Abstract
We demonstrate that the mitogen-activated protein kinases extracellular signal-regulated kinase (ERK)-1 and ERK-2 have a central role in mediating T-cell receptor-dependent induction of IL4 expression in human CD4+ T cells. Significantly, this involved a novel mechanism wherein receptor cross-linking induced activated ERK to physically associate with a promoter element on the IL4 gene. The proximally localized ERK then facilitated recruitment of the key transcription factors necessary for initiating IL4 gene transcription. Although both ERK-1 and ERK-2 bound to the promoter, recruitment of either one alone was found to be sufficient. We thus identify a novel mode of function for ERK wherein its physical association with the promoter serves as a prerequisite for enhanceosome assembly. This unusual pathway is also indispensable for human Th2-cell differentiation.
Keywords: Th-2 differentiation, IL-4 transcription, MAP kinase
The process of Th2 differentiation has been broadly divided into three separate stages, the first of which is initiation.1 Here, cooperativity between T-cell receptor (TCR) and interleukin (IL)-4 receptor-dependent signals activates various transcription factors (TFs) including signal transducers and activators of transcription (STAT)6 and nuclear factor of activated T cells (NFAT), which collectively induce transcription of both IL4 and the gene encoding the TF GATA3 (GATA binding protein 3).2 GATA3 then functions as the major regulator of Th2 lineage commitment.3 The second stage of reinforcement follows initiation, and primarily consists of an IL-4-dependent feedback loop that further reinforces its own expression in an autocrine and paracrine manner. Maintenance, the final stage, represents the culmination of epigenetic events that imprint the lineage choice in a heritable manner. In addition to ensuring locus accessibility of genes that characterize the Th2 phenotype, this also involves silencing of expression of Th1 cytokine genes.1, 4 The driving force for cells through each of these individual stages is provided by IL-4, and several studies have speculated that this derives from an external source, although the identity of the latter is still in question.1 Nonetheless, concomitant reports indicating that Th2 responses are optimally generated only when T cells produce IL-4 also argue for at least a significant role for the endogenously expressed cytokine by activated T cells.5, 6
Stimulation of naive CD4+ T cells through the TCR induces activation of two major signaling pathways, namely the Ras/mitogen-activated protein kinase (MAPK) pathway and the calcium-dependent calcineurin pathway.7, 8 Of these, the downstream pathways initiated by the canonical MAPKs, extracellular signal-regulated kinase (ERK)-1 and ERK-2 (ERK), have been implicated in regulating diverse cellular functions such as differentiation, division, movement and apoptosis of cells.9, 10 Importantly, ERK has also been shown to regulate differentiation of naive CD4+ T cells. Thus, for example, the TCR-activated Ras/ERK pathway mediates enhancement of IL-4R signaling, thereby contributing towards the generation of Th2 cells.11 In addition to this, related studies have also demonstrated the existence of crosstalk between the Ras/ERK and IL-4R-signaling pathways, which then regulates IL4 transcription.12 At least one of the mechanisms through which this is achieved seems to be through altering the composition of the AP-1 complex, in favor of those composed of Jun–Jun homodimers.13 Most significant in this context perhaps is the observation that genetic disruption of the MAPK pathway impairs Th2 differentiation in mice,14 with a consequent bias towards Th1 immune responses.15
Thus, although there is sufficient evidence to suggest that ERK serves as an important regulator of Th2-cell differentiation, the precise molecular mechanisms by which this influence is exerted remains ill-resolved. Therefore, we investigated this specific aspect in this study. Although our results confirm an obligatory role for ERK activation during Th2-cell differentiation, they further establish that this action is primarily mediated through the regulation of IL4 transcription. Importantly, an unusual mechanism was found to be involved wherein both ERK-1 and ERK-2 proteins were directly recruited at the proximal promoter of the IL4 gene. This association then served to nucleate assembly of the enhanceosome, thereby facilitating recruitment of the pre-initiation complex, and the consequent initiation of IL4 gene transcription. Thus, in addition to providing further mechanistic insights into Th2-cell differentiation, our results also reveal a novel mode of function for ERK during T-cell differentiation.
RESULTS
ERK selectively regulates Th2-cell differentiation
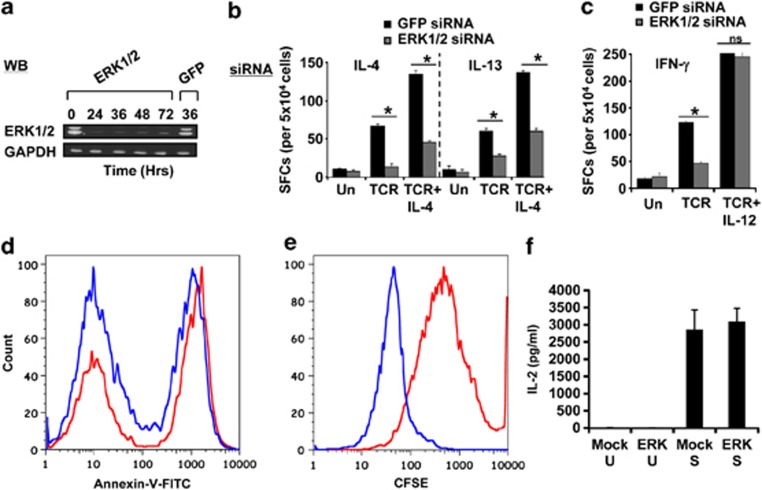
The experiments described in this study were conducted in naive CD4+ T cells that were purified to a level of >99% purity, with no detectable contamination from NKT cells from human cord blood (Supplementary Figure 1). We first determined whether levels of the ERK proteins could be depleted by small-interfering RNA (siRNA). Treatment of cells with a combination of siRNAs targeted against both ERK-1 and ERK-2 led to a substantial reduction in intracellular concentrations of these proteins by 24 h, which persisted up to at least 72 h (Figure 1a). This effect was specific as parallel transfection of cells with green fluorescent protein (GFP)-specific siRNA had no detectable effect (Figure 1a). Therefore, we next subjected GFP- or ERK-silenced cells to activation, Th1 polarization or Th2 polarization, and the frequency of either IL-4- or interferon-γ (IFN-γ)-producing cells was determined in an ELISPOT assay.
Figure 1.
ERK selectively regulates Th2-cell differentiation. (a) Results of a western blot analysis, at indicated times, for ERK-1/2 in naive CD4+ T cells transfected with ERK-specific siRNA. The effect of treatment of GFP-specific siRNA on ERK levels, at 36 h later, is also included, with GAPDH as an internal loading control. (b, c) Effect of treatment of naive CD4+ T cells with siRNA specific for either ERK or GFP on the generation of either IL-4 and IL-13 (panel b) or IFN-γ (panel c) cytokine-producing cells as measured by an ELISPOT assay at day 7 of culture. The stimulation conditions tested here were activation (TCR), Th2 polarization (TCR+IL-4) and Th1 polarization (TCR+IL-12). The corresponding profiles obtained in mock (GFP)-siRNA-treated cells cultured under similar conditions are also shown. *P⩽0.05 (one-way analysis of variance (ANOVA) test); NS=not significant. Each bar represents the mean (±s.d.) of three biological replicates in which each replicate represented a pool of cells isolated from 3 to 5 individuals. In a separate experiment, we stimulated either mock- (red line) or ERK-silenced (blue line) cells under Th2-polarizing conditions for 7 days and then measured the extent of apoptotic cell death by staining with Annexin-V, followed by flow-cytometric analysis. The profiles obtained are shown in d and represents one of three separate experiments. For e, a similar experiment was performed, except that cells were first labeled with CFSE before stimulation and the extent of cell proliferation was determined 7 days later (n=3). f compares the concentration of IL-2 present in the supernatant of these cultures at day 7. Here, U indicates cells that were not activated, whereas S denotes the stimulated groups. Values are the mean (±s.d.) of three separate experiments.
Silencing of ERK in cells stimulated through the TCR alone led to a reduction in IL-4, IL-13 and IFN-γ secretion (Figures 1b and c), suggesting a mediatory role for the MAPK pathway in regulating expression/secretion of these signature cytokines for Th-cell polarization. However, whereas ERK depletion continued to suppress generation of IL-4 and IL-13 producing cells under conditions of Th2 polarization (Figure 1b), its inhibitory effect on IFN-γ production was abolished by addition of the Th1-polarizing cytokine, IL-12 (Figure 1c). To rule out contributions from ‘off-target' effects of the siRNA, we compared the effects of ERK-1/2-specific siRNA obtained from two independent commercial sources, targeting distinct sequences of the ERK transcripts (Supplementary Table 1a). Similar results were obtained in both these cases. Thus, although our results implicate a role for ERK in regulating IFN-γ production, this role, however, was rendered functionally redundant by the addition of IL-12. In contrast, generation of Th2 cells, both under conditions of only either naive T-cell activation or under that of Th2 polarization consistently displayed an obligate requirement for the presence of ERK.
