Abstract
RECQ1 is the most abundant RecQ homolog in humans but its functions have remained mostly elusive. Biochemically, RECQ1 displays distinct substrate specificities from WRN and BLM, indicating that these RecQ helicases likely perform non-overlapping functions. Our earlier work demonstrated that RECQ1-deficient cells display spontaneous genomic instability. We have obtained key evidence suggesting a unique role of RECQ1 in repair of oxidative DNA damage. We show that similar to WRN, RECQ1 associates with PARP-1 in nuclear extracts and exhibits direct protein interaction in vitro. Deficiency in WRN or BLM helicases have been shown to result in reduced homologous recombination and hyperactivation of PARP under basal condition. However, RECQ1-deficiency did not lead to PARP activation in undamaged cells and nor did it result in reduction in homologous recombination repair. In stark contrast to what is seen in WRN-deficiency, RECQ1-deficient cells hyperactivate PARP in a specific response to H2O2 treatment. RECQ1-deficient cells are more sensitive to oxidative DNA damage and exposure to oxidative stress results in a rapid and reversible recruitment of RECQ1 to chromatin. Chromatin localization of RECQ1 precedes WRN helicase, which has been shown to function in oxidative DNA damage repair. However, oxidative DNA damage-induced chromatin recruitment of these RecQ helicases is independent of PARP activity. As other RecQ helicases are known to interact with PARP-1, this study provides a paradigm to delineate specialized and redundant functions of RecQ homologs in repair of oxidative DNA damage.
Keywords: RecQ, helicase, PARP-1, oxidative DNA damage, DNA repair
1. Introduction
Delineating the functional specialization of human RecQ helicases has attracted considerable interest in recent years. The RecQ helicases are a highly conserved family of DNA-unwinding enzymes that play key roles in protecting the genome integrity in all kingdoms of life [1]. Of the five human RecQ homologs, mutations in the genes encoding BLM, WRN and RECQL4 are associated with rare diseases of Bloom syndrome (BS), Werner syndrome (WS), and Rothmund-Thomson syndrome (RTS), respectively. Increased genomic instability and predisposition to cancer is hallmark of RecQ-related diseases, however they are each distinguished by clinical features ranging from growth defects (BS and RTS) to premature aging (WS). It is likely that despite being similar in their catalytic core and overlapping biochemical properties in vitro, individual RecQ proteins participate in distinct aspects of DNA metabolism in human cells under basal and genotoxic stress conditions.
Although a disease phenotype has not yet been linked to the mutations in RECQ1 and RecQ5β, they may be responsible for as yet unidentified cancer predisposition disorders. RECQ1 is overexpressed in transformed cells [2]. Depletion of RECQ1 using small inhibitory RNA results in reduced cellular growth and proliferation [3], and has been shown to exert anticancer effects in cells [4] and xenografts [5]. Furthermore, a single nucleotide polymorphism in RECQ1 has been associated with reduced survival of pancreatic cancer patients [6–7]. Thus, RECQ1 exhibits a predictive/ prognostic potential in tumor progression and may also serve as an important target for cancer therapy.
RECQ1 resides on chromosome 12p12 [8–9] and encodes the most abundant RecQ protein in humans [2, 10]. Several observations suggest an important role of RECQ1 in the repair of DNA damage during cellular replication [11]. RECQ1 was recovered as the major Holliday junction branch migration activity from human embryonic kidney cells [12]. Knockout of RECQ1 in mice or suppression of its expression in HeLa cells resulted in cellular phenotypes that include chromosomal instability, increased sister chromatid exchanges and sensitivity to ionizing radiation (IR) [3, 11–13]. Moreover, RECQ1-mutant cells display increased DNA double strand breaks (DSBs) and accumulation of unresolved recombination intermediates [13]. These observations imply that similar to the prominent RecQ proteins such as WRN and BLM, RECQ1 also plays a role in the processing of DNA recombination and repair intermediates. However, biochemically, RECQ1 displays substrate specificities distinct from WRN and BLM, indicating that these helicases are likely to perform non-overlapping functions [14–18]. Precise functions of RECQ1 in genome maintenance are poorly defined.
