sites of BARD1 using 5′RACE from different exons (2, 3, and 6), but we did not observe any other start sites than those already described (Li et al, 2007).
Interestingly, all BARD1 isoforms that were recently identified in NSCLC (Zhang et al, 2012), including isoforms κ, lacking exon 3, and π with a partial deletion (408 bp) of exon 4, were expressed in colorectal cancer (Figure 1A and C).
BARD1 expression in colon cancer is not regulated by oestrogen or methylation
As oestrogen signalling was associated with colon cancer initiation and progression (Hogan et al, 2009), and expression of BARD1 and BARD1 isoforms can be modulated by oestrogen through ERα (Niell et al, 2004; Russo et al, 2009), we also examined the expression of ERα mRNA in colon peri-tumour and tumour tissues from males and females. MCF-7 cells were used as a positive control. We found no ERα expression in colorectal tissues in these samples (Figure 1B). Consistent with this result, similar profiles were observed for FL BARD1 and isoforms expression levels and frequency in colorectal tumours and corresponding peri-tumour tissue from males and females (P>0.05) (data not shown).
As methylation was reported for the BRCA1 promoter (Esteller et al, 2001), we investigated whether the BARD1 promoter was methylated in colon cancer. Methylation analysis of 109 tumour samples (Table 1) by MSP revealed only one methylation-positive case (data not shown). A similar negative result was obtained for BARD1 promoter methylation in breast/ovarian cancer (Li et al, 2007).
BARD1 isoforms are upregulated in colon tumours and expression correlates with clinical variables
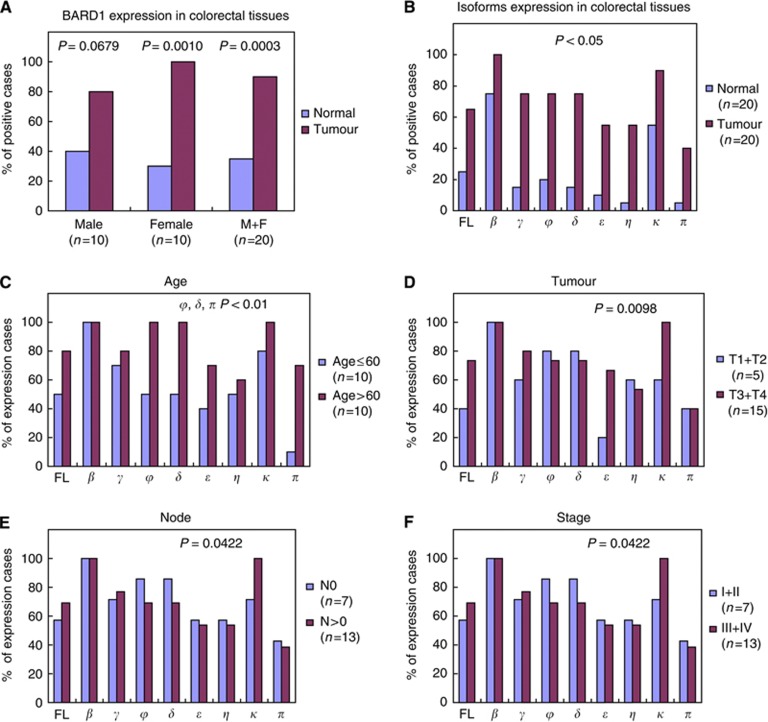
On the contrary to what was observed in NSCLC (Zhang et al, 2012), FL BARD1 and BARD1 isoforms were significantly more expressed in tumour than in peri-tumour tissues (P<0.05) (Figure 1A and 2A, B). Full-length BARD1 and isoforms were expressed in 90%, of the tumour samples (18 of 20), whereas in peri-tumour tissue BARD1 expression was much less frequent (35%, 7 of 20 cases). In seven samples, only FL BARD1 or FL BARD1 and few isoforms were expressed. The difference was statistically significant (P=0.0003) (Figure 2A). Similar results were obtained in males (8 out of 10 in tumour tissues vs 4 out of 10, in peri-tumour, P=0.0679) and in females (10 out of 10 vs 3 out of 10, respectively, P<0.001).
Figure 2.
Correlation of BARD1 mRNA isoform expression with clinicopathological variables of the patients with colorectal cancer. (A) Comparison of BARD1 expression in peri-tumour and tumour tissues from males, females, or both, based on absence or presence of any form of BARD1. BARD1 expression significantly was more frequent and more abundant in tumours than peri-tumour tissues. The P-value was obtained by the χ2 test. (B) Comparison of FL BARD1 and isoform expression in peri-tumour and tumour tissues. All forms were upregulated in tumours with statistical significance (P<0.05 for all). The P-value was obtained by the χ2 test. (C) Comparison of FL BARD1 and isoform expression in younger (⩽60 years) and older (>60 years) patients. Full-length BARD1 and all isoforms, except isoform β, were more upregulated in older than in younger patients. Specially, expression of isoforms φ, δ, and π were significantly associated with older patients (P<0.01). The P-value was obtained by the χ2 test. (D–F) Comparison of FL BARD1 and isoforms expression with primary tumour and lymph node status, and tumour stage and grade. BARD1 isoform κ expression was significantly associated with large tumour size or nearby tissue invasion (D), lymph node involvement (E), and advanced stage (F) (stage III and IV). The P-value was obtained by the χ2 test in all cases.
