Recent advances in fluorescence microscopy technologies have revolutionized our views on bacterial cell organization. A prolific area of this revolution in tiny cell visualization has been cell division. Most bacteria divide in their middle, implying that they are aware of at least one particular site of themselves, the midcell division site. This raises the fundamental question: How do bacterial cells identify midcell? It all started with the immunolocalization studies of the FtsZ division protein, which showed that FtsZ assembles at midcell in a ring-like structure (1). Since then, additional cell division proteins have been shown to localize to midcell (2). In a recent issue of the Proceedings, Raskin and de Boer (3) extend this concept of site-specific protein targeting by describing the unique dynamic properties of the localization of the MinD division inhibitor. This discovery adds a new dimension to our exploration of the unexpected complexity in the spatial organization of bacterial cells.
One cannot talk about bacterial cell division without considering the FtsZ protein, which shares properties with cytoskeletal molecules. FtsZ, which binds GTP and has a GTPase activity (4–6), plays a central role in cytokinesis as a major component of a contractile ring (1). The assembly of the FtsZ ring at midcell occurs well before the constriction is initiated (1). In addition to ftsZ (7), many more genes are specifically involved in cell division (2, 8). The other cell division proteins are later recruited to the FtsZ ring to form the membrane-associated septal ring that mediates septation. The answer to the problem of midsite selection is still largely unknown, but important insights have come from studies on minicell-producing (min) mutants (9). These mutants often divide at the poles (¼ and ¾ sites on the cell), generating small, chromosomeless minicells. Here, we discuss the roles of the Min proteins in cell division and assess the different strategies used by two rod-shaped bacteria, the Gram-negative Escherichia coli and the Gram-positive Bacillus subtilis, to solve the problem of positioning the midcell site.
In E. coli, the minicell genetic locus has three genes: minC, minD, and minE (10). By analyzing the effect of expressing various combinations of the min genes, it was found that MinC is a division inhibitor and that MinD stimulates MinC activity (10, 11). Deletions of minC, minD, or all three min genes give rise to a minicell phenotype, whereas inactivation of the minE gene alone results in the formation of long filamentous cells. MinD has an ATPase activity that is required for MinC function (12). MinCD acts either directly or indirectly to interfere with the function of the FtsZ protein (13, 14). Overexpression of MinCD prevents formation of the FtsZ ring at midcell. Conversely, MinCD inhibitory function can be overcome by increasing FtsZ concentration, leading to minicell production (15, 16). These observations led to the hypothesis that in the absence of MinE, the inhibitory MinCD complex blocks division at all potential division sites (PDSs) (10, 17). When the ratio of MinD to MinE is normal, the division inhibitory activity of MinCD is restricted to the polar PDSs, leaving the midcell PDS free for septum formation. MinE therefore would impart the topological specificity, ensuring medial cell division. Experiments using the yeast two-hybrid system confirmed the MinC-MinD interaction and its inhibition by MinE (18).
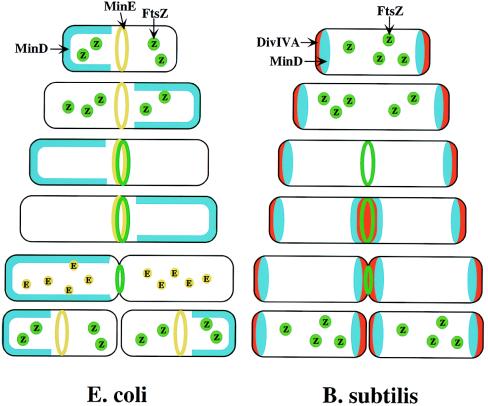
If MinE confers topological activity, does it localize to a particular site in the cell? By using a biologically active MinE derivative that was fused to green fluorescent protein (GFP), it was shown that MinE-GFP accumulates at or near midcell in a ring-like structure that appears to depend on a functional MinD protein, but is independent of FtsZ ring assembly (19). MinE rings were found in very young cells but not in deeply constricting cells, suggesting that the MinE ring disassembles before the completion of cytokinesis. Thus, MinE appears to localize and delocalize as a function of the cell cycle. An astonishing result came from studies of MinD localization (3). Visualization of a functional GFP-MinD fusion protein in living cells revealed that the protein rapidly oscillates from one pole to the other. Time-lapse experiments showed that GFP-MinD resides for 10–30 sec in only one of the cell halves (along one polar cap up to approximately the middle of the cell) then moves to the opposite cell half, only to shift back to its original cell half 10–30 sec later. In between polar shifts, the protein appears to dissociate from the membrane periphery to accumulate in the cytoplasm. The GFP-MinD oscillatory pattern was observed in newborn cells as well as in constricting cells, suggesting that this dynamic behavior lasts for most, if not all, of the cell cycle. GFP-MinD oscillation was independent of FtsZ. However, the polar localization of GFP-MinD was lost in a minE mutant, as visualized by a diffuse static fluorescence in the cytoplasm (3). These results predict a model in which the localization of the Min proteins directs the division machinery to midcell (Fig. 1, Left). Early in the division cycle, MinE localizes to a ring-like structure at or near the middle of the cell. MinD accumulates alternately at the membrane periphery on either side of the MinE ring. The rapid relocation of MinD ensures that no FtsZ ring is assembled at either the ¼ or ¾ sites in the cell halves. The presence of MinE at midcell prevents the MinCD inhibitory activity at this site, allowing assembly of the FtsZ ring. Additional division proteins then are recruited to the FtsZ ring (not shown in the Fig. 1, Left), resulting in septal ring formation.