Human RecQ proteins have also been associated with repair of oxidative damage, prompting selective investigation of the role of RECQ1 in cellular response to this type of DNA damage [19]. This is also supported by the fact that RECQ1-deficient cells are sensitive to IR which in addition to DSBs, also results in oxidative DNA damage [20]. Moreover, the chemistry of DNA lesions produced by hydrogen peroxide (H2O2; this study) and IR is shown to be very similar [21]. If left unrepaired, these DNA lesions can block essential processes such as transcription or replication [22], or can induce mutations, the accumulation of which can lead to cancer [23]. Results presented here provide evidence for the functional specialization of human RecQ homologs and identify a novel involvement of RECQ1 in cellular response to oxidative DNA damage.
2. Materials and Methods
2.1. Cell lines, culture conditions and DNA damage treatment
Human HeLa cells (ATCC) and U2OS cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone Laboratories), 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen). Cells were grown in a humidified 5% CO2 incubator at 37°C. For hydrogen peroxide (H2O2) treatment, cells were grown to near confluence, the growth medium was changed to serum free OptiMEM (Invitrogen) containing various concentrations of H2O2 (Sigma-Aldrich), and cells were incubated at 37°C for 20 minutes before harvesting. For recovery experiments, H2O2 containing media was removed; cells were washed and grown in regular complete medium for indicated times. Hydroxyurea (HU) (Sigma-Aldrich) treatment was as indicated. Stock solutions of the PARP inhibitor (ANI (4-amino-1,8-naphthalimide)) (Acros Organics), and AZD2281 (Olaparib) (Selleck Chemicals) were made in DMSO and the inhibitors were added to the medium at indicated concentration 24 h or 1 h before treatment with H2O2, respectively. Same volume of DMSO was added to the control plates.
2.2. Immunoprecipitation
HeLa nuclear extract was prepared as previously described [24]. Extracts were incubated with Protein G-Dynal beads coupled with polyclonal antibody against human RECQ1 (Bethyl), RPA32 (Santa Cruz Biotech) or normal rabbit IgG (Vector Labs) at 4°C for 90 min, and the immunecomplexes were eluted with 2X SDS-sample buffer following three washes with lysis buffer. Where indicated, nuclear extract was preincubated with benzonase (Sigma, 50 U/ml, 2 hr at 4°C) or ethidium bromide (EtBr) (50µg/ml) was present during the immunoprecipitation reaction. Proteins were resolved by 8–16% SDS-PAGE, transferred onto PVDF membrane and subjected to Western detection of RECQ1 (1:750, Santa Cruz Biotech), RPA32 (1:500, Santa Cruz Biotech) and PARP-1 (1:500, BD Pharmingen).
2.3. Preparation of samples for mass spectrometry
Following staining with SYPRO Ruby (Invitrogen), gel slices containing specific protein bands from RECQ1 and corresponding IgG control immunoprecipitation were excised and transferred to microfuge tubes. Gel pieces were washed twice with 50% acetonitrile containing 0.1% TFA, incubated in 0.2 ml 100 mM NH4HCO3 dissolved in 50% acetonitrile, dehydrated in 100% acetonitrile, and dried in a SpeedVac. The gel slices were rehydrated overnight at 37°C in 40 mM NH4HCO3, 10% acetonitrile containing sequencing-grade trypsin (20 µg/ml), and the supernatant was collected. Gel slices were re-extracted with 50% acetonitrile containing 5% TFA for 1 h. The extracts were dried in SpeedVac. ZipTips were prewashed with 100% acetonitrile followed by a wash with 0.1% TFA. Extracted peptides were dissolved in 0.1% TFA, loaded into ZipTip, washed with 0.1% TFA, and eluted with 70% acetonitrile, 0.1% TFA.
