Abstract
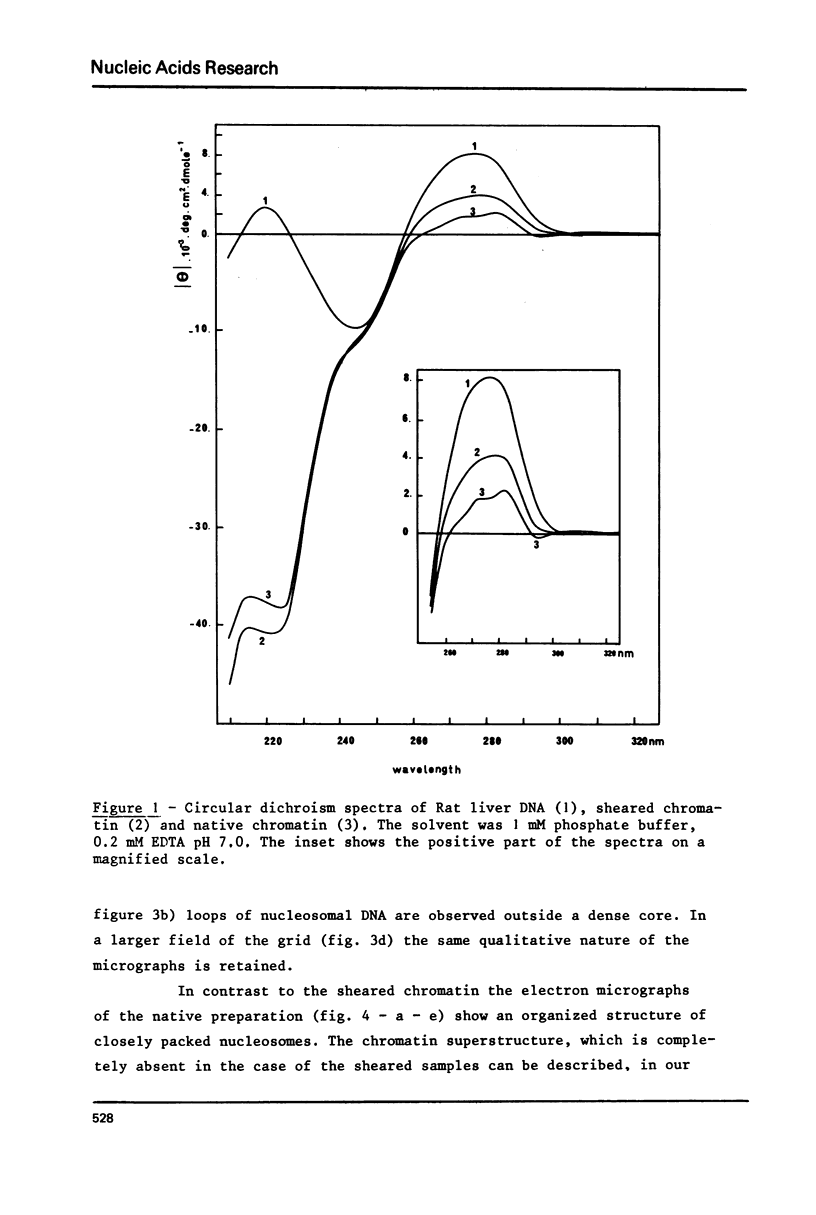
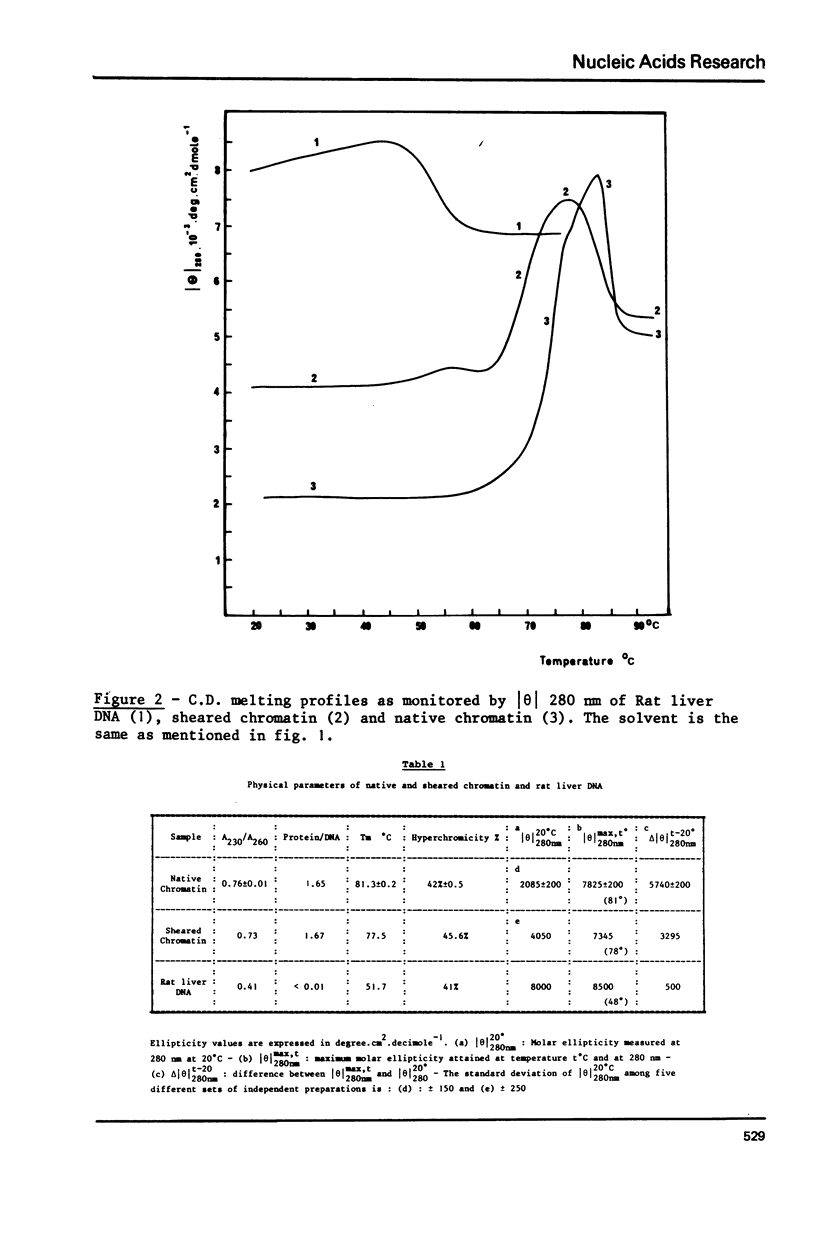
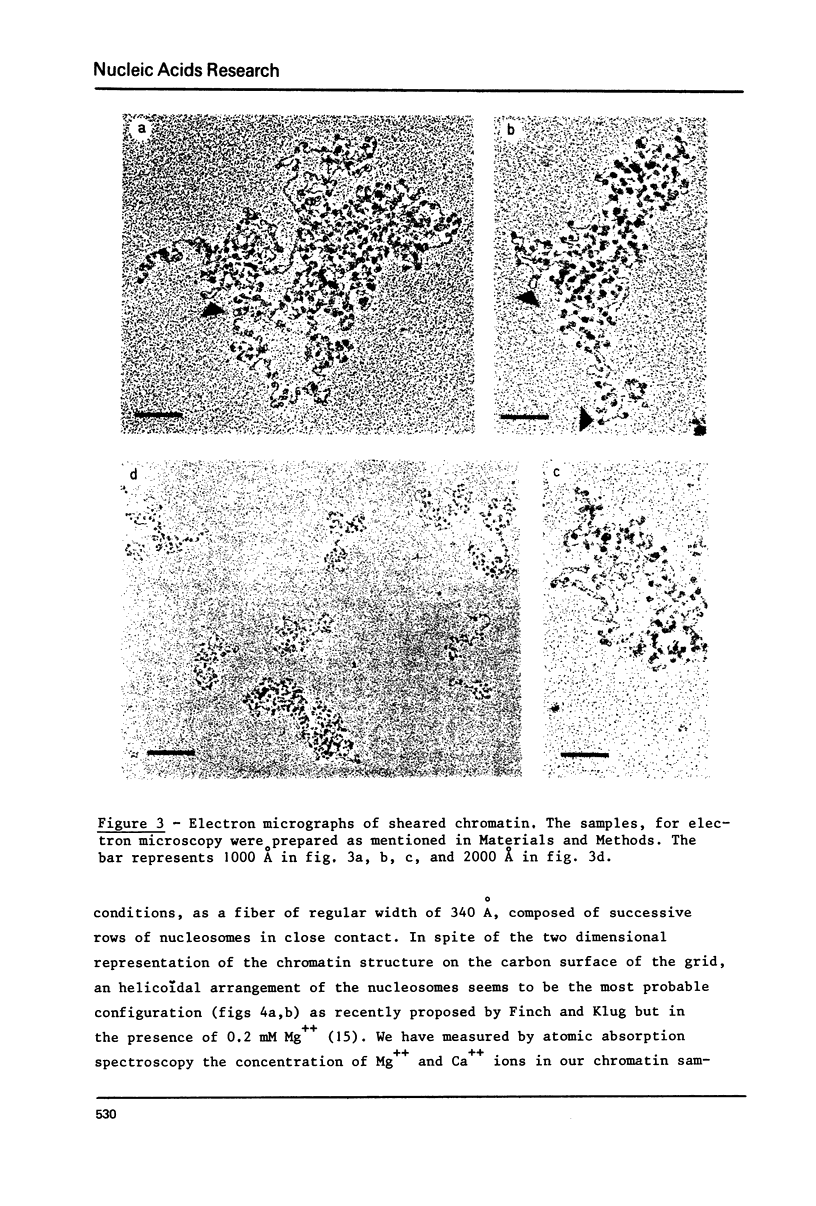
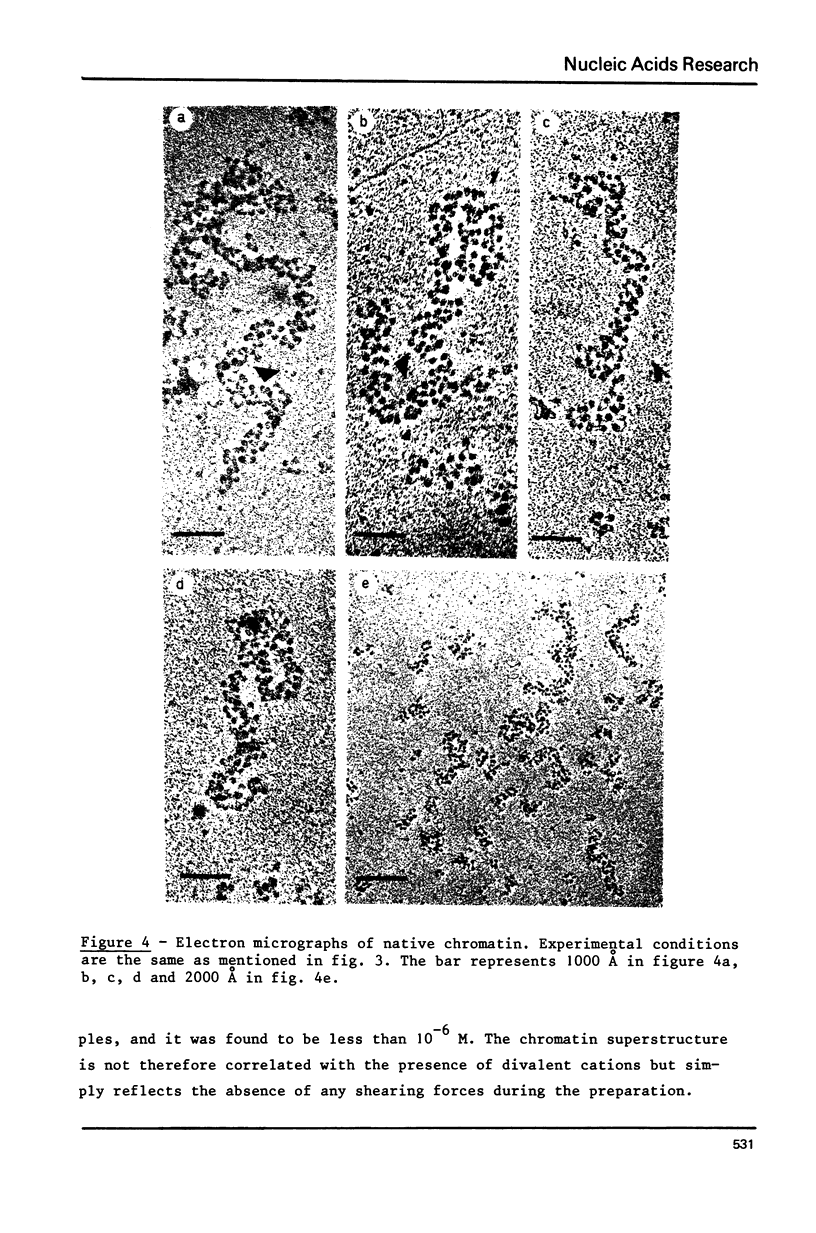
Two types of chromatin were extracted from the same stock of rat liver nuclei by a short exposure to micrococcal nuclease and by shearing respectively. These two materials which are identical in their protein/DNA content and by the presence of the five histones, were compared by means of circular dichroism and electron microscopy. Under the electron microscope and in absence of any divalent cation a superstructure of the unfixed chromatin fiber can be viewed only with native material but is no more present in sheared one. The increase of CD signal at 280 nm (from 2000 to about 4000 cm2 deg.dmole-1) in the case of sheared chromatin is not related to the loss of superstructure but to the structural changes of DNA inside the nucleosomal core which are always produced by shearing. These two correlated observations offer new sensitive probes of the integrity of any native or reconstituted chromatin.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Doenecke D., McCarthy B. J. Movement of histones in chromatin induced by shearing. Eur J Biochem. 1976 May 1;64(2):405–409. doi: 10.1111/j.1432-1033.1976.tb10316.x. [DOI] [PubMed] [Google Scholar]