To determine whether BARD1 isoform expression correlated with patients’ clinicopathological characteristics, we compared expression of FL BARD1 and isoforms with clinicopathological variables based on the presence or absence of their expression in tumour tissues. Full-length BARD1 and BARD1 isoforms were more frequently expressed in patients older than 60 years (Figure 2C). In particular, frequencies of BARD1 isoforms φ, δ, and π expression were significantly associated with older age (P<0.01). The frequency of isoform BARD1κ expression was significantly associated with large tumour size or with nearby tissue invasion (T3 and T4; P=0.0098), lymph node involvement (N1 and N2, P=0.0422), and advanced tumour stages (stage III and IV, P=0.0422) (Figure 2D–F). No correlation was observed between BARD1 expression and histopathological tumour grade.
Only few BARD1 isoforms are likely to influence tumorigenesis
We compared our BARD1 isoform expression pattern with 19 isoforms reported by Sporn et al (2011). Only those isoforms that were previously reported by us (Li et al, 2007; Zhang et al, 2012) contained an open reading frame (ORF) and are likely to be translated (Supplementary Figure 1). Oncogenic functions have been attributed previously to isoforms BARD1β, κ, and π (Li et al, 2007; Ryser et al, 2009; Zhang et al, 2012). Isoforms φ, δ, ε, and η were found in tumours with poor outcome in ovarian and breast cancer (Li et al, 2007). The oncogenic roles of BARD1β and δ are based on antagonising the functions of FL BARD1 (Ryser et al, 2009; Dizin and Irminger-Finger, 2010; Bosse et al, 2012).
Isoform γ could be translated in two ways: either translation of ORF common to FL BARD1 from exon 1 through exon 3, ending in a stop codon in exon 4, or an alternative ORF and translation start in exon 3 and translation of exons 4 through 11. We used γ-specific siRNA to repress γ expression in cell cultures and investigated the resulting BARD1 protein profile on western blots (Supplementary Figure 2). We thus identified BARD1γ as a protein translated from exons 1 through 3. BARD1γ comprises the RING domain and could potentially bind to BRCA1 as well as FL BARD1. Consistent with this view, the level of FL BARD1 dropped in the cells treated with γ-specific siRNA (Supplementary Figure 2). This suggests that a BARD1γ-encoded protein is able to interact and stabilise FL BARD1 and possibly BRCA1.
Our data suggest that only few of the BARD1 mRNA isoforms are translated into stable proteins. These few protein isoforms therefore could be reflected in the antibody staining patterns of colon cancer tissues.
BARD1 expression in colorectal cancer samples
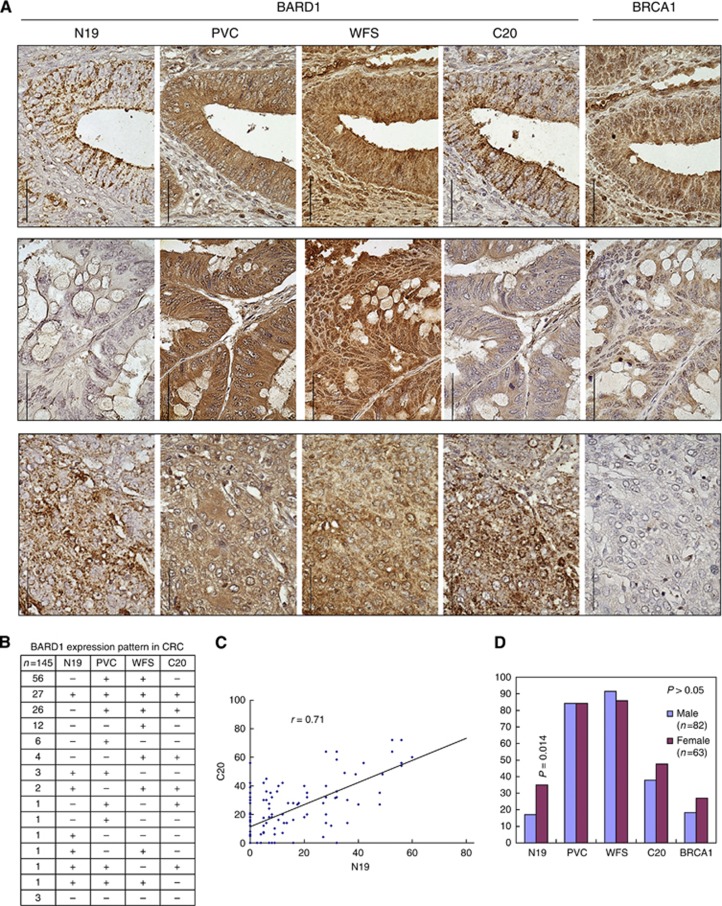
To investigate BARD1 expression in colorectal cancer, we performed IHC on 168 colon cancer cases, including 20 paired tumour and peri-tumoral normal tissue sections and 148 colorectal tumours presented as tissue microarray with tetramerous for each case (Table 1). To distinguish the expression of different exons of BARD1, we used four previously characterised antibodies (Irminger-Finger et al, 1998; Wu et al, 2006; Li et al, 2007; Zhang et al, 2012) (N19, PVC, WFS, and C20) recognising regions in exon 1 (N-terminus), end of exon 3 (after RING region), beginning of exon 4, and exon 11 (C-terminus) (Figure 1C), on adjacent tissue sections. We also investigated BRCA1 expression using an antibody against BRCA1. Staining was mostly cytoplasmic with all BARD1 antibodies. However, different antibodies stained different sub-cellular compartments and/or different tumour regions (Figure 3A). Typically, BARD1 N19 and C20 showed a granular staining, whereas PVC and WFS showed diffuse staining and they were co-localised to the same cells or the same regions, respectively.