Figure 1.
Models for division-site selection in E. coli (Left) and B. subtilis (Right). MinD is in blue, MinE in yellow, FtsZ in green and DivIV in red. Shown are different stages of the cell cycle, beginning with a newborn cell and finishing with cell division that produces two daughter cells. (Left) In E. coli, MinE localizes to a ring-like structure at or near the middle of the cell early in the division cycle. MinD accumulates alternately at the membrane periphery on either side of the MinE ring (3). The alternation of MinD localization from one pole to the other occurs at a frequency of the order of tens of seconds. The rapid relocation of MinD ensures that no FtsZ ring is assembled at either the ¼ or ¾ sites in the cell halves. The presence of MinE at midcell prevents the MinD inhibitory activity at this site, allowing assembly of the FtsZ ring at this site. The MinE ring disassembles before completion of constriction. (Right) In B. subtilis, DivIVA and MinD are localized to the cell poles in a newborn cell, and therefore the presence of the MinD inhibitor prevents the formation of the FtsZ ring at these sites. Later, presumably after completion of DNA replication, a new potential division site is created at midcell. The sequestration of the MinD inhibitor to the poles allows assembly of the FtsZ ring at midcell and recruitment of other cell division proteins. At this point, the division machinery presumably becomes resistant to the MinD inhibition. DivIVA and MinD proteins then are recruited to the midcell. Constriction then is initiated. When constriction is completed, the FtsZ ring disassembles, but DivIVA and MinD remain at the newly formed poles, preventing further divisions from taking place in these polar sites.
B. subtilis has homologues of MinC and MinD. The function of the MinD homologue has been conserved, as minD mutants of B. subtilis have the typical minicell phenotype (20–22). However, the B. subtilis genome lacks a minE homologue. The topological specificity of the MinCD division inhibitor in B. subtilis appears to be mediated by the product of the unrelated divIVA gene (23, 24). Mutations in divIVA lead both to minicell production and the formation of long nonseptate filaments (25). Depletion of DivIVA proteins results in disruption of FtsZ ring assembly, indicating that the control of division inhibition by MinCD-DivIVA of B. subtilis, like the Min system in E. coli, acts at the level of FtsZ. However, the mechanism of DivIVA action appears to be fundamentally different from that of MinE. DivIVA-GFP localizes to both cell poles as well as the midcell site (24). Measurements of DivIVA location as a function of cell length suggests that DivIVA is targeted to midcell late in the cell cycle, before septal constriction begins, and remains at the new poles that arise from midcell division, and forever after (26). That contrasts with E. coli MinE that arrives at midcell relatively early and disappears before constriction is complete. Furthermore, DivIVA localization at midcell does not require MinD but depends on the formation of the FtsZ ring and on later assembling components of the division apparatus (26). The topological target for DivIVA may be a component of the division apparatus. Thus, the requirements for targeting DivIVA to midcell are different from those of the E. coli MinE. The B. subtilis MinD accumulates at both the septal ring and the polar caps with a cell cycle-dependent pattern that is similar to DivIVA (26). No oscillation of MinD was observed. These results favor a mechanism for the division site selection in B. subtilis (Fig. 1, Right) in which DivIVA and MinD are localized to the cell poles in a newborn cell both, and therefore the presence of the MinD inhibitor prevents the formation of the FtsZ ring at these sites. After completion of DNA replication, a new potential division site is created at midcell. The sequestration of the MinD inhibitor to the poles allows assembly of the FtsZ ring at midcell and recruitment of other cell division proteins. At this point, the division machinery presumably becomes resistant to the MinCD inhibition, perhaps because the presence of other cell division proteins stabilizes the FtsZ ring. DivIVA then is recruited to the midcell, possibly by a later-assembling division protein. Assembly of DivIVA promotes the targeting of some MinD proteins to midcell. Constriction then is initiated. When constriction is completed, the division apparatus disassembles, but DivIVA and MinD remain at the newly formed poles. Thus, both daughter cells have the MinD inhibitor at their poles, preventing further divisions from taking place in these polar sites.