- Dubochet J., Ducommun M., Zollinger M., Kellenberger E. A new preparation method for dark-field electron microscopy of biomacromolecules. J Ultrastruct Res. 1971 Apr;35(1):147–167. doi: 10.1016/s0022-5320(71)80148-x. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lawrence J. J., Chan D. C., Piette L. H. Conformational state of DNA in chromatin subunits. Circular dichroism, melting, and ethidium bromide binding analysis. Nucleic Acids Res. 1976 Nov;3(11):2879–2893. doi: 10.1093/nar/3.11.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loening U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem J. 1967 Jan;102(1):251–257. doi: 10.1042/bj1020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel R., Fasman G. D. Chromatin and nucleosome structure. Nucleic Acids Res. 1976 Aug;3(8):1839–1855. doi: 10.1093/nar/3.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P., Kendall F., Nicolini C. Thermal denaturation of sheared and unsheared chromatin by absorption and circular dichroism measurements. Nucleic Acids Res. 1976 Aug;3(8):1875–1881. doi: 10.1093/nar/3.8.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolini C., Baserga R., Kendall F. DNA structure in sheared and unsheared chromatin. Science. 1976 May 21;192(4241):796–798. doi: 10.1126/science.1265482. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Noll M., Thomas J. O., Kornberg R. D. Preparation of native chromatin and damage caused by shearing. Science. 1975 Mar 28;187(4182):1203–1206. doi: 10.1126/science.187.4182.1203. [DOI] [PubMed] [Google Scholar]