2.4. Mass spectrometry analysis
Peptides eluted from ZipTips were loaded to C18 column prepared in-house (30 cm long with volume of 4 µl). Samples were eluted for 60 min with 2% to 30% gradient of acetonitrile and flow rate 300 nL/min using Shimadzu Prominence Nano HPLC. The column was coupled online through nanospray to an ion trap mass spectrometer Thermo LTQ Orbitrap XL. The mass spectrometer was operated in a data-dependent MS/MS mode using normalized collision-induced dissociation (CID) energy of 35%. The CID spectra were compared against those of the EMBL non-redundant protein database. Only peptides having cross-correlation (Xcorr) cutoffs of 2.6 for [M + 2H]2+, 3.0 for [M + 3H]3+ and higher charge state were considered. These SEQUEST criteria thresholds resulted in a 1–2% of False Discovery Rate. The proteome analysis of the spectra was made by Proteome Discoverer 1.2 software (Thermo Fisher Scientific).
2.5. GST-pull down
Wild-type and truncated fragments of GST-RECQ1 fusion proteins were overexpressed in BL21(DE3) pLysS by 1 mM isopropyl-B-d-thiogalactopyranoside induction for 8 h at 23°C. The bacterial cell pellet was sonicated in lysis buffer (phosphate-buffered saline (PBS), 10% glycerol and 0.4% Triton X-100) and the lysate was clarified by centrifugation at 35,000 rpm for 1 h at 4°C. Approximately 1 ml of the resulting supernatant was incubated with 100 µl of glutathione S-transferase beads (50% v/v) for 1 h at 4°C. The beads were washed three times with 1 ml lysis buffer, and split into two aliquots, one for binding experiments and one for determination of expression by Coomassie Blue staining. For binding experiments, proteinbound beads were incubated for overnight at 4°C with 500 µg of lysate from untreated HeLa cells. The beads were subsequently washed five times with 1 ml of lysis buffer and eluted by boiling with 2X SDS-sample buffer. Eluted proteins were electrophoresed on 8–16% polyacrylamide SDS gels and either stained with Coomassie Blue to demonstrate protein loading or transferred onto PVDF membranes for Western detection of PARP-1 bound to GST-RECQ1 proteins using anti-PARP-1 antibody.
2.6. ELISA
Enzyme-linked immunosorbent assay (ELISA) was performed similarly to previously described [3]. Wells were coated with 50 µl (1 ng/µl) of either BSA or RECQ1 protein diluted in carbonate buffer (0.016 M Na2CO3, 0.034 M NaHCO3 [pH 9.6]), and incubated overnight at 4°C. Wells were blocked for 2 h at 30°C with blocking buffer (PBS, 0.5% Tween-20, 3% BSA) and washed with PBS containing 0.5% Tween-20. Serial dilutions of PARP-1 (0–20 nM, Active Motif) in blocking buffer were then added to the appropriate wells of the microtitre plate and incubated for 1 h at 30°C. DNaseI (100 U/ml) or EtBr (50 µg/ml) was included during the binding step in the corresponding wells to test for DNA mediated protein interaction. Wells were washed five times and incubated with mouse anti-PARP-1 antibody (1:500, BD Pharmingen) 30°C for 1 h. Following three washes, wells were incubated with HRP-conjugated anti-mouse secondary antibody (1:5000) for 30 min at 30°C. After washing five times, any PARP-1 bound to the RECQ1 was detected using OPD substrate (Sigma) and recording absorbance at 490 nm.
2.7. siRNA transfection
Depletion of various RecQ helicases was achieved by transfecting HeLa and U2OS cells with a scrambled control, RECQ1, WRN, or BLM siRNA (siGenome smartpool, Dharmacon) at a final concentration of 10 nM using Lipofectamine 2000 transfection reagent as per the manufacturer’s instructions (Invitrogen). Following transfection, cells were cultured in regular growth medium for 24 h before replating for survival assays or for 36 h before harvesting for biochemical fractionation, lysate preparation for PAR analyses, and COMET assay with or without H2O2 treatment. When needed, treatment with inhibitors began 24 h after transfection.