ERK silencing does not compromise either cell viability or IL-2 production by activated T cells
Given the diverse cellular functions that ERK is known to control, it was important for us to determine whether the inhibitory effects of its silencing on Th2-cell differentiation were in fact due to an increase in cell death. Alternatively, ERK suppression could also potentially affect IL-2 production by activated T cells, thereby limiting the extent of their proliferation.
To assess this, we first compared the proportion of apoptotic cells obtained in cultures of either mock-transfected or ERK siRNA-transfected CD4+ T cells that had been stimulated under Th2-polarizing conditions for 7 days. As shown in Figure 1d, a significant fraction of apoptotic cells (presumably reflecting activation-induced cell death) was detected in cultures of mock-transfected cells. However, in ERK-silenced cells, this fraction was reduced by a small but significant amount (Figure 1d). These findings support that the observed inhibition of Th2 polarization in ERK-silenced cells is not the result of any enhancement in cell death.
Interestingly, a parallel experiment wherein cells were labeled with carboxyfluorescein succinimidyl ester (CFSE) revealed that depletion of ERK caused a reduction in population doubling time of CD4+ T cells. Thus, in comparison with mock-transfected cells, cells treated with ERK siRNA had undergone between 3 and 4 more doublings over the 7-day period of culture (Figure 1e). Thus, during Th2-cell polarization, ERK seems to at least partially act by regulating the proliferation rate of cells. This function may, in turn, relate to its role in facilitating Th2-cell differentiation.
Finally, the level of IL-2 in culture supernatants from both mock- and ERK-silenced cells was comparable (Figure 1f), suggesting that the presence of ERK was not obligatory for secretion of this cytokine at least under Th2-polarizing conditions. Therefore, the cumulative results in Figures 1d–f support that the inhibitory effects of ERK depletion on Th2-cell differentiation was not due either to an enhancement in death of activated cells or to an inhibition in production and secretion of the mitogenic stimulus IL-2. The only detectable effect of ERK silencing was an acceleration in proliferation rate, which may at least partially rationalize the observed reduction in the extent of Th2-cell differentiation.
ERK regulates IL-4 transcription in activated T cells
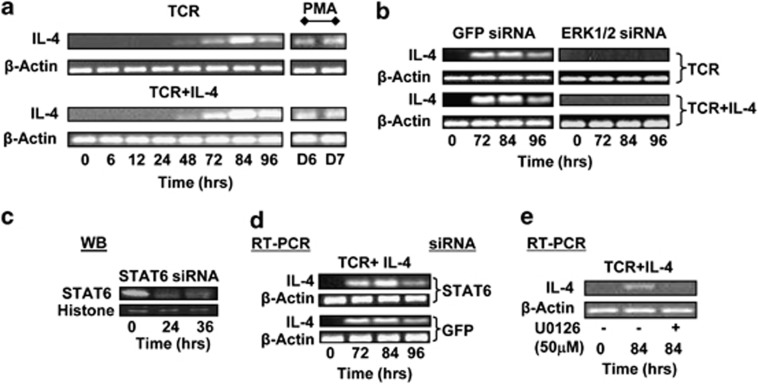
Subsequent to activation of naive CD4+ T cells, Th2-cell differentiation is driven by IL-4.16 Importantly, previous studies have shown that stimulation through TCR was alone sufficient to induce IL-4 production in naive CD4+ T cells.17 Therefore, we wanted to determine whether ERK had any role in mediating IL4 transcription in a TCR-dependent manner. For this, we first compared the kinetics of IL4 gene expression in human CD4+ T cells that were subjected to either activation or Th2 polarization. Figure 2a reveals that similar levels of IL-4 mRNA were induced, with similar kinetics, in both instances. The IL-4 transcript was first detected at 48 h after stimulation, with its levels peaking by 84 h. Interestingly, although treatment of cells with non-silencing siRNA had no effect, siRNA-mediated silencing of ERK resulted in a near complete inhibition of the stimulus-induced expression of IL4 under both experimental conditions used (Figure 2b). Furthermore, this inhibition was also evident when monitored at the level of the intracellular IL-4 protein by flow cytometry (Supplementary Figure 2a).
Figure 2.
ERK regulates IL4 gene transcription. (a) Kinetics of IL-4 mRNA induction in naive CD4+ T cells stimulated with TCR in the presence (TCR+IL-4) or absence (TCR) of IL-4. (b) Effects of treatment of cells with either GFP or ERK1/2-specific siRNA on induction of the IL-4 transcript in activated and Th2-polarizing cells. The time points at which transcript levels were determined by RT-PCR are indicated. Here, primers specific for β-actin were also included to provide an internal normalization control. Western blots in c confirm the silencing of STAT6 protein levels in the nuclear extract of cells treated for the indicated times with specific siRNA. Blots were also probed with anti-histone H3 antibodies to provide a loading control. The effects of STAT6- versus GFP-silencing on IL-4 mRNA levels as a function of time are shown in d. In e, naive CD4+ T cells were stimulated under Th2-polarizing conditions either in the absence (−) or presence (+) of U0126, and IL-4 transcript levels determined by RT-PCR 84 h later.
Parallel experiments revealed that ERK silencing had no effect on the activation-induced transcription of IL-13 (Supplementary Figure 2b). Furthermore, ERK silencing also did not affect mRNA levels of either the β-subunit of the IL-10 receptor or that of GATA3 (Supplementary Figures 2c and d), whereas surface expression of IL-4R also remained unchanged (Supplementary Figure 2e). Thus, at least within the context of the molecules examined here, the effect of ERK suppression seems to be specific for induction of IL-4. As expected, siRNA-mediated depletion of STAT6 (Figure 2c) did not influence activation-induced IL4 transcription (Figure 2d). Importantly, however, IL4 upregulation was also inhibited when naive CD4+ T cells were stimulated in the presence of U0126, an inhibitor of ERK activation18 (Figure 2e). While reaffirming the relevance of ERK during transcriptional activation of IL4, these latter results also underscore that this process requires ERK to be activated through phosphorylation by its upstream kinase, MEK-1/2.
The role of ERK during Th2-cell differentiation is primarily mediated through regulation of IL-4 production
In view of the known pleiotropic functions of ERK,9, 10, 19 we wanted to clarify whether regulation of IL4 expression alone accounted for its contribution towards Th2 differentiation or whether additional pathways were also involved. To assess this, we performed rescue experiments wherein we examined the ability of exogenously added IL-4 to overcome the inhibitory effects of ERK depletion on Th2 differentiation. As shown in Figure 3a, additional IL-4 supplemented at day 3 of the culture indeed resulted in a near complete rescue of ERK-silenced cells from the block in Th2-cell differentiation. The frequency of polarized cells subsequently obtained was comparable to that generated by mock-silenced cells in both instances in which cells were either only activated through the TCR or in which they were stimulated under Th2-polarizing conditions (Figure 3a). Thus, these results confirm that regulation of IL4 transcription constitutes at least the dominant mechanism by which ERK influences Th2-cell differentiation.
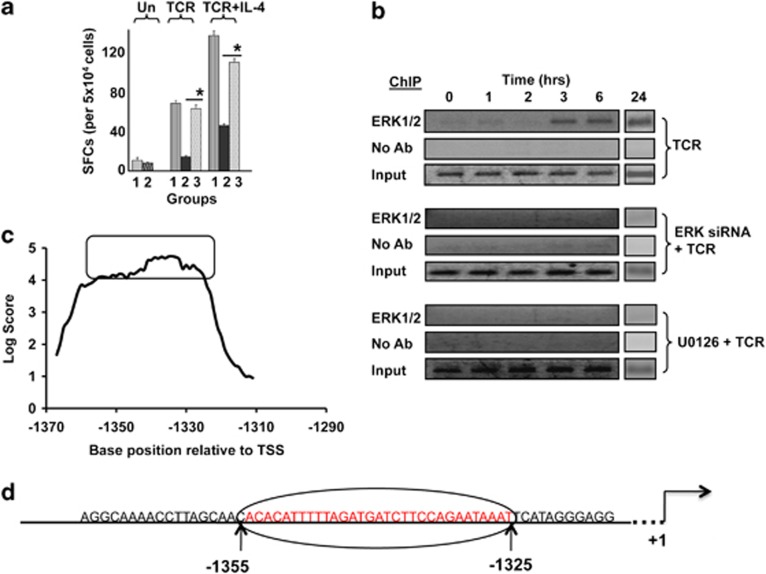
Figure 3.
