Abstract
Background:
MicroRNA (miRNA)-126 (miR-126) is an endothelial-specific miRNA located within intron 7 of epidermal growth factor-like domain 7 (EGFL7). However, the role of miR-126 in cancer is controversial.
Methods:
We examined the function of miR-126 in oral squamous cell carcinoma (OSCC) cells. Furthermore, a series of 118 cases with OSCC were evaluated for the expression levels of miR-126.
Results:
MicroRNA-126 (miR-126) was associated with cell growth and regulation of vascular endothelial growth factor-A activity, and demethylation treatment increased expression levels of miR-126 and EGFL7 in OSCC cells. A significant association was found between miR-126 expression and tumour progression, nodal metastasis, vessel density, or poor prognosis in OSCC cases. In the multivariate analysis, decreased miR-126 expression was strongly correlated with disease-free survival.
Conclusion:
The present results suggest that miR-126 might be a useful diagnostic and therapeutic target in OSCC.
Keywords: miR-126, oral squamous cell carcinoma, angiogenesis, lymphangiogenesis, VEGF-A
Head and neck cancer, including oral squamous cell carcinoma (OSCC), is the sixth most common cancer worldwide (Argiris et al, 2008). The incidence of OSCC is estimated at 263 900 cases and 128 000 deaths annually (Jemal et al, 2011), and an estimated 7850 patients die from this disease in the United States (Siegel et al, 2012). Oral squamous cell carcinoma has a high potential for local invasion and lymph node metastasis, and overall 5-year survival rates have not significantly improved during the past three decades (Dos Reis et al, 2008). The 5-year survival rate in OSCC is less than 50% (Marsh et al, 2011). Although more than 80% of early-stage OSCCs can be cured by treatment, approximately 70% of advanced-stage patients cannot be cured (Mydlarz et al, 2010). Therefore, early detection and clarification of detailed molecular mechanism of OSCC is very important.
MicroRNAs (miRNAs) are non-coding small RNAs, approximately 18–25 nucleotides in length (Bartel, 2009; Martello et al, 2010), which function as suppressors of gene expression by binding to the 3′-untranslated region (UTR) of target mRNAs (Calin and Croce, 2006; Liu et al, 2010; Martello et al., 2010). MicroRNA-126 (miR-126) is located within intron 7 of epidermal growth factor-like domain 7 (EGFL7) and is highly expressed in vascular endothelial cells (Fish et al, 2008; Wang et al, 2008a). In tumour cells, miR-126 acts as a tumour suppressor gene in many malignant tumour cells (Wang et al, 2008b; Liu et al, 2009; Saito et al, 2009; Feng et al, 2010; Miko et al, 2011; Zhu et al, 2011). MicroRNA-126 (miR-126) is significantly expressed lower in carcinoma as compared with adenoma and useful diagnostic marker after thyroid fine-needle aspiration biopsy (Kitano et al, 2011). MicroRNA-126 (miR-126) is also strongly downregulated in pancreatic cancer, with an associated elevation in K-Ras (Slaby et al, 2012), and lower expression of miR-126 is significantly correlated with short survival in non-small cell lung carcinoma (NSCLC) and renal cell carcinoma (Donnem et al, 2012; Jiao et al, 2012). Further, Yu et al (2009) have reported downregulation of miR-126 to promote oral carcinogenesis in Syrian hamster. Saito et al (2009) showed that miR-126 and EGFL7, which is the host gene for miR-126, are downregulated by DNA hypermethylation, and miR-126 and EGFL7 expression are restored after treatment with 5-aza-2′-deoxycytidine (5-Aza-dc) in urologic cancer. Silencing of miR-126 by the DNA methylation of EGFL7 was also reported in NSCLC (Watanabe et al, 2012). However, several reports have described an oncogenic role for miR-126, such as the inhibition of apoptosis in acute myeloid leukaemia and the promotion of gastric carcinogenesis (Otsubo et al, 2011). In addition, higher expression of miR-126 was shown to be a poor prognostic factor in NSCLC (Donnem et al, 2011) and promote metastasis in prostate cancer (Watahiki et al, 2011). Meister and Schmidt (2010) recently reported that higher expression of miR-126 accelerates cancer progression by activation of mitogen-activated protein kinase and Akt, whereas miR-126 suppresses tumour proliferation and invasion because of decreased Crk expression. These contradictory findings suggest that miR-126 has several functions specific to each type of malignancy.
Angiogenesis has a pivotal role in prenatal development, wound healing, chronic inflammation, tumour progression, and metastasis (Carmeliet, 2005; Sasahira et al, 2007a), and lymphangiogenesis promotes lymph node metastasis in cancer cells (Adams and Alitalo, 2007; Sasahira et al, 2010). Vascular endothelial growth factor (VEGF)-A and VEGF-C/D are one of the representative factors of angiogenesis and/or lymphangiogenesis (Adams and Alitalo, 2007), and we have also previously reported that the VEGF family induce angiogenesis and/or lymphangiogenesis in OSCC (Sasahira et al, 2007a, b, 2008, 2010). However, several reports have revealed that VEGF-A is not related with angiogenesis and VEGF-C/D are not associated with nodal metastasis in cancer (Currie et al, 2004; Nomiya et al, 2006; Miyahara et al, 2007; Donnem et al, 2009). It has also been reported that vessel numbers in nodal metastasis cases are lower than cases without metastasis (Moriyama et al, 1997). Thus, the role of tumour angiogenesis and lymphangiogenesis are still controversial.
Recently, it was reported that VEGF-A is a target gene of miR-126 and downregulation of miR-126 increases VEGF-A activity in lung (Liu et al, 2009; Zhu et al, 2012) and breast cancer (Zhu et al, 2011). However, other reports indicated that miR-126 is an inducer of angiogenesis by enhancing the proangiogenic activity of VEGF-A (Fish et al, 2008; Wang et al, 2008a). Thus, miR-126 may function differently in tumour cells and stromal cells. In this study, we examined the angiogenic role of miR-126 and confirmed the relationship between the expression of miR-126 and lymphangiogenesis in OSCC.