Figure 3.
BARD1 and BRCA1 expression in colorectal cancer. Immunohistochemistry (IHC) was performed on samples of 168 colorectal cancer cases with BARD1 antibodies N19, C20, PVC, WFS, and BRCA1. Samples were presented as tissue microarray with tetramerous for each of the cases. A total of 145 samples were eligible for analysis after IHC assay. (A) Examples of IHC using BARD1 antibodies and BRCA1 antibody. BARD1 N19 and C20 showed cytoplasmic granular staining, and co-localised to the same cells or tissue regions. BARD1 PVC and WFS stainings were diffusely cytoplasmic. BRCA1 staining was granular in both cytoplasm and nucleus. Examples of the positive staining with BARD1 antibodies and BRCA1 antibody (upper panel), the weaker staining with N19, C20, and BRCA1 antibodies (middle panel) and BARD1-positive but BRCA1-negative staining (lower panel) are shown. Scale bars correspond to 50 μm. (B) BARD1 expression patterns in colorectal cancer. Expression patterns were obtained for four BARD1 antibodies based on positive (+) and negative (−) staining for each case. PVC- and WFS-positive, but N19- and C20-negative staining was the most frequent expression pattern, ‘all four antibodies positive’ staining was the second, N19-negative but PVC-, WFS-, and C20-positive staining was the third most frequently observed expression pattern. (C) The correlation of BARD1 N19 and C20 antibodies staining. (D) BARD1 N19-positive staining was significantly associated with female gender (P=0.014). The P-value was obtained by the χ2 test.
The positivity was variable for each antibody (Figure 3B). BARD1 N19, PVC, WFS, and C20 staining were classified as positive in 36 (24.8%), 122 (84.1%), 129 (89%) and 61 (42.1%) cases, respectively. A total of 142 cases were positive for at least one antibody, and no expression of BARD1 was found in only 3 cases. Hence, 97.9% (142 of 145) of colorectal cancer samples expressed at least one epitope of BARD1.
Although there are 16 possible combinations for the expression of four BARD1 epitopes, only 3 combinations were observed (Figure 3B): simultaneous expression of epitopes PVC and WFS (after RING domain and exon 4, respectively) was the most frequent pattern (38.6%), positive staining for all four antibodies was the second most frequent (18.6%), and loss of the N-terminal epitope but expression of PVC, WFS, and C20 (17.9%) the third most frequent.
To investigate this further, we quantified and compared the expression patterns obtained for each antibody. Strong correlation was observed between expression levels of N19 and C20 (ρ=0.71, P=0.001) (Figure 3C). Other comparisons showed only weak correlations, namely for PVC and WFS (ρ=0.39, P=0.001), PVC and C20 (ρ=0.36, P=0.001), and WFS and C20 (ρ=0.27, P=0.001), or no correlation for N19 and PVC, and N19 and WFS staining (data not shown).
Correlated expression of N19 and C20 is consistent with expression of isoforms δ, φ, and ε. All other patterns of expression might reflect a combination of expression of isoforms β, κ, and π. Based on these analyses, none of the observed expression patterns is compatible with expression of FL BARD1.