ERK associates with the IL4 promoter in activated T cells. (a) IL-4 spot-forming cells (SFCs) from an ELISPOT assay in which naive CD4+ T cells were either transfected with GFP siRNA (Group 1), ERK siRNA (Group 2) or ERK siRNA, followed by supplementation of recombinant IL-4 (Group 3) in the culture. ‘Un' represents unstimulated cells, whereas TCR and TCR+IL-4 represent activation and polarizing conditions, respectively. *P⩽0.0001. Each bar represents the data from three independent experiments (mean±s.d. value), performed on three biological replicates in which each replicate represented a pool of cells isolated from 3–5 individuals. (b) Naive CD4+ T cells were either left untreated (top panel) or treated with either ERK1/2 siRNA or U0126 (lower two panels). These cells were then stimulated through the TCR, and time-dependent recruitment of ERK to the IL4 promoter was monitored by ChIP analysis. The corresponding profiles obtained without anti-ERK antibody (no Ab) and input are also shown. (c) ERK-specific chromatin-immunoprecipitated samples from either TCR-activated (3 h) or unstimulated cells were hybridized against a tiled array that was generated around the 60-nucleotide long IL-4 promoter region from −1374 to −1315 nt. This region included the ERK-binding site. The first probe in this array was a 60 nt sequence spanning from nt positions −1404 to −1345. All subsequent probes—also 60 nt long—then represented successive one-nucleotide shifts in sequence, terminating with a probe extending from −1344 to −1285 nt. The fold enhancement in hybridization intensity—in terms of log2 values—obtained for each of these probes with sample from TCR-activated cells, relative to that obtained with a parallel sample from unstimulated cells, is plotted here. The individual probes are identified on the X axis on the basis of the sequence position of the central nucleotide. (d) Parent 60 nt region where the optimal ERK-binding domain is highlighted in red.
ERK associates with the IL4 promoter in activated T cells
Extracellular signal-regulated kinase is the terminal kinase of the canonical MAPK signaling pathway, and it primarily regulates cellular function through the phosphorylation-dependent activation of various TFs.10 Several different TFs are activated by ERK, thus accounting for the broad spectrum of regulatory activities displayed by this kinase.10 However, more recent studies have revealed that regulation of gene expression by ERK may also involve its direct binding to regions within its target genes. This property was first observed in yeast in which MAPK orthologs were found to physically associate with the genes that they regulate.20 A subsequent examination of the human protein–DNA interactome further identified that ERK-2 could bind DNA in a sequence-specific manner. Importantly, at least one of the consequences of this interaction was the suppression of the expression of several of the IFNγ-induced genes.21
In view of the non-redundant role of ERK in regulating TCR-dependent IL4 transcription, we wanted to explore whether this process was also mediated through direct interactions between the kinase and regulatory elements within the IL4 promoter. For this, we performed chromatin immunoprecipitation (ChIP) experiments in naive CD4+ T cells that had been stimulated for various times with a combination of anti-CD3 and anti-CD28. As shown in Figure 3b, activation of T cells indeed resulted in recruitment of ERK to a region spanning from −1436 to −1274 of the IL4 promoter. Binding at this site was detectable by 3 h of activation, and the association persisted in a stable manner up to at least the 24-h time point (Figure 3b). As expected, no such binding could be detected in cells in which ERK expression had been first silenced by siRNA (Figure 3b). Furthermore, ERK binding to the IL4 promoter was also inhibited in cells that were stimulated in the presence of the inhibitor of ERK activation, U0126 (Figure 3b). These latter results suggest that phosphorylation-dependent activation of ERK is a prerequisite for its engagement by the promoter DNA. However, it remains to be determined whether phosphorylation merely enables access of ERK to its target DNA sequence by facilitating cytoplasmic-to-nuclear translocation or whether the binding activity of ERK—at least in the context of the IL4 promoter—is specific to its phosphorylated form.
Identification of the ERK-binding site within the IL4 promoter
In a separate study, we were also exploring the larger question of the genome-wide association of ERK with promoter sequences in activated human CD4+ T cells. For this, we used a ChIP-on-ChIP approach wherein samples chromatin immunoprecipitated with ERK-specific antibody from primary CD4+ T cells, stimulated either in the presence or absence of U0126, were hybridized against a promoter array consisting of 60-nt long probes derived from −3 kb to +2 kb regions relative to the transcription start sites of all known human open reading frames (ORFs). In these experiments, we noted that a single probe corresponding to the sequence spanning from −1315 to −1374 nt of the IL4 promoter was occupied by ERK in cells that had been stimulated in the absence, but not in the presence, of U0126. At one level, these results provided additional confirmation of our findings in Figure 3b that TCR-dependent activation of naive human CD4+ T cells leads to recruitment of ERK at the IL4 promoter. However, in addition, these findings also localized the ERK-binding site to reside between −1315 and −1374 nt of the promoter sequence.
To further resolve the ERK-binding site, we next used a tiled array in which the sequence of the above 60-mer probe was scanned through a series of overlapping probes that were shifted by one nucleotide at a time. The chromatin-immunoprecipitated sample generated from stimulated cells (3 h) with anti-ERK antibody was then hybridized against this tiled array and the results obtained are shown in Figure 3c. As is evident, a significant proportion of the probes tested displayed high levels of ERK binding, implying that this activity involved a relatively broader domain within the parent, −1374 to −1315 nt region. To identify the optimal ERK-binding site, we chose a cutoff score for the ratio of the log value of the hybridization intensity of >4.0 and compared the sequence of the overlapping probes that met this criteria. This exercise identified that the optimal ERK-binding site consisted of a 31-nt stretch that extended from −1325 to −1356 nt of the IL4 promoter (see Figure 3d).
In a previous study, Schones et al.22 had mapped the genome-wide distribution of nucleosome positions in resting human CD4+ T cells. A careful inspection of these results revealed that the IL4 promoter region between −1310 and −1370, which includes the ERK-binding site, was devoid of nucleosomes. This then suggests that the association of ERK with the IL4 promoter was not mediated by interactions with a nucleosome. Rather, it more likely involved either direct binding of the proteins to DNA or was facilitated through engagement by one or more of TFs that were already associated with the target promoter sequence. However, these results do not permit a distinction between either of these two possibilities.
Association of ERK is necessary for IL4 promoter activity
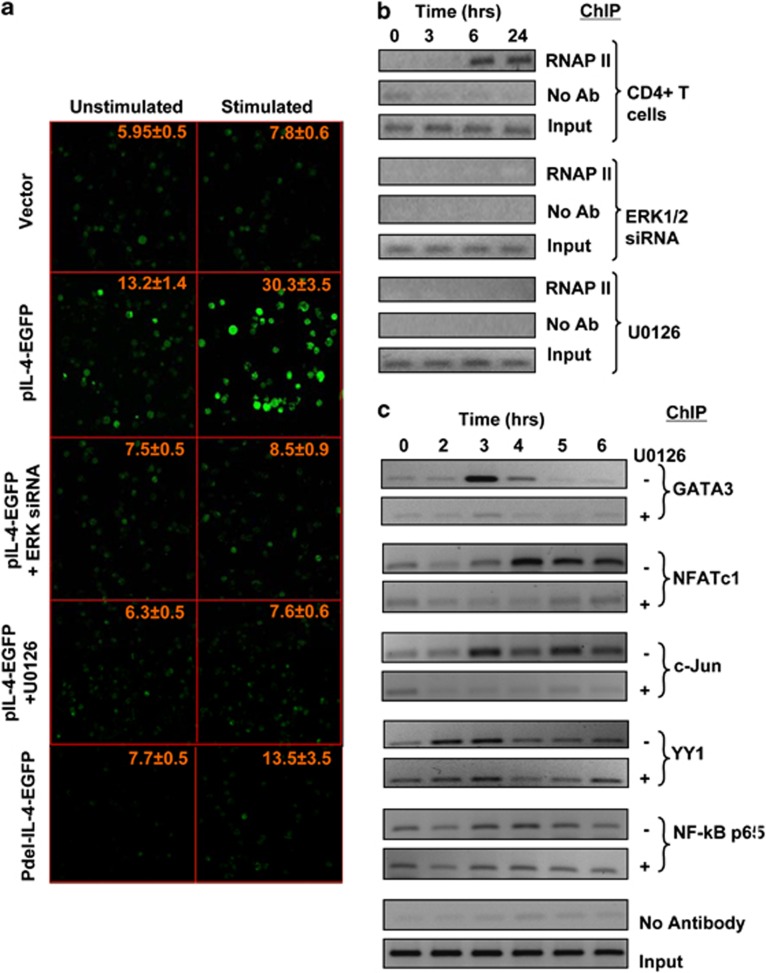
To further probe the mechanism for ERK-induced IL4 transcription, we cloned the IL4 promoter, from −1374 to +224 nt relative to the transcription start site, upstream of EGFP in the pEGFP1 vector (pIL-4-EGFP). The resulting construct was transfected into Jurkat cells, and expression of EGFP was monitored by confocal microscopy. Cells transfected either with the promoter-less or with the pIL-4-EGFP vector showed low levels of background fluorescence in the absence of any stimulation (Figure 4a). However, cross-linking of TCR led to a marked increase in EGFP expression in cells containing the pIL-4-EGFP vector (Figure 4a), confirming that the cloned IL4 promoter sequence retained sensitivity to TCR-dependent signaling. Significantly, either siRNA-mediated silencing of ERK expression or inhibition of its activation with U0126, led to a substantial reduction in the TCR-induced expression of EGFP (Figure 4a). Thus, in addition to providing further evidence to support that TCR-dependent activation of ERK constitutes an obligatory step in the induction of IL4 expression, these results also provide direct confirmation for the presence of an ERK-responsive element within the promoter region of the IL4 gene.
Figure 4.