Materials and methods
Cell culture
Human OSCC cell lines, HSC3 and HSC4 cells, were obtained from the Health Science Research Resources Bank and maintained in Dulbbeco’s modified Eagle’s medium (DMEM; Wako Pure Chemical industries, Ltd, Osaka, Japan) supplemented with 10% fetal bovine serum (Sigma Chemical, St Louis, MO, USA) under the conditions of 5% CO2 in air at 37 °C. The HSC3 cells have high metastatic potential and HSC4 cells have low metastatic ability (Sasahira et al, 2010).
RNA isolation and quantitative reverse-transcription PCR
Total RNA and small RNA were extracted using RNeasy Mini Kit (Qiagen, Valencia, CA, USA) or mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA), and total RNA (1 μg) and small RNA (10 ng) were synthesised with the ReverTra Ace qRT Kit (Toyobo, Osaka, Japan) or TaqMan MicroRNA RT Kit (Applied Biosystems, Foster City, CA, USA), respectively. Quantitative reverse-transcription PCR (qRT–PCR) were performed on StepOne Plus Real-Time PCR Systems (Applied Biosystems) using EXPRESS qPCR Supermixes (Invitrogen, Carlsbad, CA, USA) and analyse the relative standard curve quantification method. The PCR condition was according to the manufacturer’s instructions and 18S (eukaryotic 18S rRNA) expression level was amplified for internal control. TaqMan MicroRNA assays of hsa-miR-126 and TaqMan Gene Expression Assays of VEGF-A, VEGF-C, VEGF-D, EGFL7, and 18S were purchased from Applied Biosystems. All PCRs were performed in triplicates.
Methylation-specific PCR
For the bisulfite modification of DNA, 2 μg of genomic DNA extracted using the QIAamp DNA Mini kit (Qiagen) was treated with the Epitect Bisulfite kit (Qiagen) according to the provider’s manual. For analysis of DNA methylation of EGFL7, we carried out methylation-specific PCR (MSP) using an Epitect MSP kit (Qiagen). The PCR products were separated by 6% non-denaturing polyacrylamide gels, stained with ethidium bromide (Sigma Chemical), and visualised under UV light.
The sequences of the primers methylated or unmethylated of EGFL7 are as follows (Saito et al, 2009): 5′-GTG GTG GTG GTG TGT GTG TGT TT-3′ and 5′-CTC AAC CCA ACC CAA ACA ACA ACC A-3′ for unmethylated EGFL7; 5′-GCG GCG CGT GCG CGT TT-3′ and 5′-CCA ACC CGA ACG ACG ACC G-3′ for methylated EGFL7.
DNA demethylation treatment
Previous results raise the possibility that miR-126 expression is regulated by DNA methylation (Saito et al, 2009). Therefore, we performed the demethylation treatment in OSCC cells. Each cell was seeded at a density of 1 × 106 cells ml−1. After 24 h, cells were treated with 1 μℳ 5-Aza-dc (Sigma Chemical) for 4 days. Treatment with 300 nℳ trichostatin A (TSA; Sigma Chemical) was also performed for 24 h. Cells then were harvested for miRNA extractions.
Transient transfection
Pre-miR-126 precursor, anti-miR126 inhibitor, pre-miR negative control 1, and anti-miR negative control 1 were purchased from Ambion. Pre-miR (10 nℳ and anti-miR (30 nℳ were transfected with Lipofectamine 2000 (Invitrogen) according to the provider’s recommendations.
Cell growth assay
The cells were seeded at density of 2000 cells per well of 96-well tissue culture plates and incubated for 48 h at 37 °C. Cell growth was assessed by MTT assay using the incorporation of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma Chemical). The experiments were performed in triplicates.
In vitro invasion assay
A modified Boyden chamber assay was performed using the BD BioCoat Cell Culture Inserts glued to type IV collagen (Becton-Dickinson, Bedford, MA, USA) as described previously. Cells were suspended in 500 μl of DMEM and placed in the insert. After 48 h incubation at 37 °C, the filters were stained with haematoxylin. The stained cells were counted in whole inserts at × 100 magnification. Each experiment was repeated at least three times.
Apoptosis assay
Apoptotic cells were detected by the terminal deoxynucleotidyl trsnsferase (TdT)-mediated dUTP-biotin nick and labelling (TUNEL) assay using the In Situ Cell Death Detection Kit, POD (Roche Diagnostics, Indianapolis, IN, USA). We also confirmed the activation of caspase-3 was detected using CaspACE Assay system, Colorimetric (Promega, Madison, WI, USA) according to the manufacturer’s protocol. The experiments were performed in triplicates.
Oral squamous cell carcinoma samples
One-hundred and eighteen patients (68 men, 50 women; 46–91 years of age, mean: 67.4 years) of primary OSCCs, who were treated at Nara Medical University Hospital, Kashihara, Japan, from February 2000 to October 2008, were randomly selected. Tumours were staged according to the UICC TNM classification system, 7th edition, and histopathological grading was in accordance with the World Health Organization criteria. None of the patients was treated before surgery and sample preparation. Medical records and prognostic follow-up data were obtained from the patient database maintained by the hospital. The median follow-up period was 3.4 years (range 0.5–5.9). For strict privacy protection, identifying information for all samples was removed before analysis. This procedure was in accordance with the Ethical Guidelines for Human Genome/Gene Research of the Japanese Government. To determine the relation between miR-126 expression and clinicopathological characteristics, we divided into two groups according to expression levels those with values higher the mean value for the entire group and those with lower than the group mean value. Ten samples of normal oral mucosa (six man, four woman; 20–47 years of age, mean: 37.9 years) were used as control.
Laser-capture microdissection
As previous reports suggested that functions of miR-126 are different in cancer cells and stromal cells (Fish et al, 2008; Wang et al, 2008a; Liu et al, 2009; Zhu et al, 2011), we performed the laser-capture microdissection (LCM) to selectively extract small RNA from OSCC cells. Tissue sections (7 μm) were prepared from each paraffin blocks and stained using haematoxylin and eosin. Slides were transferred for microdissection using a Pix Cell II laser capture microscope (Arcturus, Mountain View, CA, USA) according to the manufacturer’s instructions. Approximately 5000 tumour cells were microdissected from each tissue. MicroRNA was extracted using the mirVana miRNA Isolation Kit (Ambion).