2.8. Survival assay
Cells (24 h after siRNA transfection) were seeded in quadruplicate at a density of 300 cells/well in 96-well plates and allowed to adhere for 16 h. Cells were then exposed to increasing doses of H2O2 (0–12.5 mM; Sigma-Aldrich) in cold serum-free OptiMEM at 4°C for 10 min, washed twice, and incubated for 72 h in complete DMEM medium. To determine the toxicity to PARP inhibitor, siRNA-depleted cells were grown in the presence or absence of increasing dose of the ANI (0–20 µM) or AZD2281 (0–10 µM) for a continuous period of 72 h. Cell survival was determined by CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (MTS) (Promega). For colony formation assay, siRNA transfected cells were seeded in triplicate at various densities in 60 mm plates and incubated overnight in the presence or absence of ANI (10 µM) before treatment with 1 mM H2O2 as above. Following removal from H2O2, cells grown in the presence or absence of ANI were allowed to form colonies for 14 days, stained with crystal violet and colonies containing >50 cells were counted.
2.9. Comet assay
The alkaline comet assays were performed using a Comet Assay kit (4250-050-K, Trevigen) according to the manufacturer’s recommendations. HeLa cells were mock-treated or treated with ANI (100 µM) for 18 h in complete growth medium at 37°C in a CO2 incubator. Cells were trypsinized, untreated or treated in suspension with 10 µM H2O2 for 5 min on ice and embedded on a microscope slide in agarose. Slides were incubated for 0–1 h at 37°C in a CO2 incubator. Cells on slides were lysed for 1 h at 4°C and the slides were then incubated in the dark for 30 min in cold electrophoresis buffer (300 mM NaOH, 1 mM EDTA, 1% (v/v) DMSO, pH 13) to allow the DNA to unwind prior to electrophoresis at 25 V for 25 min. After neutralization with 0.5 M Tris-HCl (pH 8.0), DNA was visualized using SYBR Green I fluorescent staining. 100 cells per sample were documented and experiment was repeated three times. Data was analyzed using CometScore (TriTek Corp).
2.10. Western detection of poly(ADP)ribose
Untreated or H2O2 -treated cells were lysed in RIPA buffer (50 mM Tris-HCl [ph 8.0], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing cocktail of protease and phosphatase inhibitors (Roche). Equal amounts of total protein for each sample was run on 8–16% SDS-PAGE and transferred to PVDF membrane for immunoblotting with mouse monoclonal antibody against poly(ADP)ribose (PAR) (1:1000, Genetax). Total PAR signal in Western blots were quantified using ImageJ to determine the mean and S.E.M. from three independent experiments.
2.11. PARP activity assay
PARP activity was assayed using the Universal colorimetric PARP assay kit according to the manufacturer's instructions (Trevigen). Cells grown overnight in the presence or absence of ANI (100 µM) were washed with PBS, incubated in 50 µl of PARP buffer containing 0.5 M NaCl, 1% NP40 and protease inhibitors (Roche) on ice for 30 min, and lysates were collected after centrifugation at 14,000 rpm for 10 min at 4°C. Lysate (30 µg total protein/well) was added in duplicates to the wells containing PARP buffer and PARP cocktail, followed by incubation at room temperature for 1 h. Activated DNA was added only to the standards but not to the extracts. Wells were washed thrice with PBS and thrice with PBS containing 0.1% Triton X-100 before and after incubation with streptavidin-horseradish peroxidase (1:1000 in strep diluent buffer) for 1 h. Colorimetric detection of PARP activity was performed at 450 nm.