Association of ERK is necessary for IL4 promoter activity. (a) Jurkat cells were transfected with the pEGFP1 vector (vector, top two panels), with the pIL-4-EGFP construct (from second to the fourth row of panels) or with a derivative of the pIL-4-EGFP construct in which the ERK-binding region was partially deleted (Pdel-IL-4-EGFP, bottom panels). This included two additional experimental groups in which pIL-4-EGF-transfected cells were also treated with either ERK-specific siRNA (panels in the third row from top) or with the MEK inhibitor U0126 (panels in the fourth row from top) as indicated. Figure shows the level of fluorescence obtained in each of these groups of cells when they were either left unstimulated or stimulated through the TCR for 24 h. Values indicated in each panel are the mean (±s.e.m) intensity of fluorescence measured for at least 40 cells from different fields in 3 separate slides for each group. (b) Naive CD4+ T cells were either left untreated (top panel) or treated with either ERK1/2 siRNA (KD) or U0126 (lower two panels). These cells were then stimulated through the TCR, and time-dependent recruitment of RNA polymerase II (RNAP II) to the IL4 promoter was monitored by ChIP analysis. The corresponding profiles obtained without anti-ERK antibody (no Ab) and input are also shown. (c) Time-dependent recruitment of indicated transcription factors, as determined by ChIP, to the IL-4 promoter after stimulation of cells through the TCR either in the presence (+) or absence (−) of U0126.
Significantly, partial deletion of the putative ERK-binding site within the promoter (that is, from −1374 to −1335 nt) in the pIL-4-EGFP construct also abrogated the sensitivity of EGFP expression to TCR-dependent stimulation (Figure 4a). Although marginal levels of EGFP expression were detected in cells transfected with this construct, no further increase was however observed upon TCR cross-linking. In contrast, the extent of TCR-stimulated phosphorylation of ERK obtained was unaffected when compared with that seen either in untransfected cells or in cells transfected with the deleted pIL-4-EGFP construct. That is, in addition to ERK activation, expression of IL4 promoter activity was also contingent upon the presence of the ERK-interacting domain within its sequence. This would then support that binding of ERK to the IL4 promoter was a prerequisite for induction of IL4 gene expression.
Recruitment of ERK at the IL4 promoter facilitates initiation of gene transcription
To explore the mechanism by which the interaction between ERK and the promoter influences IL4 gene transcription, we performed ChIP experiments to compare the recruitment kinetics of ERK with that of RNA polymerase II (RNAPII) to the IL4 promoter. Figure 4b reveals that recruitment of RNAPII to the IL4 promoter in cells stimulated through the TCR was first detected at 6 h after stimulation, which then remained stable up to at least the time point of appearance of the IL4 transcript. Significantly, however, this process was completely abrogated in cells in which ERK expression was silenced by siRNA, and also in cells in which ERK activation was inhibited by stimulation in the presence of U0126 (Figure 4b). Thus, in conjunction with our earlier findings that promoter activity was dependent on the presence of the complete ERK-binding sequence (see Figure 4a), these latter results support that TCR-induced association of ERK with the proximal promoter is an obligatory step in the process that leads to recruitment of the pre-initiation complex, and the consequent initiation of IL4 gene transcription.
The observed time lag between RNAPII recruitment (6 h) and first detection of the IL-4 transcript (48 h, Figure 2a) likely reflects sensitivity of the PCR method used for transcript detection. In addition although it is possible that post-transcription initiation mechanisms also come into play to regulate synthesis of the complete transcript.
ERK activation mediates enhanceosome assembly on the IL4 promoter
Transcriptional regulation of the IL4 gene has been extensively studied in human CD4+ T cells, and a series of regulatory elements located in the 5′ proximal promoter region have been characterized.23 Six regions (designated P0–P5) with sequence homology to the binding sites for the TF, NFAT have been identified within the proximal 400 bp of the core promoter region. Interestingly, the majority of these sites are flanked by sequences with high affinity for the AP-1 family of proteins, and the activation of IL4 transcription requires collaborative binding of both NFAT and AP-1 at these composite elements. Other positive regulatory elements encoded by the proximal promoter sequence include binding sites for the TFs nuclear factor-κB (NF-κB), YY1, GATA3 and nuclear factor of interleukin 6 (NF-IL6). All of these TFs actively participate during TCR-induced upregulation of IL4 transcription.23, 24, 25 In addition, this region also includes negative regulatory elements that are bound by interferon-regulatory factors IRF1 and IRF2.
To further probe the mechanism by which ERK mediates initiation of IL4 transcription, we examined activation-induced recruitment of a panel of key TFs to the IL4 proximal promoter. Figure 4c shows that TCR stimulation of naive CD4+ cells indeed induced the recruitment of NFATc, NF-κB, YY1, GATA3 and AP-1 (as detected at the level of its c-Jun subunit) at the IL4 proximal promoter. However, the kinetics and stability of recruitment differed for the individual TFs. Enhanced association of the TF YY1 was first detected at 2 h after stimulation, with a subsequent decrease in band intensity by 4 h. GATA3 was only transiently recruited at 3 h after stimulation, which was then followed by a progressive disappearance from this site at the later time points (Figure 4c). In contrast, whereas NFATc1 recruitment also occurred at the same time, the resulting association was stable and persisted over the duration of the experiment. This was also true for the c-Jun subunit of AP-1 (Figure 4c). The p65 subunit of NF-κB was found to be constitutively associated with the IL4 promoter. However, TCR-mediated stimulation resulted first in its dissociation from the target site at 2 h, followed by a re-recruitment that was detectable by 3 h (Figure 4c). Although the mechanism underlying this bi-phasic process is presently unclear, this could likely represent an exchange between distinct, p65-containing, heterodimeric complexes of NF-κB. However, such a possibility remains to be experimentally verified.
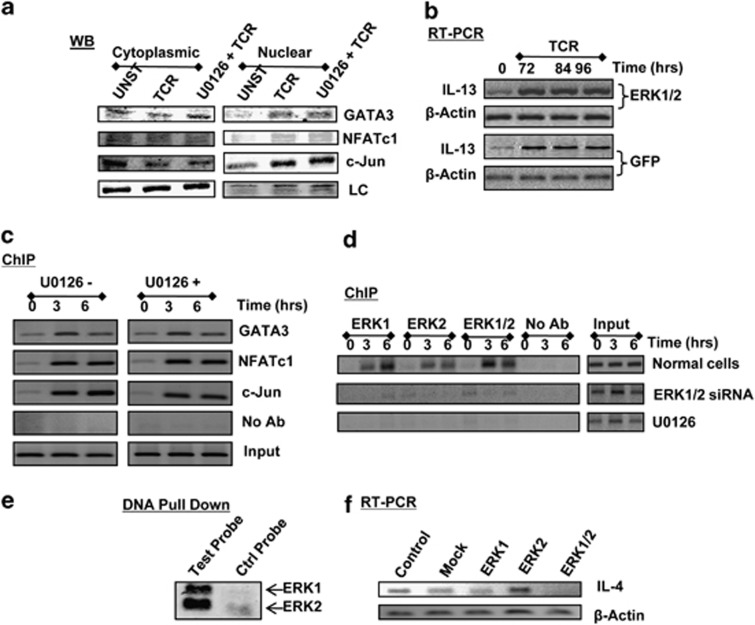
Importantly, inhibition of ERK activation with U0126 also selectively inhibited the recruitment of NFATc, AP-1 and GATA3, whereas both the kinetics and the extent of NF-κB and YY1 accumulation at this region remained relatively unaltered (Figure 4c). In parallel experiments, we established that although phosphorylation of ERK was markedly inhibited (Supplementary Figure 3a), the inclusion of U0126 had no detectable effect on TCR-induced activation—when measured in terms of translocation from the cytoplasmic to the nuclear compartment—of the former three TFs (Figure 5a). This observed redundancy of ERK activation, during the TCR-dependent induction of GATA3, NFATc and c-Jun activation, is consistent with the literature. Thus, nuclear translocation of GATA3 is mediated through phosphorylation on serine residues by the p38 MAPK,24 whereas activation of the NFAT family of TFs involves dephosphorylation of these proteins by the Ca2+-activate phosphatase calcineurin.8 On the other hand, activity of c-Jun is primarily regulated through phosphorylation by the JNK family of MAPKs.26, 27
Figure 5.