Immunohistochemistry
Immunohistochemical analysis was performed with the EnVision+ DualLink (DAKO, Carpinteria, CA, USA) system and antigen retrieval was performed with microwaving in citrate buffer at 95 °C for 45 min. After endogeneous peroxidase block by 3% H2O2-methanol, anti-CD34 antibody (a marker for vascular endothelial cells; DAKO) and anti-LYVE-1 antibody (a marker for lymphovascular endothelial cells; Abcam, Tokyo, Japan) were used for primary antibody for 2 h and incubated with the secondary antibody for 30 min at room temperature. The specimens were colour-developed with diaminobenzidine solution (DAKO) and specimens were counterstained with Mayer’s haematoxylin (Sigma Chemical). The microvessel density (MVD) and lymphovessel density (LVD) were measured on anti-CD34 and anti-LYVE-1 antibody immunopositive specimens, respectively. To quantify MVD or LVD, five maximum vessel-density fields were selected from around of the tumour cells (the ‘hot spot’) and examined under a 200-fold magnification by a microscope and averaged. We divided the tissue samples into two groups according to MVD levels: those with values higher than the mean value for the entire group, and those with lower values than the group mean value. The same was applied based on the LVD values.
Statistical analysis
The correlation of variables was performed by Fischer’s exact test, one-factor ANOVA test, and Mann–Whitney U-test. Disease-free survival analysis was calculated using the Kaplan–Meier method, and differences between groups were tested by means of a log lank test. Univariate analysis for disease-free survival was calculated by log lank test. For multivariate analysis, Cox proportional hazards model was used (described as risk ratio with 95% confidence intervals, together with the P-value). All statistical analysis was carried out with JMP8 (SAS Institute, Cary, NC, USA). P-values less than 0.05 were regarded as statistically significant.
Results
Expression of miR-126 and EGFL7 in OSCC cells
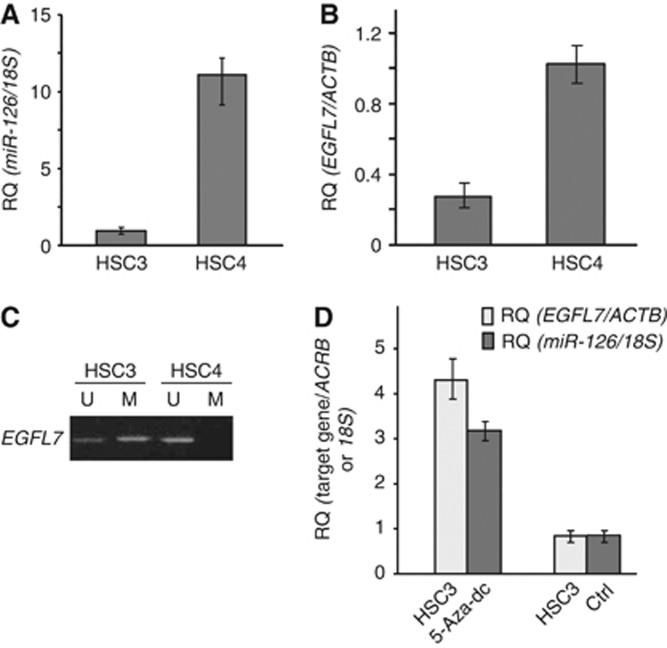
The expressions of miR-126 and EGFL7, which is the host gene of miR-126, were examined by qRT–PCR in the HSC3 and HSC4 cells. The expression levels of miR-126 and EGFL7 in the HSC3 cells with high metastatic potential were approximately 10 times lower than HSC4 cells with low metastatic potential (Figures 1A and B). Examination of the methylation status of EGFL7 by MSP showed the presence of a methylated EGFL7 band in the HSC3 cells that was absent in the HSC4 cells (Figure 1C). We next investigated the possibility that demethylation treatment could upregulate the miR-126 and EGFL7 expressions in the HSC3 cells. The treatment with 5-Aza-dc restored the expressions of miR-126 and EGFL7 (Figure 1D), whereas the expression levels of miR-126 was not changed by TSA treatment (data not shown).
Figure 1.
Expression levels of miR-126 (A) and EGFL7 (B) measured using qRT–PCR in the OSCC cells. The 18S or beta-actin (ACTB) transcript levels were used as internal controls for miR-126 and EGFL7 expressions, respectively. The methylation status of EGFL7 by MSP in the OSCC cells is shown in (C). The changes of the miR-126 and EGFL7 expressions in the 5-Aza-dc-treated HSC3 cells are shown in (D). The expression levels of miR-126 and EGFL7 in the untreated HSC3 cells were set to 1.
Functional analysis of miR-126 in the OSCC cells
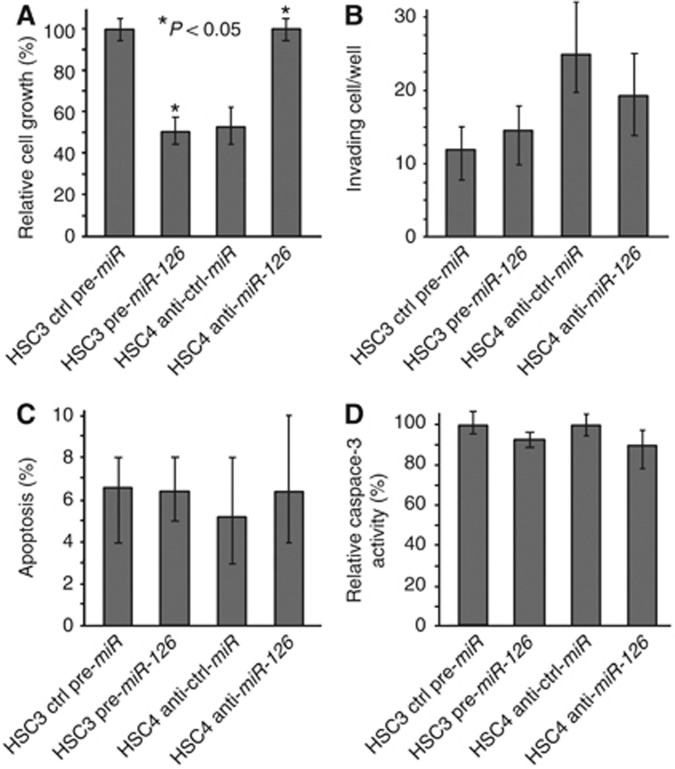
The role of miR-126 in OSCC progression was investigated using a functional analysis. The growth of HSC3 cells treated with pre-miR-126 was inhibited in comparison with that of cells treated with control pre-miR. However, the growth of the HSC4 cells treated with an anti-miR-126 inhibitor was restored compared with that of the cells treated with the control anti-miR inhibitor (Figure 2A). The number of invading cells was not affected by the treatment with pre-miR-126 in the HSC3 cells or anti-miR-126 in the HSC4 cells, in contrast to the number of cells treated with the control miR (Figure 2B). No significant changes in TUNEL-positive cells and caspase-3 activity were observed between the pre-miR-126-treated HSC3 cells or the anti-miR-126-treated HSC4 cells and the control-treated HSC3 or HSC4 cells (Figures 2C and D).