Although the actual biochemical mechanisms of Min/DivIVA-mediated site selection in E. coli and B. subtilis have yet to be resolved, the localization of these proteins provides a valuable lesson; the importance of the spatial cellular coordinates of the molecules involved in the selection of the division site. It is noteworthy that protein localization in bacteria is not a phenomenon restricted to cell division. The transmembrane chemoreceptors in E. coli accumulate at the cell poles, recruiting the cytoplasmic proteins CheA and CheW to these sites (27). The virulence factor ActA, which is required for nucleating actin polymerization, localizes at one pole of Listeria monocytogenes (28). Several B. subtilis proteins that are involved in sporulation provide further examples of subcellular localization (29). Recently, the histidine kinase CckA that is involved in the cell cycle control of Caulobacter crescentus was found to exhibit a dynamic pattern of polar localization that is cell cycle dependent (30). In light of the accumulating evidence of proteins with specific addresses in the cell, the concept of bacteria as amorphous bags of enzymes is most definitively passé.
Footnotes
The companion to this commentary begins on page 4971 in issue 9 of volume 96.
References
- 1.Bi E F, Lutkenhaus J. Nature (London) 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 2.Bramhill D. Annu Rev Cell Dev Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 3.Raskin D M, de Boer P A J. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.RayChaudhuri D, Park J T. Nature (London) 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 5.de Boer P, Crossley R, Rothfield L. Nature (London) 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee A, Dai K, Lutkenhaus J. Proc Natl Acad Sci USA. 1993;90:1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutkenhaus J F, Wolf-Watz H, Donachie W D. J Bacteriol. 1980;142:615–620. doi: 10.1128/jb.142.2.615-620.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirota Y, Ryter A, Jacob F. Cold Spring Harb Symp Quant Biol. 1968;33:677–693. doi: 10.1101/sqb.1968.033.01.077. [DOI] [PubMed] [Google Scholar]
- 9.Adler H I, Shekhtman E M, Zechiedrich E L, Schmid M B, Cozarelli N R. Proc Natl Acad Sci USA. 1967;57:321–326. [Google Scholar]
- 10.de Boer P A, Crossley R E, Rothfield L I. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 11.de Boer P A, Crossley R E, Rothfield L I. J Bacteriol. 1988;170:2106–2112. doi: 10.1128/jb.170.5.2106-2112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Boer P A, Crossley R E, Hand A R, Rothfield L I. EMBO J. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Boer P A, Crossley R E, Rothfield L I. J Bacteriol. 1992;174:63–70. doi: 10.1128/jb.174.1.63-70.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi E, Lutkenhaus J. J Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bi E, Lutkenhaus J. J Bacteriol. 1990;172:5610–5616. doi: 10.1128/jb.172.10.5610-5616.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Boer P A, Crossley R E, Rothfield L I. Proc Natl Acad Sci USA. 1990;87:1129–1133. doi: 10.1073/pnas.87.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothfield L I, Zhao C R. Cell. 1996;84:183–186. doi: 10.1016/s0092-8674(00)80971-x. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, Cao C, Lutkenhaus J. J Bacteriol. 1996;178:5080–5085. doi: 10.1128/jb.178.17.5080-5085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raskin D M, de Boer P A. Cell. 1997;91:685–694. doi: 10.1016/s0092-8674(00)80455-9. [DOI] [PubMed] [Google Scholar]
- 20.Levin P A, Margolis P S, Setlow P, Losick R, Sun D. J Bacteriol. 1992;174:6717–6728. doi: 10.1128/jb.174.21.6717-6728.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varley A W, Stewart G C. J Bacteriol. 1992;174:6729–6742. doi: 10.1128/jb.174.21.6729-6742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S, Price C W. Mol Microbiol. 1993;7:601–610. doi: 10.1111/j.1365-2958.1993.tb01151.x. [DOI] [PubMed] [Google Scholar]
- 23.Cha J H, Stewart G C. J Bacteriol. 1997;179:1671–1683. doi: 10.1128/jb.179.5.1671-1683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards D H, Errington J. Mol Microbiol. 1997;24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- 25.Reeve J N, Mendelson N H, Coyne S I, Hallock L L, Cole R M. J Bacteriol. 1973;114:860–873. doi: 10.1128/jb.114.2.860-873.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marston A L, Thomaides H B, Edwards D H, Sharpe M E, Errington J. Genes Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddock J R, Shapiro L. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 28.Kocks C, Hellio R, Gounon P, Ohayon H, Cossart P. J Cell Sci. 1993;105:699–710. doi: 10.1242/jcs.105.3.699. [DOI] [PubMed] [Google Scholar]
- 29.Shapiro L, Losick R. Science. 1997;276:712–718. doi: 10.1126/science.276.5313.712. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs C, Domian I J, Maddock J R, Shapiro L. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]