The effect of ERK on IL4 gene expression is specific and involves both ERK-1 and ERK-2. (a) compares the distribution of three transcription factors (GATA3, NFATc1 and c-Jun) between the cytoplasmic and nuclear fractions in unstimulated (UNST), TCR-triggered (TCR, stimulation time of 30 min) and U0126-treated cells, followed by TCR triggering (U0126+TCR, stimulation time of 30 min). LC denotes the loading control, which was PLCγ2 for cytoplasmic fractions and Histone 1 for the nuclear fractions. (b) Effects of treatment of cells with either GFP or ERK1/2-specific siRNA on induction of the IL-13 transcript in activated cells. The time points at which transcript levels were determined by RT-PCR are indicated. Here, primers specific for β-actin were also included to provide an internal normalization control. (c) Results from a ChIP assay that monitored the recruitment of NFATc, c-jun and GATA3 to the region spanning −76 to −234 nt of the IL-13 promoter. Here, either untreated cells (U0126−) or cells treated with U0126 (U0126+) were stimulated through the TCR as indicated. Input and no Ab controls are also shown. (d) Results of experiments in which cells were activated through the TCR for indicated time and then subjected to a ChIP analysis using antibodies that were either specific only to ERK-1 or to ERK-2. For the purposes of comparison, an additional group was also included in which antibodies to both ERK-1 and ERK-2 were combined in the ChIP experiment (ERK1/2). Results shown are a representative of three separate experiments. (e) gives the results of a DNA pull-down experiment in which nuclear extracts from CD4+ T cells, stimulated through the TCR for 3 h, were incubated either with a 20-mer double-stranded DNA probe extracted from Figure 3d or with a control probe of similar length. In both cases, the probes were biotinylated. After incubation, the probes were separated by affinity chromatography (streptavidin-agarose), and the bound proteins eluted and analyzed for the presence of ERK-1 and ERK-2 by western blot analysis (see the ‘Methods' section). Results shown are from one of three separate experiments. (f) Results obtained in an RT-PCR experiment in which cells transfected separately with ERK1 siRNA (ERK1), ERK2 siRNA (ERK2) or with a combination of both (ERK1/2) were stimulated through the TCR. At the end of 84 h, cells were harvested, the total RNA was isolated and analyzed by RT-PCR for the IL-4 transcript. For the purposes of comparison, results obtained in cells that were either not treated with any siRNA (Control) or those treated with siRNA specific for GFP (Mock) are also shown. In all cases, primers specific for β-actin were also included to provide an internal normalization control.
To further confirm that ERK inhibition did not significantly interfere with activation of transcriptional regulatory function of these three TFs, we also examined their recruitment to target sites in the promoter region of the IL-13 gene. As previously noted, siRNA-mediated silencing of ERK expression had no detectable effect on the TCR-dependent induction of IL-13 gene expression (Figure 5b). Consistent with this, we found that although all three TFs were recruited to the IL-13 promoter in a TCR-dependent manner, this process, however, was insensitive to the inclusion of U0126 (Figure 5c). These findings imply that the earlier observed dependence of recruitment of these TFs on ERK activation (see Figure 4c) was a feature that was specific for the IL4 promoter.
Therefore, the results in Figures 5a–c confirm that although TCR-dependent ERK activation was essential for IL4 promoter-targeted recruitment of key TFs its involvement—however—occurred subsequent to the step of activation of these TFs. That is, an unusual mechanism for ERK-mediated regulation of IL4 transcription was likely involved wherein assembly of the NFATc/AP-1 complex and GATA3 on the promoter required that activated ERK be first recruited in the vicinity. Such an interpretation would also be consistent with the fact that promoter binding of these latter three TFs does not precede ERK recruitment. Thus, the primary function of ERK during the initiation of IL4 transcription seems to be to respond to TCR activation by associating with an upstream element in the proximal promoter and, thereby, facilitating the coordinated recruitment of key TFs.
Both ERK-1 and ERK-2 are involved in the regulation of IL4 gene expression and binding to its promoter
Our studies so far had examined the effects of simultaneous inhibition, or silencing, of both ERK-1 and ERK-2. Therefore, it was pertinent to next discriminate between the individual functional roles of these two proteins. For this, we performed ChIP experiments, in TCR-stimulated cells, using antibodies that were specific either to ERK-1 or to ERK-2. Interestingly, as shown in Figure 5d, both ERK-1 and ERK-2 were found to be associated with the IL4 proximal promoter in a stimulus-dependent manner. Importantly, the kinetics of recruitment of the individual proteins was similar, as also was the sensitivity of each of these processes to either ERK silencing or inhibition (Figure 5d). To further verify the co-association of both ERK-1 and ERK-2, we next generated a double-stranded, biotinylated, probe that was derived from the sequence between −1354 and −1335 nt of the IL4 promoter. Nuclear extracts from TCR-stimulated CD4+ T cells were incubated with this probe, and the bound proteins were then separated by affinity chromatography. These proteins were subsequently examined for the presence of either ERK-1 or ERK-2 by western blot analysis. Figure 5e reveals that both of these proteins were indeed present in this fraction. In contrast, neither of these kinases could be detected to any significant extent in a parallel fraction generated from a control probe that was of comparable length, but derived from the −1510 to −1531 nt segment of the murine CD80 promoter (Figure 5e). Collectively then, these results support that stimulation of primary human CD4+ T cells through the TCR leads to the recruitment of both ERK-1 and ERK-2 to the IL4 proximal promoter.
To next compare the relative functional roles of these two MAPK proteins, we separately silenced either only ERK-1 or ERK-2, by siRNA in primary CD4+ T cells (see Supplementary Figure 3b). These cells were then stimulated through the TCR, and the consequent effects on induction of the IL4 transcript were determined. Surprisingly, we found that specific depletion of either ERK-1 or ERK-2 alone had no significant effect on the IL4 transcript levels obtained (Figure 5f). Instead, inhibition of IL4 induction was only obtained when both of the MAPK proteins were silenced together (Figure 5f). This result suggests the existence of functional redundancy between ERK-1 and ERK-2, at least in the context of regulation of IL4 gene expression. That is, although both of these proteins were recruited at the proximal promoter in a TCR-dependent manner, each of them was—however—individually capable of inducing IL4 gene transcription. Therefore, the latter process was only inhibited under conditions in which both ERK-1 and ERK-2 were prevented from associating with the IL4 promoter.
Therefore, our cumulative results support that stimulation of naive human CD4+ T cells through the TCR induces recruitment of ERK to the IL4 proximal promoter. This constitutes a critical step in that it is the promoter-bound ERK that then facilitates completion of enhanceosome assembly by actively ensuring engagement of at least three of the key TFs involved. The functional end point of this process is the recruitment of RNAPII at the core promoter, and the consequent activation of IL4 gene transcription.
Stochastic variability in ERK levels influence individual-specific variations in susceptibility to Th2-cell differentiation
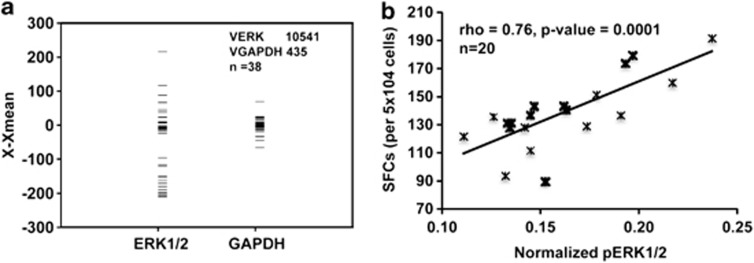
In the course of our experiments, we were intrigued by the fact that naive CD4+ T cells purified from different individuals displayed significant variations in the amount of ERK proteins that were present. Figure 6a shows the results obtained in cells isolated from a representative set of 38 individuals in whom variance, across samples, of the combined concentrations of ERK-1 and ERK-2 were compared by using GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as the internal control (see Supplementary Figure 4a). The significantly greater heterogeneity in ERK levels, relative to that of GAPDH, is clearly evident even within this relatively small-sized group. Such differences could potentially arise from differences in either the genetic/epigenetic make-up of the individuals, or from stochastic variations at the level of either gene expression or protein turnover. However, independent of the explanation, it was reasonable to suspect that the observed variability in basal ERK protein levels should also affect the relative efficiency with which Th2-cell differentiation can be induced from the respective naive T-cell populations. For example, simple mass action dynamics would dictate that ERK protein concentrations would serve as an important determinant of the extent of its sequestration in a complex with the upstream kinase MEK-1/2.28 This in turn would influence both quantitative and qualitative (that is, kinetic) aspects of ERK phosphorylation. In this connection, several recent studies have shown that ERK activation response is highly sensitized to the overall systems properties of the MAPK-signaling module, with concentration of the individual MAPK constituents serving as important regulatory parameters for defining the eventual output.29, 30, 31
Figure 6.
Stochastic variability in ERK levels influences individual-specific variations in susceptibility to Th2-cell differentiation. (a) Extent of variability in ERK1/2 protein levels in unstimulated naive CD4+ T cells. ERK1/2 levels were determined from western blot analysis using antibodies specific for both ERK-1 and ERK-2. Here, each bar depicts the results for one individual (n=38). For the purposes of comparison, GAPDH levels were also monitored in each of these samples and the panel shows the comparison of the relative variability of total ERK1/2 levels, in comparison with that of GAPDH. The Y axis gives the normalized intensity of the protein band in each sample, after subtracting the mean value of the band intensities in all the samples (X−Xmean). The corresponding values for variance in X−Xmean for ERK (VERK) and GAPDH (VGAPDH) are indicated. (b) Results of a regression analysis between p-ERK/ERK (that is, ratio of phospho-ERK1/2 to ERK1/2 protein) and the frequency of IL-4-producing cells in naive CD4+ T cells obtained from different individuals (n=20) as described in the text. Here, the frequency of IL-4-producing cells was determined by an ELISPOT assay. Each value represents the mean of triplicate sets.