Non-coordinate expression of BARD1 epitopes and BRCA1
As mentioned, BARD1 is important for stability and subcellular localisation of BRCA1. Unlike BARD1, BRCA1 showed both cytoplasmic and nuclear granular staining within the same cell (Figure 3A). BRCA1-positive staining was observed in 22.1% (32 of 145) of colorectal cancer cases, similar to BARD1 N19-positive staining, which was observed in 24.8% (36 of 145) of the cases. However, BRCA1 expression was not correlated with expression of any BARD1 epitope (data not shown).
BARD1 expression pattern correlates with patients’ prognosis but not with other clinicopathological characteristics
Immunohistochemistry analysis of BARD1 N19, PVC, WFS, C20, and BRCA1 expression was compared with clinical variables of 145 cases eligible for the statistical analysis.
No significant correlation was observed between the expression of BRCA1 or BARD1 epitopes and clinicopathologic variables, such as tumour grade, primary tumour, lymph node and distant metastasis status, or tumour stage. We also analysed the correlation between the three major expression patterns of BARD1 (Figure 3B) with clinicopathological variables. No significant correlation was observed in this case either (data not shown).
To assess the correlation of BARD1 and BRCA1 expression with survival, we compared the individual expression of the four BARD1 epitopes, as well as BRCA1 epitope, and the different BARD1 expression patterns (Figure 3B) with survival data for 75 colorectal cancer cases with follow-up data (Tables 2 and 3). For the individual BARD1 epitopes, we found that patients with BARD1 N19 (N-terminal epitope)-positive staining had significantly higher 1-year, 2-year, and 3-year survival rates, while the BARD1 C20 (C-terminal epitope)-positive patients had significantly higher 1-year and 3-year survival rates when compared with the patients with the negative staining for the corresponding epitopes (Table 2). Interestingly, the frequency of N19-positive staining was also significantly associated with female sex (P=0.014, Figure 3D). No conclusion could be made from the comparison of BARD1 PVC- and WFS-positive patients’ survival rates as the number of negative staining cases was not sufficient for the analysis. No significant difference was observed for the comparison of BRCA1-positive and -negative cases with corresponding survival rates (data not shown).
Table 2. Correlation of distinct epitopes of BARD1 and BRCA1 expression with survival in 75 colorectal cancer patients.
|
1-year survival
|
2-year survival
|
3-year survival
|
||||||
|---|---|---|---|---|---|---|---|---|
| Expression pattern | No. of patients | Median survival | % | P -value | % | P -value | % | P -value |
| N neg vs pos | 55 vs 20 | 11 vs 26 | 47.3 vs 85.0 | 0.0035 | 21.8 vs 50.0 | 0.0178 | 12.7 vs 40.0 | 0.009 |
| P neg vs pos | 14 vs 61 | 14 vs 15 | 57.1 vs 57.4 | 0.9873 | 28.6 vs 29.5 | 0.9446 | 21.4 vs 19.7 | 0.8822 |
| W neg vs pos | 4 vs 71 | |||||||
| C neg vs pos | 42 vs 33 | 9 vs 17 | 45.2 vs 72.7 | 0.0169 | 21.4 vs 39.4 | 0.0898 | 11.9 vs 30.3 | 0.048 |
| BRCA1 neg vs pos | 60 vs 15 | 16 vs 12 | 60.0 vs 46.7 | 0.3504 | 31.7 vs 20.0 | 0.3747 | 23.3 vs 6.7 | 0.1489 |
Note: ‘neg’, negative staining; ‘pos’, positive staining.
Antibody abbreviations: N - N19, P - PVC, C - C20, W - WFS. For WFS, negative staining cases were not enough for further analysis.
Table 3. Correlation of BARD1 expression patterns with survival in 75 colorectal cancer patients.
|
1-year survival
|
2-year survival
|
3-year survival
|
||||||
|---|---|---|---|---|---|---|---|---|
| Expression pattern | No. of patients | Median survival | % | P -value | % | P -value | % | P -value |
| ++++ vs −++− | 17 vs 31 | 27 vs 9 | 88.2 vs 41.9 | 0.0019 | 52.9 vs 16.1 | 0.0073 | 41.2 vs 6.5 | 0.0032 |
| ++++ vs −+++ | 17 vs 11 | 27 vs 12 | 88.2 vs 45.5 | 0.0144 | 52.9 vs 18.2 | 0.0659 | 41.2 vs 9.1 | 0.0664 |
| ++++ vs others | 17 vs 58 | 27 vs 12 | 88.2 vs 48.3 | 0.0034 | 52.9 vs 22.4 | 0.0151 | 41.2 vs 13.8 | 0.0131 |
| −++− vs −+++ | 31 vs 11 | 9 vs 12 | 41.9 vs 45.5 | 0.8390 | 16.1 vs 18.2 | 0.8750 | 6.5 vs 9.1 | 0.7702 |
| −++− vs others | 31 vs 44 | 9 vs 16.5 | 41.9 vs 68.2 | 0.0236 | 16.1 vs 38.6 | 0.0350 | 6.5 vs 29.5 | 0.0138 |
| −+++ vs others | 11 vs 64 | 12 vs 15.5 | 45.5 vs 59.4 | 0.3885 | 18.2 vs 31.3 | 0.3792 | 9.1 vs 21.9 | 0.3274 |
Note: ++++, four antibodies positive staining; −++−, only PVC and WFS positive staining; −+++, only N19-negative staining; others, other than the expression pattern that is compared.