Figure 2.
Functional analysis of miR-126 in the OSCC cells. Cell growth (A), invasion (B), ratio of TUNEL-positive cells (C), and activity of caspase-3 (D) in the HSC3 or HSC4 cells treated with a miR-126 precursor or anti-miR-126, respectively. Statistical significance was calculated by the Mann–Whitney U-test.
Changes in the VEGF expression levels in the OSCC cells treated with anti-miR-126 or pre-miR-126
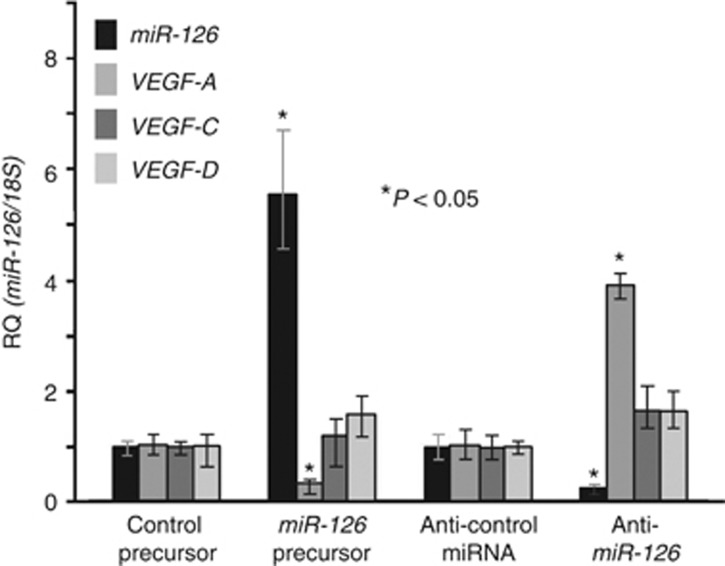
As VEGF-A has been suggested to be negatively or positively regulated by miR-126 (Fish et al, 2008; Liu et al, 2009; Zhu et al, 2011), we examined the effect of miR-126 on the VEGF-A, VEGF-C, and VEGF-D expressions in the OSCC cells (Figure 3). Exposure to pre-miR-126 increased miR-126 expression but did not affect the VEGF-C and VEGF-D expression levels in the HSC3 cells, whereas the treatment with anti-miR-126 inhibitor decreased the miR-126 expression but not the VEGF-C and VEGF-D expressions in the HSC4 cells. However, induction of miR-126 in the HSC3 cells and reduction of miR-126 in the HSC4 cells were associated with the downregulation and the upregulation of VEGF-A expression, respectively. This was verified by the co-transfection of the VEGF-A wild-type 3′-UTR with pre-miR-126, which decreased luciferase activity, whereas this effect was completely ablated by deletion of the miR-126-binding site in the VEGF-A 3′-UTR (data not shown). These results suggested that miR-126 acts as a negative regulator of VEGF-A, but not of VEGF-C and VEGF-D.
Figure 3.
Effect of the miR-126, VEGF-A, VEGF-C, and VEGF-D expression levels in the OSCC cells treated with anti-miR-126 or the miR-126 precursor. The expression levels in the control miRNA precursor-treated HSC3 cells and the control anti-miRNA-treated HSC4 cells were set to 1. Statistical significance was determined using the Mann–Whitney U-test.
Relationship between the miR-126 expression and the clinicopathological characteristics in the OSCC specimens
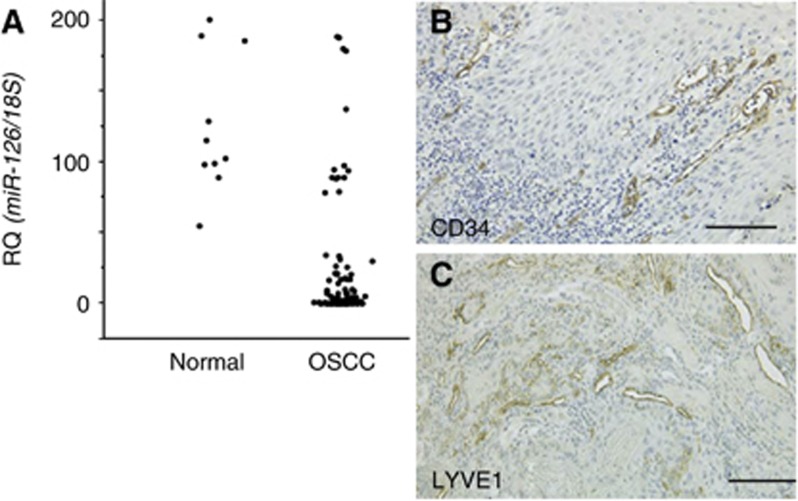
MicroRNA-126 (MiR-126) expression was examined by qRT–PCR in the 118 cases with OSCC. The expression levels of miR-126 in almost all the OSCC cases were low in comparison with the normal oral mucosa (Figure 4A). The miR-126 expression and the clinicopathological characteristics in the OSCC specimens are summarised in Table 1. Ninety-four of the 118 OSCC samples (79.7%) showed low expression of miR-126. In the cases with local progression (T3 and T4), the expression levels of miR-126 were significantly lower than those in the T1 and T2 cases (P=0.036). Low miR-126 expression was detected in 52 (70.3%) of the 74 cases with early clinical-stage disease (stages I and II) and in 42 (95.5%) of the 44 cases with advanced clinical-stage disease (stages III and IV; P=0.0006). Although downregulation of miR-126 was observed in 100% (34 out of 34) of the nodal metastasis-positive cases, 71.4% (60 out of 84) of the cases without nodal metastasis showed low expression of miR-126 (P<0.0001). No significant correlation was found between the expression levels of miR-126 and age, sex, site, or histological differentiation.