We purified naive CD4+ T cells from 20 different individuals and separately stimulated them under Th2-polarizing conditions. At 3 h after initiation of stimulation, an aliquot was removed for determination of both phospho-ERK and ERK protein levels by western blot analysis. The remainder of cells was continued in culture for the subsequent enumeration of Th2 effectors by an ELISPOT assay. We then compared the resulting frequency of Th2 cells obtained, with the ratio of phospho-ERK to total ERK levels within each sample (Supplementary Figure 4b). Here, our choice of phospho-ERK/ERK ratio as the parameter for comparison was based on the assumption that this ratio would encapsulate individual-specific differences in the extent of ERK activation that may arise from differences in intracellular concentrations of either ERK and/or that of other cellular proteins that influence functioning of the MAPK pathway. Figure 6b presents the results from these experiments in the form of a regression analysis. As is evident, a good correlation was clearly obtained between these two variables. At one level, these findings further support the functional relevance of ERK activation, in terms of regulating Th2-cell differentiation from naive, human CD4+ T cells. More importantly although, these results also suggest that variability in intracellular ERK concentrations may represent one of the parameters that accounts for the observed individual-specific differences in the propensity for Th2-cell generation against external antigens and/or allergens.32 In addition to influencing the frequency of Th2 cells generated, it is likely that variations in ERK concentrations would also correlate with the amount of IL-4 produced by each cell. Unfortunately, however, the limited number of purified cells obtained from each individual did not allow for an additional measurement of intracellular IL-4 levels. Nonetheless, recent studies have shown increased levels of both ERK mRNA and phosphorylated ERK in lymphocytes from asthmatic rats, relative to that obtained in corresponding cells from normal controls.33
DISCUSSION
Induction of naive CD4+ T cells into the Th2 differentiation pathway is, at least to a significant extent, dependent on IL-4 that is endogenously produced upon TCR cross-linking.6 This IL-4 establishes a positive feedback loop through IL-4R that further reinforces IL4 expression, while silencing the IFNg locus at the same time.1, 16 The highlight of this report is the demonstration that, although dispensable for Th1-cell differentiation, ERK has a key role during the polarization of Th2 cells. Furthermore, these studies also clarify that this role of ERK is primarily achieved through activation of the early phase of TCR-dependent IL-4 production. Importantly, the functional relevance of this kinase could be further highlighted by experiments demonstrating that intrinsic variations in the extent of its activation, in CD4+ T cells from diverse individuals, is a good predictor of the magnitude of the Th2 response that is eventually generated. Thus, in addition to STAT6 and GATA3,2, 34, 35, 36, 37 ERK constitutes yet another addition to the list of obligatory mediators of the Th2-cell differentiation pathway.
Perhaps the most intriguing aspect of our study is the delineation of the mechanism by which ERK mediates IL4 gene transcription. Traditionally, ERK is believed to function primarily as a signaling molecule that influences cellular function through regulating the activity of a diverse array of TFs. Both ERK-1 and ERK-2 are proline-directed protein kinases that phosphorylate proline-neighboring serine and threonine residues on target substrates. In the cytoplasm, activated ERK regulates the activity of various growth factor-responsive targets, which include scaffolding proteins and also those involved in the protein translation machinery.10 In addition although, activated ERK also translocates to the nucleus where it phosphorylates a number of TFs that regulate gene expression.10 Some representative examples include ternary complex factors such as Elk-1 and other members of the Ets family, the proto-oncogene product c-myc and the BRF1 subunit of TFIIIB.10 Nuclear ERK also phosphorylates, and activates, the mitogen- and stress-activated protein kinases, which in turn influence an additional array of TFs including CREB.38
A novel facet to the regulatory pathways used by ERK was first revealed in the yeast system, when it was found that activated orthologs of mammalian MAPKs physically occupied specific sites on subsets of genes that they regulated.20 Interestingly, distinct patterns of association were noted suggesting that these kinases adopted different mechanisms for accessing these sites.20 More recent studies have confirmed that, at least in the context of ERK-2, this feature is also conserved in higher eukaryotes. Thus, a recent profiling of the human proteome–DNA interactome revealed sequence-specific DNA-binding properties for ERK-2.21 The consensus sequences for ERK-2 binding were enriched in the promoter regions of several genes, and DNA-binding activity was independent of the kinase activity. Interestingly, at least one of the physiological consequences of ERK-2 biding to its target DNA was the suppression of expression of IFNγ-induced genes.21 However, given the earlier findings in yeast, the similar existence of additional mechanisms that promote the access of ERK to sites on genes in higher eukaryotes cannot be ruled out.
Significantly, our results support that TCR-dependent activation of IL4 gene transcription in naive human CD4+ T cells was contingent upon the direct association of activated ERK to the proximal promoter. This physical engagement was essential for facilitating recruitment of at least three of the key TFs—GATA3, NFATc and AP-1—that are required for inducing IL4 gene transcription.23 This mechanism was revealed in experiments in which either silencing of ERK expression or inhibition of its activation prevented assembly of these TF complexes on the IL4 promoter. As a result, the subsequent recruitment of RNAPII was also inhibited. The dependence of this process on the physical association of activated ERK with a proximal promoter element was first indirectly suggested from observations that inhibition of ERK phosphorylation did not interfere either with the activation of the three TFs or with their recruitment to consensus sites on alternate genes. Complementary findings that the integrity of the ERK-associating promoter element was indispensable for TCR-induced expression of the IL4 gene provided further support for such an inference.
Importantly, our results also indicate that although TCR cross-linking induces the simultaneous recruitment of both ERK-1 and ERK-2 at the IL4 promoter, the latter process was associated with functional redundancy. That is, recruitment of either one of the two ERK proteins alone was sufficient to activate IL4 transcription. The observed functional equivalence between ERK-1 and ERK-2 is not surprising given the extensive sequence identity (∼85%) that they share.39 Indeed, an overlap in properties between these two proteins has also been documented in the ERK-dependent regulation of other biological processes.40, 41 In view of the key role that the MAPK pathway has in regulating diverse cellular processes, a redundant expression of the two highly analogous proteins presumably contributes to conferring robustness against mutational perturbations. Such an explanation then could also potentially extend to the regulation of Th2-cell differentiation.
Clearly, several mechanistic aspects of ERK-dependent regulation of IL4 transcription remain to be resolved. First is the mode of recruitment of the kinase pair at the proximal promoter. In principle, this could be achieved either through direct binding to the DNA or, alternatively, through interactions with a pre-existing protein–protein complex at this site. In separate experiments, we were unable to detect any binding of either purified or recombinant ERK to DNA probes derived for the target region of the IL4 promoter. Although this finding undoubtedly requires further confirmation, the implication that ERK does not directly bind to this sequence would also be consistent with our related observation that the ERK-interacting domain encompasses a relatively large (∼31 nt) segment of the promoter sequence. In this connection, it is also pertinent to note that, unlike ERK-2, a DNA-binding domain on ERK-1 has not so far been identified. On the other hand, bioinformatic analysis of the DNA sequence constituting the ERK-responsive element revealed the presence of consensus binding sites for about nine TFs. These included proteins that are known to interact with both ERK-1 and ERK-2, such as members of the Ets and STAT family of TFs.11 Experiments are currently underway to resolve whether any of these proteins are actively involved in mediating ERK engagement at the IL4 promoter.
The other outstanding question is the mechanism by which promoter-associated ERK facilitates recruitment of the three TFs, namely GATA3, NFATc and AP-1. This issue is particularly intriguing given that this dependency on ERK was observed only in the case of the IL4 promoter. A further analysis of the genome-wide distribution of nucleosome positions in resting CD4+ T cells described by Schones et al. revealed that the promoter region encoding the binding sites for these TFs (that is, up to 400 nt upstream of the transcription start site) was rich in nucleosomes. Consequently, ERK association could potentially initiate a nucleosome re-modeling program that renders the TF-binding sites accessible. Alternatively, ERK may also mediate the release of a repressive factor such as IRF1 or IRF2, thereby enabling subsequent binding of the transcriptional activators. Future investigations will need to distinguish between these and other possibilities.
Thus, in summary, these studies identify ERK as a key mediator of human Th2-cell differentiation. This influence is primarily exerted through regulation of the early IL-4 production in response to TCR cross-linking in naive cells. Significantly, a novel mechanism was found to be involved wherein activated ERK was first required to physically occupy a proximal promoter element of the IL4 gene. This association then facilitated completion of assembly of the enhanceosome, and the consequent initiation of IL4 transcription. It would clearly be of future interest to elaborate the finer details of this mechanism. Also pertinent here would be to more definitively characterize the functional relevance of ERK, in terms of defining susceptibility to asthma and other atopic allergies in humans.