When BARD1 epitope expression patterns were used for the correlation studies, we found that the simultaneous expression of all four BARD1 epitopes correlated with higher 1-, 2-, and 3-year survival rates, when compared with the expression of only two middle epitopes or other expression patterns. However, the expression of only two middle epitopes (PVC and WFS) was correlated with lower 1-, 2-, and 3-year survival rates as compared with all other expression patterns including the all four antibodies positive staining pattern (Table 3).
Taken together our data suggest that the simultaneous expression of all four BARD1 epitopes is a positive prognostic factor, as well as the expression of BARD1 N and C-terminal epitopes. Inversely, the simultaneous expression of only two middle epitopes (PVC and WFS) is a negative prognostic factor. These data can only be explained with the simultaneous expression of a combination of isoforms (Figure 1C). Isoforms expressing PVC and WFS epitopes are clearly correlated with poor prognosis in colon cancer, and were correlated with decreased survival in lung cancer (Zhang et al, 2012). Which isoforms are contributing to a positive prognosis has to be determined.
Discussion
In the present study, we demonstrate that BARD1 is differentially spliced in colon cancer, that protein products of splice isoforms might affect BRCA1 localisation, and that the splice isoforms might have oncogenic functions by themselves. Alternative splicing of tumour suppressor genes can produce proteins with dominant negative functions, which are often found associated with cancer (Srebrow and Kornblihtt, 2006). Antagonistic functions were reported for BARD1 isoform β (Li et al, 2007; Bosse et al, 2012) and for isoform δ (Dizin and Irminger-Finger, 2010).
By using antibodies against the differentially expressed regions, we found at least one of the respective epitopes expressed in each of 168 samples of the colorectal cancer. The pattern of positive epitopes excluded any relevant expression levels of FL BARD1. Overexpression of oncogenic forms rather than repression of FL BARD1 is consistent with lack of promoter methylation of BARD1, as observed in all colorectal cancer samples tested (N=98). Similarly, no methylation of BARD1 promoter was found in ovarian cancer (Li et al, 2007). In vitro repression experiments demonstrated that BARD1 isoform expression is essential for cell proliferation (Li et al, 2007; Ryser et al, 2009; Bosse et al, 2012). Our results therefore suggest that rather than loss of BARD1 expression, it is the expression of at least one form of BARD1 that might be essential for tumour growth.
There is evidence for a role of BRCA1 in hereditary as well as sporadic colon cancer (Garcia-Patiño et al, 1998; Lin et al, 1999; Mohamad and Apffelstaedt, 2008; Russo et al, 2009; Suchy et al, 2010). BRCA1 expression in colon cancer might be affected by the aberrant expression of BARD1. The un-coordinated expression of BARD1 epitopes excludes expression of FL BARD1 and suggests that the E3 ubiquitin ligase functions of the BRCA1–BARD1 heterodimer (Baer and Ludwig, 2002) are jeopardised in colorectal cancer. Dysfunction of the BRCA1-BARD1 ubiquitin ligase can affect repair functions and lead to genomic instability.
There is evidence that differentially spliced BARD1 isoforms might be themselves drivers of tumorigenesis. Their expression was correlated with poor prognosis in breast, ovarian, and lung cancer (Wu et al, 2006; Li et al, 2007; Zhang et al, 2012) and as shown here, in colon cancer. Especially, isoforms that express epitopes mapping to exons 3 and 4, present on BARD1 β, κ, π, are correlated with short survival in colorectal cancer, as well as lung cancer (Zhang et al, 2012). In vitro experiments support the notion that BARD1 isoforms may be drivers of tumorigenesis, as they have transforming activity (Bosse et al, 2012) and are required for cancer cell proliferation (Li et al, 2007; Ryser et al, 2009; Bosse et al, 2012).
We found that BARD1 mRNA isoforms were generally more expressed in females (Figure 1A), and N19-positive staining was significantly associated with female gender in colorectal cancer (P=0.014). However, their expression cannot be driven by oestrogen and ERα, as no ERα mRNA expression was found in 20 cancer cases that we analysed.