Figure 4.
Expression levels of miR-126 in OSCC compared with that in the normal oral mucosa (A) and MVD (B) or LVD (C) by immunohistochemistry. MVD and LVD were assessed according to the number of anti-CD34 and anti-LYVE1-positive vessels. Scale bar, 100 μm.
Table 1. Relationship between the miR-126 expressions and the clinicopathological parameters.
| miR-126 | |||
|---|---|---|---|
| Parameters | Low | High | P -value |
| Sex | |||
| Male | 52 | 16 | |
| Female | 42 | 8 | 0.1573 |
| Age | |||
| ⩽65 | 34 | 12 | |
| >65 | 60 | 12 | 0.221 |
| Site | |||
| Tongue | 48 | 16 | |
| Other | 46 | 8 | 0.1268 |
| Histological differentiation | |||
| Well | 56 | 14 | |
| Mod, Poor | 38 | 10 | 0.5451 |
| T classification | |||
| T1–T2 | 68 | 22 | |
| T3–T4 | 26 | 2 | 0.036 |
| Clinical stage | |||
| I, II | 52 | 22 | |
| III, IV | 42 | 2 | 0.0006 |
| Nodal metastasis | |||
| Negative | 60 | 24 | |
| Positive | 34 | 0 | <0.0001 |
Abbreviations: miR-126=microRNA-126; Well=well-differentiated squamous cell carcinoma; Mod=moderately differentiated squamous cell carcinoma; Poor=poorly differentiated squamous cell carcinoma.
The statistical analysis was performed by Fischer’s exact t-test. The T classification and the clinical stage were performed according to the TNM classification.
On the basis of the in vitro results showing that miR-126 is a negative regulator of VEGF-A, we verified the relationship between miR-126 and angiogenesis and lymphangiogenesis in OSCC (Table 2). A significant inverse correlation was observed between the miR-126 expression levels and MVD (P<0.0001; Figure 4B) or LVD (P<0.0001; Figure 4D).
Table 2. Relationship between the miR-126 expressions and angiogenesis or lymphangiogenesis.
| miR-126 | |||
|---|---|---|---|
| Parameter | Low | High | P -value |
| MVD | 25.5761±10.1091 | 11.0231±4.6017 | <0.0001 |
| LVD | 25.4468±10.0379 | 10.3167±4.014 | <0.0001 |
Abbreviations: ANOVA=analysis of variance; LVD=lymphovessel density; miR-126=microRNA-126; MVD=microvessel density.
The statistical analysis was performed by one-factor ANOVA. Means±s.d., each s.d. was less than 10%.
Relation between the miR-126 expression and the disease-free survival, and the prognosis of OSCCs
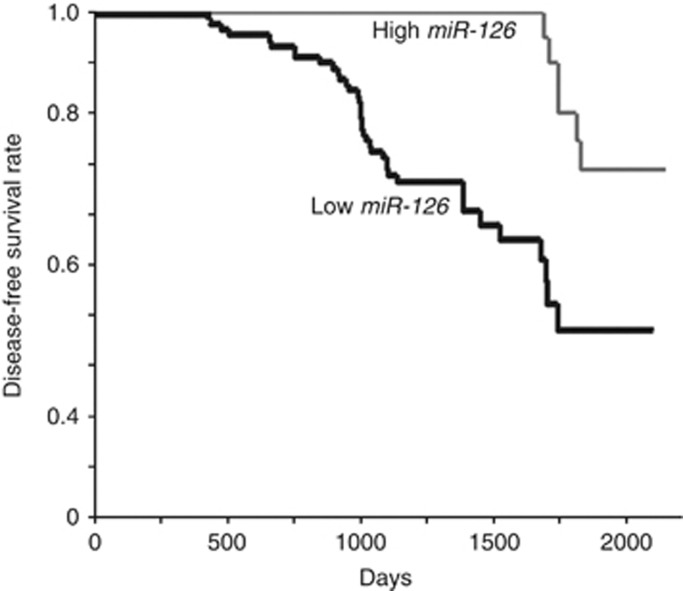
Local and nodal recurrence occurred in 44 of the 118 cases. The disease-free survival analysis of the OSCC patients revealed that a poor prognosis was associated with low miR-126 expression compared with high miR-126 expression cases (P=0.0013; Figure 5). The univariate analysis using the log-rank test indicated that the histological differentiation (P=0.0022), the clinical stage (P=0.0015), the nodal metastasis (P<0.0001), and the miR-126 expression levels (P=0.0006) were associated with the poor outcome in OSCCs (Table 3). The multivariate analysis using the Cox proportional hazards model showed that nodal metastasis (P=0.0093) and miR-126 expression levels (P=0.048) were prognostic factors for disease-free survival periods (Table 3).
Figure 5.
Disease-free survival curves of the OSCC patients calculated using the Kaplan–Meier method. Disease-free survival was analysed in correlation to the expression levels of miR-126. The miR-126-high cases (n=24, event and censored data were 6 and 18, respectively), miR-126-low cases (n=94, event and censored data were 38 and 56, respectively).