METHODS
Reagents and cell culture
All cell culture experiments were performed in RPMI 1640 supplemented with penicillin (100 U ml−1)/streptomycin (100 μg ml−1) and 10% fetal calf serum (Life Technologies, Carlsbad, CA, USA). CD4+ T cell-positive isolation kits were obtained from Dynal Biotech, Carlsbad, CA, USA. Recombinant human IL-4, IL-12, IL-2 cytokines, anti-human IL-4 and IL-4R-neutralizing antibodies, human IL-4 and IFN-γ ELISPOT kits were procured from R&D Systems, Minneapolis, MN, USA. ERK1/2, STAT6 and GFP siRNAs were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA), Sigma-Aldrich (St Louis, MO, USA) and Qiagen (Hilden, Germany), respectively. PMA, ionomycin and CFSE were purchased from Sigma. Ficoll paque was obtained from Amersham Pharmacia Biotech, Piscataway, NJ, USA. Anti-human CD3 and CD28, IL-4PE, IL-4R phycoerythrin antibodies and IC staining fixation/permeabilization kit were obtained from BD Biosciences, Franklin Lakes, NJ, USA. Phospho ERK1/2, GAPDH and MEK-1/2 inhibitor (U0126) were obtained from Cell Signaling Technology Inc., Danvers, MA, USA. Flurochrome-labeled secondary antibody was purchased from LI-COR Biosciences, Lincoln, NE, USA. OptiMEM nucleofection medium was procured from Gibco (Invitrogen Corporation, Carlsbad, CA, USA). Duo set IL-2 ELISA kit was purchased from R&D Systems and annexin-V-fluorescein isothiocyanate from BD Pharmingen, Franklin Lakes, NJ, USA.
Isolation of human CD4+ lymphocytes from umbilical cord blood and their stimulation
We obtained approval for this study by the institutional ethics committee, and human blood samples were obtained with written informed consent. Purified CD4+ T cells were isolated from cord blood mononuclear cells using Ficoll paque isolation using CD4+ T-cell isolation kit as described earlier.42 The resulting populations were >98% viable, >98% CD3+, CD4+ and CD45RA+ T cells (Supplementary Figure 1). These cells were stimulated with plate-bound anti-CD3 (2.5 μg ml−1 in phosphate-buffered saline) and 500 ng ml−1 soluble anti-CD28 MoAb in the presence or absence of polarizing cytokines, rhIL-12 (2.5 ng ml−1) and rhIL-4 (10 ng ml−1). In the polarization groups, we deliberately avoided the inclusion of antibodies against the opposing cytokine (such as anti-IFN-γ antibodies during Th2 polarization) so as to maximize the sensitivity to any possible effects of ERK depletion. Furthermore, we also avoided any additional supplementation with the inductive cytokine in the later stages of the cultures for the same reason. In all cases, rhIL-2 (17 ng ml−1) was added 48 h after the initiation of polarization. For long-term cultures (over a period of 7 days), the cell concentration was maintained in the range of 0.5–2 × 106 cells per ml. For experiments examining the consequences of inhibiting ERK activation, cells were incubated with U0126 (50 μM) for 60 min before stimulation and subsequent analysis.
Transfection of CD4+ T cells with siRNA
Cells were transfected with siRNA duplexes (2 μg each/5 × 106 cells) against the appropriate target using the Amaxa Nucleofection II system from Lonza, Basel, Switzerland, and following the protocol recommended by the manufacturer. Aliquots of cells were collected at various times to confirm downregulation of the target gene by western blot analysis for the corresponding protein. Cell experiments were initiated at 24 h (for STAT-6 siRNA treatment) and at 36 h (for ERK1/2 and GFP siRNA treatment) to obtain optimal knockdown.
ELISPOT assay
ELISPOT assays for IL-4, IL-13 and IFN-γ cytokines was performed using a human ELISPOT kit. For this, polarized cells were re-stimulated after 7 days of culture with a combination of PMA (Phorbol 12-myristate 13-acetate) (5 ng ml−1) and ionomycin (500 ng ml−1) for 18–20 h. Spots were counted and analyzed by using an immunospot reader (CTL Analyzers, LLC, Cleveland, OH, USA).
Reverse transcriptase-PCR
Total RNA was prepared using Trizol from Invitrogen, Life Technologies (Invitrogen Corporation) according to the manufacturer's instructions. A one- or two-step reverse transcriptase-PCR was performed using primers given in Supplementary Table 1b. For two-step reverse transcriptase-PCR, the Fermentas kit (Fermentas, Canada Inc., Burlington, ON, Canada) was used for first-strand cDNA synthesis followed by PCR. For the one-step reverse transcriptase-PCR, a Qiagen kit (Qiagen) was used.
Western blotting
Lysates were resolved on 10% SDS-PAGE gel and transferred to HyBond C transfer membrane (Amersham Biotech) with a semi-dry transfer unit (Hoefer Inc., Holliston, MA, USA). The membranes were probed with respective specific primary antibodies, followed by detection using a fluorochrome-labeled secondary antibody. Blots were visualized on a LI-COR Odyssey Infrared Imaging System, and band intensities were quantified using the ‘Odyssey V3.0' software from Licor Biosciences, Lincoln, NE, USA, after normalizing against respective loading controls.
Flow cytometry
Cell surface expression of receptors was measured using respective primary antibodies labeled with fluorescein isothiocyanate or phycoerythrin. A minimum of 10 000 total events were acquired and analyzed using CellQuestPro of BD biosciences software. For determination of intracellular IL-4, we used the BD Cytofix/Cytoperm Plus kit (with Golgi Stop) from BD Biosciences, as per the protocol recommended by the manufacturer. The analysis was performed either in FlowJo (Tree Star, Inc., Ashland, OR, USA) or CellQuest Pro. In all experiments, unstained and stained cells without any activation, as well as those stained with isotype controls were used as appropriate controls.
ChIP assay
The ChIP procedure was carried out following the manufacturer's instructions (Upstate Biotechnology Inc., Lake Placid, NY, USA) with some modifications.43 In brief, after specific stimulus, 3 × 106 cells were fixed with 1% formaldehyde and quenched with 0.125 M glycine (both for 10 min at 37 °C). Chromatin was sheared to an average size of 500 bp and pre-cleared with protein A-agarose beads. The soluble chromatin was incubated overnight with 2 μg of antibody, followed by incubation with blocked beads. The immune complexes were collected by centrifugation and washed following the manufacturer's protocol. Input and immunoprecipitated chromatin samples were reverse cross-linked by incubating at 65 °C overnight in the presence of 200 mM NaCl. This was followed by proteinase K digestion and DNA was extracted with phenol/chloroform and precipitated with ethanol. Precipitated DNA was analyzed by PCR consisting of 28–30 amplification cycles, and resolved on an agarose gel. The antibodies and primer sequences used have been listed in Supplementary Table 1c. All experiments were repeated at least three times.
Tiled array
Extracellular signal-regulated kinase-specific chromatin-immunoprecipitated samples from either TCR-activated (3 h) or unstimulated cells were hybridized against a tiled array that was generated around the 60-nt long IL-4 promoter region from −1374 to −1315 nt. This region included the ERK-binding site. The first probe in this array was a 60 nt sequence spanning from nt positions −1404 to −1345. All subsequent probes—also 60 nt long—then represented successive one-nucleotide shifts in sequence, terminating with a probe extending from −1344 to −1285 nt.
Chromatin-immunoprecipitated samples were amplified by ligation-mediated PCR before being subjected to labeling by Agilent genomic DNA labeling kit (Agilent, Santa Clara, CA, USA). This kit uses random primer labeling using Exo-Klenow. Agilent's in situ hybridization kit 5188–5220 was used for hybridization. For this, equal amounts of Cy3-labeled Input were mixed with Cy5-labeled IP DNA. Hybridization was carried out at 65 °C for 40 h as per the protocol recommended by the manufacturer. After hybridization, the slides were washed and scanned at 5-μm resolution using the Agilent Microarray Scanner. Normalization methods used were Median Blanks Subtraction, Inter-array median normalization and dye bias median normalization. Fold enrichment of hybridization signal was then calculated based on the signal obtained from two samples
Cloning of the IL4 promoter and analysis of activity
The IL4 proximal promoter (−1374 to +224) was cloned into the Kpn1/BamH1 site of the promoter-less reporter vector pEGFP1. Purified plasmid DNA was transfected into Jurkat cells by electroporation. Transfected cells were cultured for an additional 36 h. At the end of this period, depending on requirement, cells were either left unstimulated or stimulated with a combination of anti-CD3 and anti-CD28 for a period of 24 h. A parallel set of cells was also stimulated in the presence of 50 μM U0126 (30 min). For experiments involving ERK silencing, we transfected the ERK-specific siRNA simultaneously with plasmid DNA. Reporter gene expression was monitored at room temperature by confocal microscopy on Nikon TE 2000E microscope (Nikon Corporation, Tokyo, Japan) equipped with × 60/1.4 PlanApochromat objective lens. Excitation was at 488 nm and emission was recorded at 515/530 nm. Mean fluorescence intensity of images was quantified using the ImageJ version 1.41 (National Institute of Health, USA) and to reduce background noise, images were treated with two-dimensional filter (Gaussian filtering).