Alternatively spliced BARD1 isoforms in colon cancer have also been reported by others (Sporn et al, 2011), based on 15 and 99 colon tumours, analysed by RT–PCR and IHC, respectively. Of 19 mRNA isoforms that were characterised, only few are protein coding and likely to affect tumorigenesis; all of these have been reported previously by us and others (Supplementary Figure 1). Immunohistochemistry analysis of 99 colon tumours was only based on one monoclonal antibody directed against an undefined epitope within the first 300 amino acids of BARD1 (Sporn et al, 2011). The expression of other regions and isoforms lacking the N-terminus were not investigated with this method. Thus, the conclusion of the authors, that lack of FL BARD1 is a negative prognostic and prospective marker is only partially true. The N-terminal epitope detected with this antibody could, in addition to FL BARD1, detect isoforms π, γ, δ, φ, and ε. Thus, the Sporn et al (2011) study identified isoforms on the mRNA level, but their relevance for tumorigenesis was not completely addressed at the protein level.
We found that a positive staining pattern for four antibodies was significantly associated with longer survival in colorectal cancer, so were N19- and C20-positive staining. The expressions of N- and C-terminal epitopes are significantly correlated and are consistent with isoform δ or φ expression, suggesting an inhibitory effect of isoform δ or φ on tumorigenesis. The positivity for four antibodies most likely reflects a combination of isoforms and not FL BARD1.
We found that PVC- and WFS-positive staining was strongly correlated with shorter survival in colon cancer. This staining is consistent with isoforms β, κ, and π expression. N19 and C20 epitopes might be blocked in these isoforms, as they are located in structured regions of the RING and BRCT domains, respectively (Figure 1C). To support the hypothesis that the C20 epitope is present, but not accessible in most isoforms, we have used an antibody against a different sequence in exon 11, as used in a study of lung cancer (Zhang et al, 2012) and compared it to the C20 staining pattern (Supplementary Figure 3). Similarly, we have performed IHC with a commercial antibody BL (exon 4) and compared its staining pattern with that of WFS (exon 4) on a selected number of colon cancer tissue samples (Supplementary Figure 3). These staining patterns demonstrate that WFS, BL, and P8 antibodies show the identical signal distribution indicating their specificity, whereas C20 staining is much weaker supporting our hypothesis that the C20 epitope may be present but not accessible in some isoforms.
Comparison of mRNA isoform expression in colon tumour tissues showed that specifically the expression of BARD1 isoforms κ was significantly correlated with advanced tumour stage and invasiveness (Figure 2D–F). Upregulation of the same mRNA isoform was also observed by Sporn et al (2011) in colon cancer.
In summary, our data strongly suggest that BARD1 isoforms κ, β, and π are involved in colon cancer tumorigenesis and progression and might be promising specific prognostic markers, while isoforms δ and φ might have an inhibitory effect. Further studies are needed for defining the use of BARD1 isoforms as prognostic markers for response to treatment regimens.
Acknowledgments
We are grateful to Professor A Roessner, University of Madgeburg, Germany, for use of TMA, Professor GJ Laurent for critical discussions, and Dr Gavino Faa for sample characterisation and selection, Dr JY Wu for help with IH scoring, Drs P-A André and M Chibi for expertise in statistical analysis and immunohistochemistry, and D Burdevet for excellent technical help. This work was supported by grant SNSF 3100A0-122353 and EU7-Bardiag-262318 to IIF.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
IIF and YQZ have a conflict of interest. Both have authored a patent on BARD1 isoforms. The other authors declare no conflict of interest.
Supplementary Material
References
- Baer R, Ludwig T (2002) The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr Opin Genet Dev 12: 86–91 [DOI] [PubMed] [Google Scholar]
- Bosse KR, Diskin SJ, Cole KA, Wood AC, Schnepp RW, Norris G, Nguyen LB, Jagannathan J, Laquaglia M, Winter C, Diamond M, Hou C, Attiyeh EF, Mosse YP, Pineros V, Dizin E, Zhang Y, Asgharzadeh S, Seeger RC, Capasso M, Pawel BR, Devoto M, Hakonarson H, Rappaport EF, Irminger-Finger I, Maris JM (2012) Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res 72(8): 2068–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose MS, Rebbeck TR, Calzone KA, Stopfer JE, Nathanson KL, Weber BL (2002) Cancer risk estimates for BRCA1 mutation carriers identified in a risk evaluation program. J Natl Cancer Inst 94: 1365–1372 [DOI] [PubMed] [Google Scholar]