Table 3. Univariate and multivariate analysis of disease-free survival.
| Parameters | Risk ratio | 95% CI | P -value |
|---|---|---|---|
| Univariate analysis | |||
| Age | |||
| ⩽65 | 1 | ||
| >65 | 0.7672 | 0.4227–1.3998 | 0.3839 |
| Gender | |||
| Male | 1 | ||
| Female | 0.8843 | 0.4873–1.6304 | 0.6885 |
| Site | |||
| Tongue | 1 | ||
| Other | 0.9075 | 0.4927–1.6449 | 0.750 |
| Histology | |||
| Well | 1 | ||
| Mod/Poor | 2.4833 | 1.3989–4.7785 | 0.0022 |
| T classification | |||
| T1-T2 | 1 | ||
| T3-T4 | 1.4582 | 0.7206–2.759 | 0.2805 |
| Clinical stage | |||
| I, II | 1 | ||
| III, IV | 2.6417 | 1.4539–4.8528 | 0.0015 |
| Nodal metastasis | |||
| Negative | 1 | ||
| Positive | 4.7203 | 2.5653–8.7999 | <0.0001 |
| MVD | |||
| Low | 1 | ||
| High | 1.0094 | 0.9827–1.0357 | 0.4872 |
| LVD | |||
| Low | 1 | ||
| High | 0.8492 | 0.4444–1.5712 | 0.607 |
| miR-126 | |||
| High | 1 | ||
| Low | 4.0492 | 1.7558–11.0409 | 0.0006 |
| Multivariate analysis | |||
| Histology | |||
| Well | 1 | ||
| Mod, Poor | 0.6673 | 0.3493–1.2557 | 0.2101 |
| Clinical stage | |||
| I, II | 1 | ||
| III, IV | 1.8972 | 0.9434–3.8782 | 0.0725 |
| Nodal metastasis | |||
| Negative | 1 | ||
| Positive | 2.6265 | 1.2658–5.6413 | 0.0093 |
| miR-126 | |||
| High | 1 | ||
| Low | 2.631 | 0.9886–7.9851 | 0.048 |
Abbreviations: CI=confidence intervals; miR-126=microRNA-126; Mod=moderately differentiated squamous cell carcinoma; Poor=poorly differentiated squamous cell carcinoma; Well=well-differentiated squamous cell carcinoma.
The univariate analysis was performed by log-rank test, and the multivariate analysis was performed by the Cox proportional hazard model.
Discussion
The present results show that miR-126 is a negative regulator of VEGF-A and promotes cell growth in OSCC cells. In addition, the decreased miR-126 expression was associated with the induction of tumoural angiogenesis and lymphangiogenesis, tumour progression, nodal metastasis, and poor prognosis in the OSCC cases. The HSC3 cells are human OSCC-derived metastatic cells, whereas the HSC4 cells have low metastatic potential (Momose et al, 1989; Sasahira et al, 2007a; Sasahira et al, 2008; Sasahira et al, 2010). The HSC3 cells are characterised by adhesion to type-IV collagen, colony formation in a type-I collagen matrix (Momose et al, 1989), high heparanase activity (Ikuta et al, 2001), reduction of nm23H1 expression and upregulated matrix metalloproteinase (MMP)-2/MMP-9 (Khan et al, 2001), and higher expression of VEGF-A/C/D in comparison with HSC4 cells. In this study, the HSC3 cells showed lower expression of miR-126 compared with the HSC4 cells, and the decrease of miR-126 activity might be associated with a higher capacity for lymph node metastasis in these cells. Further investigation of the expression levels of miR-126 in OSCC may help predict the tendency for lymph node metastasis development in this type of malignancy. Vascular endothelial growth factor (VEGF)-A is a potent trigger for tumoural angiogenesis (Stockmann et al, 2008); however, we previously reported that VEGF-A induces not only angiogenesis but also lymphangiogenesis, and VEGF-A-dependent angiogenesis and lymphangiogenesis accelerate local progression, nodal metastasis, and recurrence of OSCC (Sasahira et al, 2010). This is in agreement with previous studies that demonstrated a strong correlation between VEGF-A expression and tumour progression (Takahashi et al, 1995), worse prognosis (Maeda et al, 1996), and lymph node metastasis via induction of lymphangiogenesis (Nagy et al, 2002; Hirakawa et al, 2005). Further examination will help reveal the mechanism underlying the role of miR-126 and VEGF-A-related angiogenesis and lymphangiogenesis in tumour progression.
MicroRNA-126 (MiR-126) is an endothelial-specific miRNA that is located within intron 7 of EGFL7 (Fish et al, 2008; Wang et al, 2008a). An intronic miRNA tends to be co-expressed with its host gene (Baskerville and Bartel, 2005; Saito et al, 2009), and a previous report showed that miR-126 and its host gene, EGFL7, are downregulated by DNA methylation, with restoration of expression levels by epigenetic treatment (Saito et al, 2009). We also observed decreased expression and methylation of EGFL7 in the highly metastatic OSCC cell line HSC3 compared with the HSC4 cell line with lower metastasis. In addition, EGFL7 and miR-126 recovered normal expression levels in response to the 5-Aza-dc treatment in the HSC3 cells. Further examination of miR-126 might reveal this molecule as a useful target for epigenetic therapy in progressive and metastatic OSCC.
MicroRNAs are well preserved in formalin-fixed, paraffin-embedded (FFPE) specimens owing to the small size of the RNA (Xi et al, 2007; Donnem et al, 2011). Most of the previous reports on miR-126 expression were based on the analysis of frozen tissues containing non-tumour cells or tumour stromal cells including vessels (Wang et al, 2008b; Saito et al, 2009; Liu et al, 2009; Feng et al, 2010; Otsubo et al, 2011; Zhu et al, 2011). However, miR-126 is a positive regulator of angiogenesis in normal endothelial cells (Fish et al, 2008; Wang et al, 2008a; Liu et al, 2009; Miko et al, 2011; Zhu et al, 2011). In vasculogenesis, which is not associated with cancer, such as in wound healing, overexpression of miR-126 in endothelial cells enhances VEGF-A activity and promotes vessel formation by repressing the expression of sprouty-related protein-1 (Spred-1; Wang et al, 2008a). Sprouty-related protein-1 (spred-1) is an intracellular inhibitor of angiogenic signals (Wang et al, 2008a). As VEGF-A is not a direct target of miR-126 in endothelial cells, the use of fresh frozen samples, including stromal cells, may lead to an incorrect interpretation of results. We therefore selected OSCC cells using the LCM technique in FFPE tissues and performed the expression analysis of miR-126 by real-time RT–PCR. We also confirmed the increased miR-126 expression and the decreased Spred-1 expression in the microdissected endothelial cells by qRT–PCR (data not shown).