- Paoletti J., Magee B. B., Magee P. T. The structure of chromatin: interaction of ethidium bromide with native and denatured chromatin. Biochemistry. 1977 Feb 8;16(3):351–357. doi: 10.1021/bi00622a002. [DOI] [PubMed] [Google Scholar]
- Rees A. W., Debuysere M. S., Lewis E. A. Soluble nucleohistone of compact configuration. Biochim Biophys Acta. 1974 Aug 15;361(1):97–108. doi: 10.1016/0005-2787(74)90212-3. [DOI] [PubMed] [Google Scholar]
- Seidman M. M., Cole R. D. Chromatin fractionation related to cell type and chromosome condensation but perhaps not to transcriptional activity. J Biol Chem. 1977 Apr 25;252(8):2630–2639. [PubMed] [Google Scholar]
- Sollner-Webb B., Felsenfeld G. A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry. 1975 Jul;14(13):2915–2920. doi: 10.1021/bi00684a019. [DOI] [PubMed] [Google Scholar]
- Sperling L., Klug A. X-ray studies on "native" chromatin. J Mol Biol. 1977 May 15;112(2):253–263. doi: 10.1016/s0022-2836(77)80142-3. [DOI] [PubMed] [Google Scholar]
- Spurrier M. H., Reeck G. R. Thermal denaturation and circular dichroism properties of sheared and unsheared chromatins. FEBS Lett. 1976 Nov;70(1):81–84. doi: 10.1016/0014-5793(76)80730-2. [DOI] [PubMed] [Google Scholar]