- Dizin E, Irminger-Finger I (2010) Negative feedback loop of BRCA1-BARD1 ubiquitin ligase on estrogen receptor alpha stability and activity antagonized by cancer-associated isoform of BARD1. Int J Biochem Cell Biol 42: 693–700 [DOI] [PubMed] [Google Scholar]
- Easton DF, Ford D, Bishop DT (1995) Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Am J Hum Genet 56: 265–271 [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Fraga MF, Guo M, Garcia-Foncillas J, Hedenfalk I, Godwin AK, Trojan J, Vaurs-Barrière C, Bignon YJ, Ramus S, Benitez J, Caldes T, Akiyama Y, Yuasa Y, Launonen V, Canal MJ, Rodriguez R, Capella G, Peinado MA, Borg A, Aaltonen LA, Ponder BA, Baylin SB, Herman JG (2001) DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet 10: 3001–3007 [DOI] [PubMed] [Google Scholar]
- Fabbro M, Rodriguez JA, Baer R, Henderson BR (2002) BARD1 induces BRCA1 intranuclear foci formation by increasing RING-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J Biol Chem 277: 21315–21324 [DOI] [PubMed] [Google Scholar]
- Figueredo A, Coombes ME, Mukherjee S (2008) Adjuvant therapy for completely resected stage II colon cancer. Cochrane Database Syst Rev CD005390 [DOI] [PMC free article] [PubMed]
- Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE (1994) Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet 343: 692–695 [DOI] [PubMed] [Google Scholar]
- Garcia-Patiño E, Gomendio B, Lleonart M, Silva JM, Garcia JM, Provencio M, Cubedo R, España P, Ramón y Cajal S, Bonilla F (1998) Loss of heterozygosity in the region including the BRCA1 gene on 17q in colon cancer. Cancer Genet Cytogenet 104: 119–123 [DOI] [PubMed] [Google Scholar]
- Gautier F, Irminger-Finger I, Grégoire M, Meflah K, Harb J (2000) Identification of an apoptotic cleavage product of BARD1 as an autoantigen: a potential factor in the antitumoral response mediated by apoptotic bodies. Cancer Res 60: 6895–6900 [PubMed] [Google Scholar]
- Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T (2001) The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 276: 14537–14540 [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 93: 9821–9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson P, Glasziou P, Irwig L, Towler B, Watson E (2007) Screening for colorectal cancer using the faecal occult blood test, Hemoccult. Cochrane Database Syst Rev CD001216 [DOI] [PMC free article] [PubMed]
- Hogan AM, Collins D, Baird AW, Winter DC (2009) Estrogen and its role in gastrointestinal health and disease. Int J Colorectal Dis 24: 1367–1375 [DOI] [PubMed] [Google Scholar]
- Irminger-Finger I, Jefford CE (2006) Is there more to BARD1 than BRCA1? Nat Rev Cancer 6: 382–391 [DOI] [PubMed] [Google Scholar]
- Irminger-Finger I, Soriano JV, Vaudan G, Montesano R, Sappino AP (1998) In vitro repression of Brca1-associated RING domain gene, Bard1, induces phenotypic changes in mammary epithelial cells. J Cell Biol 143: 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov V, Chen J, Fox EA, Green JB, Livingston DM (2001) Functional communication between endogenous BRCA1 and its partner, BARD1, during Xenopus laevis development. Proc Natl Acad Sci USA 98: 12078–12083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadouri L, Hubert A, Rotenberg Y, Hamburger T, Sagi M, Nechushtan C, Abeliovich D, Peretz T (2007) Cancer risks in carriers of the BRCA1/2 Ashkenazi founder mutations. J Med Genet 44: 467–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff T, Satagopan JM, Kauff ND, Huang H, Kolachana P, Palmer C, Rapaport H, Nafa K, Ellis NA, Offit K (2004) Frequency of BRCA1 and BRCA2 mutations in unselected Ashkenazi Jewish patients with colorectal cancer. J Natl Cancer Inst 96: 68–70 [DOI] [PubMed] [Google Scholar]
- Li L, Ryser S, Dizin E, Pils D, Krainer M, Jefford CE, Bertoni F, Zeillinger R, Irminger-Finger I (2007) Oncogenic BARD1 isoforms expressed in gynecological cancers. Cancer Res 67: 11876–11885 [DOI] [PubMed] [Google Scholar]
- Lin KM, Ternent CA, Adams DR, Thorson AG, Blatchford GJ, Christensen MA, Watson P, Lynch HT (1999) Colorectal cancer in hereditary breast cancer kindreds. Dis Colon Rectum 42: 1041–1045 [DOI] [PubMed] [Google Scholar]