In conclusion, the present results demonstrate that low miR-126 expression is correlated with tumour progression through the activation of angiogenesis and lymphangiogenesis in OSCC. However, the exact mechanism underlying the miR-126-mediated activation of angiogenesis and lymphangiogenesis remains to be elucidated. In vivo studies will be helpful in the future to further clarify this mechanism. Our results suggest that re-expression of miR-126 could be a useful therapeutic strategy against human OSCC.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science, Japan.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
The authors declare no conflict of interest.
References
- Adams RH, Alitalo K (2007) Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol 8(6): 464–478 [DOI] [PubMed] [Google Scholar]
- Argiris A, Karamouzis MV, Raben D, Ferris RL (2008) Head and neck cancer. Lancet 371: 1695–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2): 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP (2005) Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11(3): 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6(11): 857–866 [DOI] [PubMed] [Google Scholar]
- Carmeliet P (2005) Angiogenesis in life, disease and medicine. Nature 438(7070): 932–936 [DOI] [PubMed] [Google Scholar]
- Currie MJ, Hanrahan V, Gunningham SP, Morrin HR, Frampton C, Han C, Robinson BA, Fox SB (2004) Expression of vascular endothelial growth factor D is associated with hypoxia inducible factor (HIF-1alpha) and the HIF-1alpha target gene DEC1, but not lymph node metastasis in primary human breast carcinomas. J Clin Pathol 57(8): 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnem T, Al-Shibli K, Al-Saad S, Delghandi MP, Busund LT, Bremnes RM (2009) VEGF-A and VEGFR-3 correlate with nodal status in operable non-small cell lung cancer: inverse correlation between expression in tumor and stromal cells. Lung Cancer 63(2): 277–283 [DOI] [PubMed] [Google Scholar]
- Donnem T, Fenton CG, Lonvik K, Berg T, Eklo K, Andersen S, Stenvold H, Al-Shibli K, Al-Saad S, Bremnes RM, Busund LT (2012) MicroRNA signatures in tumor tissue related to angiogenesis in non-small cell lung cancer. PLos One 7(1): e29671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnem T, Lonvik K, Eklo K, Berg T, Sorbye SW, Al-Shibli K, Al-Saad S, Andersen S, Stenvold H, Bremnes RM, Busund LT (2011) Independent and tissue-specific prognostic impact of miR-126 in nonsmall cell lung cancer: coexpression with vascular endothelial growth factor-A predicts poor survival. Cancer 117(14): 3193–3200 [DOI] [PubMed] [Google Scholar]
- Dos Reis PP, Bharadwaj RR, Machado J, Macmillan C, Pintilie M, Sukhai MA, Perez-Ordonez B, Gullane P, Irish J, Kamel-Reid S (2008) Claudin 1 overexpression increases invasion and is associated with aggressive histological features in oral squamous cell carcinoma. Cancer 113(11): 3169–3180 [DOI] [PubMed] [Google Scholar]
- Feng R, Chen X, Yu Y, Su L, Yu B, Li J, Cai Q, Yan M, Liu B, Zhu Z (2010) miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett 298(1): 50–63 [DOI] [PubMed] [Google Scholar]
- Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D (2008) miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 15(2): 272–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa S, Kodama S, Kunstfeld R, Kajiya K, Brown LF, Detmar M (2005) VEGF-A induces tumor and sentinel lymph node lymphangiogenesis and promotes lymphatic metastasis. J Exp Med 201(7): 1089–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta M, Podyma KA, Maruyama K, Enomoto S, Yanagishita M (2001) Expression of heparanase in oral cancer cell lines and oral cancer tissues. Oral Oncol 37(2): 177–184 [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2): 69–90 [DOI] [PubMed] [Google Scholar]
- Jiao LR, Frampton AE, Jacob J, Pellegrino L, Krell J, Giamas G, Tsim N, Vlavianos P, Cohen P, Ahmad R, Keller A, Habib NA, Stebbing J, Castellano L (2012) MicroRNAs targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PLos One 7(2): e32068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MH, Yasuda M, Higashino F, Haque S, Kohgo T, Nakamura M, Shindoh M (2001) nm23-H1 suppresses invasion of oral squamous cell carcinoma-derived cell lines without modifying matrix metalloproteinase-2 and matrix metalloproteinase-9 expression. Am J Pathol 158(5): 1785–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano M, Rahbari R, Patterson EE, Xiong Y, Prasad NB, Wang Y, Zeiger MA, Kebebew E (2011) Expression profiling of difficult-to-diagnose thyroid histologic subtypes shows distinct expression profiles and identify candidate diagnostic microRNAs. Ann Surg Oncol 18(12): 3443–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Peng XC, Zheng XL, Wang J, Qin YW (2009) MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer 66(2): 169–175 [DOI] [PubMed] [Google Scholar]
- Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, Demidenko E, Korc M, Shi W, Preis M, Dragnev KH, Li H, Direnzo J, Bak M, Freemantle SJ, Kauppinen S, Dmitrovsky E (2010) MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest 120(4): 1298–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Chung YS, Ogawa Y, Takatsuka S, Kang SM, Ogawa M, Sawada T, Sowa M (1996) Prognostic value of vascular endothelial growth factor expression in gastric carcinoma. Cancer 77(5): 858–863 [DOI] [PubMed] [Google Scholar]
- Marsh D, Suchak K, Moutasim KA, Vallath S, Hopper C, Jerjes W, Upile T, Kalavrezos N, Violette SM, Weinreb PH, Chester KA, Chana JS, Marshall JF, Hart IR, Hackshaw AK, Piper K, Thomas GJ (2011) Stromal features are predictive of disease mortality in oral cancer patients. J Pathol 223(4): 470–481 [DOI] [PubMed] [Google Scholar]
- Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, Parenti AR, Daidone MG, Bicciato S, Piccolo S (2010) A MicroRNA targeting dicer for metastasis control. Cell 141(7): 1195–1207 [DOI] [PubMed] [Google Scholar]