2.12. Homologous recombination assay
DR-U2OS cells were seeded in 12-well plates [25]. 24 h later, subconfluent DR-U2OS cells were treated with control, BLM, or RECQ1 siRNA as described above. 48 h after initial siRNA treatment, cells were either harvested for Western blot analysis or treated with a second siRNA mixture in addition to either empty vector (pCAGGS) or the I-SceI expression vector (pCBASce; [26]). 48 h after the second siRNA treatment, cells were harvested for Western blot analysis and 30,000 cells were quantitated by flow cytometric analysis on a Becton Dickinson FACScan. Relative HR frequencies were determined by the % GFP positive cells in RECQ1 or BLM-depleted cells compared to control siRNA transfected cells.
2.13. Biochemical fractionation
Sequential extraction of subcellular fractions was performed as previously described [27]. Briefly, HeLa cells were extracted with CSK (10 mM PIPIES [pH 6.8], 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2) with 0.5% Triton X-100 for 5 min at 4°C (the “CSK” fraction) and extraction buffer (42.5 mM Tris-HCL [pH 8.3], 8.5 mM NaCl, 2.6 mM MgCl2, 0.5% deoxycholic acid) with 1% Tween-20 for 5 min at 4°C (the “EXT” fraction). The remaining pellet was sonicated in 2X SDS-sample buffer (the “nuclear pellet” fraction) and consists of nuclear proteins that associate with DNA and/or nuclear matrix. Proteins from equivalent cell volumes of each fraction were resolved by SDS-PAGE and probed for RECQ1 (1:750, Santa Cruz Biotech), WRN (1:500, Santa Cruz Biotech), BLM (1:1000, Abcam), RecQ5β (1:1000, Abcam), PARP-1 (1:500, BD Pharmingen), XRCC1 (1:1000, Abcam), PCNA (1:500, Santa Cruz Biotech), histone H3 (chromatin marker, 1:5000, Upstate Biotech), and GAPDH (marker for cytoplasmic, cytoskeletal and soluble nuclear proteins, 1:1000, Abcam) by immunoblotting. ImageJ was used for quantification.
3. Results
The functions of RecQ helicases have a direct impact on human health, but RECQ1 remains the least characterized in terms of its biological roles. Our earlier work identified that RECQ1 is a DNA damage responsive protein and RECQ1-deficient cells display spontaneous genomic instability. This suggested that RECQ1 is important for the repair of endogenous and/or exogenously induced DNA damage. Here we examined if RECQ1 is involved in cellular response to oxidative DNA damage.
3.1. RECQ1 exists in complex with PARP-1 in vivo and in vitro
We performed mass spectrometry to identify RECQ1-associated DNA repair proteins in human cells. Proteins identified specifically in RECQ1 immunoprecipitates included previously reported interacting proteins such as the single strand DNA-binding protein RPA [15], mismatch repair proteins MLH1, MSH2 and MSH6 [24], and importin alpha [28], providing validity to the approach (data not shown). Additionally, we reproducibly detected multiple unique peptides from PARP-1. Overall, we identified 12 unique peptides representing nearly 17% coverage of PARP-1 (Supplementary Fig. 1).