- Lynch HT, Lemon SJ, Karr B, Franklin B, Lynch JF, Watson P, Tinley S, Lerman C, Carter C (1997) Etiology, natural history, management and molecular genetics of hereditary nonpolyposis colorectal cancer (Lynch syndromes): genetic counseling implications. Cancer Epidemiol Biomarkers Prev 6: 987–991 [PubMed] [Google Scholar]
- Markowitz SD, Bertagnolli MM (2009) Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med 361: 2449–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza JE, Brzovic PS, King M-C, Klevit RE (1999) Mapping the functional domains of BRCA1. J Biol Chem 274: 5659–5665 [DOI] [PubMed] [Google Scholar]
- Mohamad HB, Apffelstaedt JP (2008) Counseling for male BRCA mutation carriers: a review. Breast 17: 441–450 [DOI] [PubMed] [Google Scholar]
- Morris JR, Solomon E (2004) BRCA1:BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet 13: 807–817 [DOI] [PubMed] [Google Scholar]
- Niell BL, Rennert G, Bonner JD, Almog R, Tomsho LP, Gruber SB (2004) BRCA1 and BRCA2 founder mutations and the risk of colorectal cancer. J Natl Cancer Inst 96: 15–21 [DOI] [PubMed] [Google Scholar]
- Ouchi M, Fujiuchi N, Sasai K, Katayama H, Minamishima YA, Ongusaha PP, Deng C, Sen S, Lee SW, Ouchi T (2004) BRCA1 phosphorylation by Aurora-A in the regulation of G2 to M transition. J Biol Chem 279: 19643–19648 [DOI] [PubMed] [Google Scholar]
- Oyake D, Nishikawa H, Koizuka I, Fukuda M, Ohta T (2002) Targeted substrate degradation by an engineered double RING ubiquitin ligase. Biochem Biophys Res Commun 295: 370–375 [DOI] [PubMed] [Google Scholar]
- Rudmik LR, Magliocco AM (2005) Molecular mechanisms of hepatic metastasis in colorectal cancer. J Surg Oncol 92: 347–359 [DOI] [PubMed] [Google Scholar]
- Russo A, Calò V, Bruno L, Rizzo S, Bazan V, Di Fede G (2009) Hereditary ovarian cancer. Crit Rev Oncol Hematol 69: 28–44 [DOI] [PubMed] [Google Scholar]
- Ryser S, Dizin E, Jefford CE, Delaval B, Gagos S, Christodoulidou A, Krause K-H, Birnbaum D, Irminger-Finger I (2009) Distinct roles of BARD1 isoforms in mitosis: full-length BARD1 mediates Aurora B degradation, cancer-associated BARD1beta scaffolds Aurora B and BRCA2. Cancer Res 69: 1125–1134 [DOI] [PubMed] [Google Scholar]
- Schneider-Stock R, Boltze C, Peters B, Höpfner T, Meyer F, Lippert H, Roessner A (2003) Differences in loss of p16INK4 protein expression by promoter methylation between left- and right-sided primary colorectal carcinomas. Int J Oncol 23: 1009–1013 [PubMed] [Google Scholar]
- Sporn JC, Hothorn T, Jung BH (2011) BARD1 expression predicts outcome in colon cancer. Clin Cancer Res 17(16): 5451–5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srebrow A, Kornblihtt AR (2006) The connection between splicing and cancer. J Cell Sci 119: 2635–2641 [DOI] [PubMed] [Google Scholar]
- Starita LM, Machida Y, Sankaran S, Elias JE, Griffin K, Schlegel BP, Gygi SP, Parvin JD (2004) BRCA1-dependent ubiquitination of gamma-tubulin regulates centrosome number. Mol Cell Biol 24: 8457–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchy J, Cybulski C, Górski B, Huzarski T, Byrski T, Dębniak T, Gronwald J, Jakubowska A, Wokołorczyk D, Kurzawski G, Kładny J, Jawień A, Banaszkiewicz Z, Wiśniowski R, Wandzel P, Starzewski J, Lorenc Z, Korobowicz E, Krokowicz P, Horbacka K, Lubiński J, Narod SA (2010) BRCA1 mutations and colorectal cancer in Poland. Fam Cancer 9: 541–544 [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang H, Guerrette S, Chen J, Mazurek A, Wilson T, Slupianek A, Skorski T, Fishel R, Greene MI (2001) Adenosine nucleotide modulates the physical interaction between hMSH2 and BRCA1. Oncogene 20: 4640–4649 [DOI] [PubMed] [Google Scholar]
- WHO (February 2009) Cancer. World Health Organization: Geneva [Google Scholar]
- Wu J-Y, Vlastos A-T, Pelte M-F, Caligo M-A, Bianco A, Krause K-H, Laurent GJ, Irminger-Finger I (2006) Aberrant expression of BARD1 in breast and ovarian cancers with poor prognosis. Int J Cancer 118: 1215–1226 [DOI] [PubMed] [Google Scholar]
- Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang M-CW, Hwang L-Y, Bowcock AM, Baer R (1996) Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet 14: 430–440 [DOI] [PubMed] [Google Scholar]
- Zhang Y-Q, Bianco A, Malkinson AM, Leoni VP, Frau G, Rosa ND, André P-A, Versace R, Boulvain M, Laurent GJ, Atzuri L, Irminger-Finger I (2012) BARD1: an independent predictor of survival in non-small cell lung cancer. International Journal of Cancer 131(1): 83–94 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.