- Meister J, Schmidt MH (2010) miR-126 and miR-126*: new players in cancer. Sci World J 10: 2090–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miko E, Margitai Z, Czimmerer Z, Varkonyi I, Dezso B, Lanyi A, Bacso Z, Scholtz B (2011) miR-126 inhibits proliferation of small cell lung cancer cells by targeting SLC7A5. FEBS Lett 585(8): 1191–1196 [DOI] [PubMed] [Google Scholar]
- Momose F, Araida T, Negishi A, Ichijo H, Shioda S, Sasaki S (1989) Variant sublines with different metastatic potentials selected in nude mice from human oral squamous cell carcinomas. J Oral Pathol Med 18(7): 391–395 [DOI] [PubMed] [Google Scholar]
- Mydlarz WK, Hennessey PT, Califano JA (2010) Advances and perspectives in the molecular diagnosis of head and neck cancer. Expert Opin Med Diagn 4(1): 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy JA, Vasile E, Feng D, Sundberg C, Brown LF, Detmar MJ, Lawitts JA, Benjamin L, Tan X, Manseau EJ, Dvorak AM, Dvorak HF (2002) Vascular permeability factor/vascular endothelial growth factor induces lymphangiogenesis as well as angiogenesis. J Exp Med 196(11): 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomiya T, Nemoto K, Nakata E, Takai Y, Yamada S (2006) Expression of thymidine phosphorylase and VEGF in esophageal squamous cell carcinoma. Oncol Rep 15(6): 1497–1501 [PubMed] [Google Scholar]
- Miyahara M, Tanuma J, Sugihara K, Semba I (2007) Tumor lymphangiogenesis correlates with lymph node metastasis and clinicopathologic parameters in oral squamous cell carcinoma. Cancer 110(6): 1287–1294 [DOI] [PubMed] [Google Scholar]
- Moriyama M, Kumagai S, Kawashiri S, Kojima K, Kakihara K, Yamamoto E (1997) Immunohistochemical study of tumour angiogenesis in oral squamous cell carcinoma. Oral Oncol 33(5): 369–374 [DOI] [PubMed] [Google Scholar]
- Otsubo T, Akiyama Y, Hashimoto Y, Shimada S, Goto K, Yuasa Y (2011) MicroRNA-126 inhibits SOX2 expression and contributes to gastric carcinogenesis. PLoS One 6(1): e16617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Friedman JM, Chihara Y, Egger G, Chuang JC, Liang G (2009) Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem Biophys Res Commun 379(3): 726–731 [DOI] [PubMed] [Google Scholar]
- Sasahira T, Kirita T, Bhawal UK, Ikeda M, Nagasawa A, Yamamoto K, Kuniyasu H (2007a) The expression of receptor for advanced glycation end products is associated with angiogenesis in human oral squamous cell carcinoma. Virchows Arch 450(3): 287–295 [DOI] [PubMed] [Google Scholar]
- Sasahira T, Kirita T, Bhawal UK, Yamamoto K, Ohmori H, Fujii K, Kuniyasu H (2007b) Receptor for advanced glycation end products (RAGE) is important in the prediction of recurrence in human oral squamous cell carcinoma. Histopathology 51(2): 166–172 [DOI] [PubMed] [Google Scholar]
- Sasahira T, Kirita T, Kurihara M, Yamamoto K, Bhawal UK, Bosserhoff AK, Kuniyasu H (2010) MIA-dependent angiogenesis and lymphangiogenesis are closely associated with progression, nodal metastasis and poor prognosis in tongue squamous cell carcinoma. Eur J Cancer 46(12): 2285–2294 [DOI] [PubMed] [Google Scholar]
- Sasahira T, Kirita T, Oue N, Bhawal UK, Yamamoto K, Fujii K, Ohmori H, Luo Y, Yasui W, Bosserhoff AK, Kuniyasu H (2008) High mobility group box-1-inducible melanoma inhibitory activity is associated with nodal metastasis and lymphangiogenesis in oral squamous cell carcinoma. Cancer Sci 99(9): 1806–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. Cancer J Clin 62(1): 10–29 [DOI] [PubMed] [Google Scholar]
- Slaby O, Redova M, Poprach A, Nekvindova J, Iliev R, Radova L, Lakomy R, Svoboda M, Vyzula R (2012) Identification of MicroRNAs associated with early relapse after nephrectomy in renal cell carcinoma patients. Genes Chromosomes Cancer 51(7): 707–716 [DOI] [PubMed] [Google Scholar]
- Stockmann C, Doedens A, Weidemann A, Zhang N, Takeda N, Greenberg JI, Cheresh DA, Johnson RS (2008) Deletion of vascular endothelial growth factor in myeloid cells accelerates tumorigenesis. Nature 456(7223): 814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Kitadai Y, Bucana CD, Cleary KR, Ellis LM (1995) Expression of vascular endothelial growth factor and its receptor, KDR, correlates with vascularity, metastasis, and proliferation of human colon cancer. Cancer Res 55(18): 3964–3968 [PubMed] [Google Scholar]
- Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN (2008a) The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell 15(2): 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM (2008b) Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One 3(7): e2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watahiki A, Wang Y, Morris J, Dennis K, O'Dwyer HM, Gleave M, Gout PW, Wang Y (2011) MicroRNAs associated with metastatic prostate cancer. PLoS One 6(9): e24950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Emoto N, Hamano E, Sunohara M, Kawakami M, Kage H, Kitano K, Nakajima J, Goto A, Fukayama M, Nagase T, Yatomi Y, Ohishi N, Takai D (2012) Genome structure-based screening identified epigenetically silenced microRNA associated with invasiveness in non-small-cell lung cancer. Int J Cancer 130(11): 2580–2590 [DOI] [PubMed] [Google Scholar]
- Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, Ju J (2007) Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA 13(10): 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Wang XY, Gong RG, Li A, Yang S, Cao YT, Wen YM, Wang CM, Yi XZ (2009) The expression profile of microRNAs in a model of 7,12-dimethyl-benz[a]anthrance-induced oral carcinogenesis in Syrian hamster. J Exp Clin Cancer Res 2009(28): 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu T, Bai Y, Shen Y, Yuan W, Jing Q, Qin Y (2011) Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem 351(1-2): 157–164 [DOI] [PubMed] [Google Scholar]
- Zhu X, Li H, Long L, Hui L, Chen H, Wang X, Shen H, Xu W (2012) miR-126 enhances the sensitivity of non-small cell lung cancer cells to anticancer agents by targeting vascular endothelial growth factor A. Acta Biochim Biophys Sin 44(6): 519–526 [DOI] [PubMed] [Google Scholar]