- Stein A., Bina-Stein M., Simpson R. T. Crosslinked histone octamer as a model of the nucleosome core. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2780–2784. doi: 10.1073/pnas.74.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro T., Kurokawa M. Chromatin conformation as affected by shearing and by the removal of a portion of non-histone proteins. FEBS Lett. 1975 Nov 15;59(2):250–253. doi: 10.1016/0014-5793(75)80386-3. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T. Influence of histone H1 on chromatin structure. Cell. 1977 Sep;12(1):101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Palter K., Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975 Sep;6(1):85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]
- Wilhelm F. X., de Murcia G. M., Champagne M. H., Daune M. P. Conformational changes of histones and DNA during the thermal denaturation of nucleoprotein. Eur J Biochem. 1974 Jun 15;45(2):431–443. doi: 10.1111/j.1432-1033.1974.tb03567.x. [DOI] [PubMed] [Google Scholar]
- Yaneva M., Dessev G. Isolation and properties of structured chromatin from Guerin ascites tumour and rat liver. Eur J Biochem. 1976 Jul 15;66(3):535–542. doi: 10.1111/j.1432-1033.1976.tb10579.x. [DOI] [PubMed] [Google Scholar]
- de Pomerai D. I., Chesterton C. J., Butterworth P. H. Preparation of chromatin. Variation in the template properties of chromatin dependent on the method of perparation. Eur J Biochem. 1974 Aug 1;46(3):461–471. doi: 10.1111/j.1432-1033.1974.tb03639.x. [DOI] [PubMed] [Google Scholar]