DNA pull-down
A minimal binding region of 20 nt was derived based on Tiled array experiment and we generated a biotinylated probe representing this sequence. The probe was first attached to streptavidin-coated beads and these beads were incubated with pre-cleared nuclear extract for 30 min. The beads were then washed extensively and bound proteins subsequently eluted by boiling in SDS. The eluted fraction was resolved on 10% SDS-PAGE and probed for the presence of ERK-1 and ERK-2 by western blot analysis.
Statistical analysis
To identify whether means of two or more samples are different, one-way analysis of variance test was performed wherever required. For Figure 6b, Spearman's rank correlation rho was estimated using ‘R' version 2.12.2 (http://www.r-project.org) (2011-02-25) and variance for Figure 6a was calculated using Microsoft Excel (Microsoft).
Acknowledgments
We are grateful to Raina Dua for help with the confocal microscopy. This study was supported by a grant from the Department of Biotechnology, Government of India (KVSR), a grant from the Academy of Finland, The Sigrid Juselius Foundation (KVSR and RL), The National Technology Agency of Finland and The European Seventh Commission Framework Grant EC-FP7-SYBILLA-201106 (RL). UU is the recipient of a Senior Research Fellowship from the Council of Scientific and Industrial Research, Government of India. Authors declare no conflict of interest.
Author contribution: PT, NS, UU and HJ performed experiments and analyzed data. AS provided blood samples, RL and KVSR analyzed data and KVSR wrote this paper.
Footnotes
The Supplementary Information that accompanies this paper is available on the Immunology and Cell Biology website (http://www.nature.com/icb)
Supplementary Material
References
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Ukai-Tadenuma M, Miyamoto T, Sugaya K, Hosokawa H, Hasegawa A, et al. Essential role of GATA3 for the maintenance of type 2 helper T (Th2) cytokine production and chromatin remodeling at the Th2 cytokine gene loci. J Biol Chem. 2004;279:26983–26990. doi: 10.1074/jbc.M403688200. [DOI] [PubMed] [Google Scholar]
- Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Theil A, Kuhn R, Rajewsky K, Muller W, Aseenmacher M, et al. Induction of interleukin 4 (IL-4) expression in T helper (Th) cells is not dependent on IL-4 from non-Th cells. J Exp Med. 1994;179:1349–1353. doi: 10.1084/jem.179.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth N, Hu-Li J, Paul WE. Conventional, naive CD4+ T cells provide an initial source of IL-4 during Th2 differentiation. J Immunol. 2000;165:3620–3625. doi: 10.4049/jimmunol.165.7.3620. [DOI] [PubMed] [Google Scholar]
- Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109 (2 Suppl. 1:S67–S79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- Pearson G, Robinson F, Gibson TB, Xu BE, Karandikar M, Berman K, et al. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24:21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Kimura M, Kubo M, Shimizu C, Tada T, Perimutter RM, et al. T cell antigen receptor-mediated activation of the Ras/mitogen-activated protein kinase pathway controls interleukin 4 receptor function and type-2 helper T cell differentiation. Proc Natl Acad Sci U S A. 1999;96:1024–1029. doi: 10.1073/pnas.96.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So EY, Oh J, Jang JY, Kim JH, Lee CE. Ras/Erk pathway positively regulates Jak1/STAT6 activity and IL-4 gene expression in Jurkat T cells. Mol Immunol. 2007;44:3416–3426. doi: 10.1016/j.molimm.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Jorritsma PJ, Brogdon JL, Bottomly K. Role of TCR-induced extracellular signal-regulated kinase activation in the regulation of early IL-4 expression in naive CD4+ T cells. J Immunol. 2003;170:2427–2434. doi: 10.4049/jimmunol.170.5.2427. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Dillon S, Denning TL, Pulendran B. ERK1-/- mice exhibit Th1 cell polarization and increased susceptibility to experimental autoimmune encephalomyelitis. J Immunol. 2006;176:5788–5796. doi: 10.4049/jimmunol.176.10.5788. [DOI] [PubMed] [Google Scholar]
- Pages G, Guerin S, Grall D, Bonino F, Smith A, Anjuere F, et al. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science. 1999;286:1374–1377. doi: 10.1126/science.286.5443.1374. [DOI] [PubMed] [Google Scholar]
- Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- Jankovic D, Kullberg MC, Noben-Trauth N, Caspar P, Paul WE, Sher A. Single cell analysis reveals that IL-4 receptor/STAT6 signaling is not required for the in vivo or in vitro development of CD4+ lymphocytes with a Th2 cytokine profile. J Immunol. 2000;164:3047–3055. doi: 10.4049/jimmunol.164.6.3047. [DOI] [PubMed] [Google Scholar]
- Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, et al. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- Holmstrom TH, Schmitz I, Soderstrom TS, Poukkula M, Johnson VL, Chow SC, et al. MAPK/ERK signaling in activated T cells inhibits CD95/Fas-mediated apoptosis downstream of DISC assembly. EMBO J. 2000;19:5418–5428. doi: 10.1093/emboj/19.20.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Weber M, Krammer PH. Regulation of IL4 gene expression by T cells and therapeutic perspectives. Nat Rev Immunol. 2003;3:534–543. doi: 10.1038/nri1128. [DOI] [PubMed] [Google Scholar]
- Maneechotesuwan K, Xin Y, Ito K, Jazrawi E, Lee KY, Usmani OS, et al. Regulation of Th2 cytokine genes by p38 MAPK-mediated phosphorylation of GATA-3. J Immunol. 2007;178:2491–2498. doi: 10.4049/jimmunol.178.4.2491. [DOI] [PubMed] [Google Scholar]
- Wierenga EA, Messer G. Regulation of interleukin 4 gene transcription: alterations in atopic disease. Am J Respir Crit Care Med. 2000;162:S81–S85. doi: 10.1164/ajrccm.162.supplement_2.ras-5. [DOI] [PubMed] [Google Scholar]
- Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, et al. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- Schilling M, Maiwald T, Hengl S, Winter D, Kreutz C, Kolch W, et al. Theoretical and experimental analysis links isoform-specific ERK signalling to cell fate decisions. Mol Syst Biol. 2009;5:334. doi: 10.1038/msb.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding A, Tian T, Westbury E, Frische E, Hancock JF. Subcellular localization determines MAP kinase signal output. Curr Biol. 2005;15:869–873. doi: 10.1016/j.cub.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Hancock JF, Kolch W. Signalling ballet in space and time. Nat Rev Mol Cell Biol. 2010;11:414–426. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhri VK, Kumar D, Misra M, Dua R, Rao KVS. Integration of a phosphatase with the mitogen-activated protein kinase pathway provides for a novel signal processing function. J Biol Chem. 2010;285:1296–1310. doi: 10.1074/jbc.M109.055863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer FJ, Olinsky A, Phelan PD. Variability of airways hyper-reactivity and allergy in cystic fibrosis. Arch Dis Child. 1981;56:455–459. doi: 10.1136/adc.56.6.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Liu XS, Liu C, Xu YJ, Xiong WN. Role of extracellular signal-regulated kinase in regulating expression of interleukin 13 in lymphocytes from an asthmatic rat model. Chin Med J (Engl) 2010;123:1715–1719. [PubMed] [Google Scholar]
- Cannons JL, Yu LJ, Hill B, Mijares LA, Dombroski D, Nichols KE, et al. SAP regulates TH2 differentiation and PKC-θ-mediated activation of NF-κB1. Immunity. 2004;21:693–706. doi: 10.1016/j.immuni.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biological functions. Annu Rev Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura SI, et al. Essential role of Stat6 in IL-4 signaling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- Kaplan MH Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating response to IL-4 and for the development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- Kaiser M, Wiggin GR, Lightfoot K, Arthur JS, Macdonald A. MSK regulate TCR-induced CREB phosphorylation but not immediate early gene transcription. Eur J Immunol. 2007;37:2583–2595. doi: 10.1002/eji.200636606. [DOI] [PubMed] [Google Scholar]
- Marchi M, D'Antoni A, Fomentini I, Parra R, Brambilla R, Ratto GM, et al. The N-terminal domain of ERK1 accounts for the functional differences with ERK2. PLoS One. 2008;3:e3873. doi: 10.1371/journal.pone.0003873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumesic PA, Scholl FA, Barragan DI, Khavari PA. Erk1/2 MAP kinases are required for epidermal G2/M progression. J Cell Biol. 2009;185:409–422. doi: 10.1083/jcb.200804038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcher JC, Nekrasova T, Paylor R, Landreth GE, Sweatt JD. Mice lacking the ERK1 isoform of MAP kinase are unimpaired in emotional learning. Learn Mem. 2001;8:11–19. doi: 10.1101/lm.37001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund R, Aittokallio T, Nevalainen O, Lahesmaa R. dentification of novel genes regulated by IL-12, IL-4, or TGF-β during the early polarization of CD4+ lymphocytes. J Immunol. 2003;171:5328–5336. doi: 10.4049/jimmunol.171.10.5328. [DOI] [PubMed] [Google Scholar]
- George AA, Sharma M, Singh BN, Sahoo NC, Rao KVS. Transcription regulation from a TATA and INR-less promoter: spatial segregation of promoter function. EMBO J. 2006;25:811–821. doi: 10.1038/sj.emboj.7600966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.