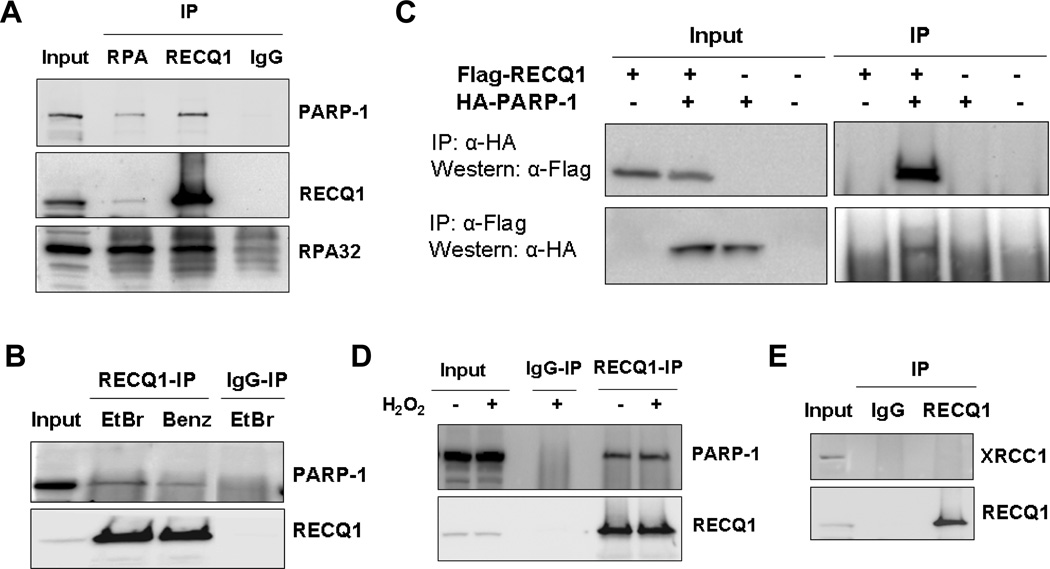
PARP-1 is implicated in DNA repair, replication, and transcriptional regulation [29]. Given their overlapping roles in DNA repair processes, we characterized the putative physical interaction of RECQ1 with PARP-1 (Fig. 1). RECQ1 was immunoprecipitated from HeLa cell nuclear extracts (Fig. 1A). RPA, a protein that has been previously shown to physically interact with RECQ1 and modulate its biochemical activities [15], was identified in the RECQ1 immunoprecipitates (Fig. 1A). Western blot analyses showed that RECQ1 immunoprecipitates also contained PARP-1 (Fig. 1A). Furthermore, immunoprecipitation of RPA resulted in coprecipitation of both RECQ1 and PARP-1 (Fig. 1A, lane 2). This observed association of RPA and PARP-1 is consistent with a recent report by Liu, et al [30]. Similar immunoprecipitation using normal rabbit IgG failed to pull down RECQ1, RPA or PARP-1 (Fig. 1A). Presence of EtBr or use of benzonase-treated extract in immunoprecipitation reaction did not abolish coprecipitation of PARP-1 and RECQ1 suggesting that the interaction is not mediated by DNA (Fig. 1B). To determine that the observed interaction is not an artifact of the RECQ1 antibody used for immunoprecipitation, HeLa cells were transiently transfected with Flag-RECQ1, HA-PARP-1 or both. Cell lysates were used for immunoprecipitation with anti-HA or anti-Flag antibody and the immune complexes were subjected to Western blot analyses with reciprocal antibodies. As shown in Fig. 1C, HA-PARP-1 co-immunoprecipitated with Flag-RECQ1 and Flag-RECQ1 co-immunoprecipitated with HA-PARP-1, thus confirming their in vivo interaction in mammalian cells.
Figure 1. RECQ1 interacts with PARP-1 in vivo.
A, Co-immunoprecipitation analysis of RECQ1 interaction with PARP-1 using HeLa nuclear extracts. Immunoprecipitations (IP) with antibodies specific for RECQ1, RECQ1 interacting protein RPA, and preimmune IgG are indicated. Eluted proteins in immunoprecipitate were analyzed by Western blotting and are indicated. Reciprocal co-IPs of RECQ1 and RPA also contained PARP-1. B, Association of RECQ1 and PARP-1 is not mediated via DNA. RECQ1 antibody co-precipitated RECQ1 and PARP-1 in the presence of EtBr or using benzonase-treated extract in IP reaction. C, Reciprocal co-IP of ectopically expressed RECQ1 and PARP-1. HeLa cells were untransfected or transiently transfected with Flag-RECQ1, HA-PARP-1 or both. Lysates of these cells were used for IP using anti-HA or anti-Flag antibody and analyzed by Western blotting as indicated. D. Association between RECQ1-PARP-1 is not modulated by H2O2-induced DNA damage. HeLa cells were untreated or treated with H2O2 (1 mM, 20 min) and harvested immediately. IP with anti-RECQ1 antibody was followed by Western analyses using anti-PARP-1 and anti-RECQ1 antibodies. E. XRCC1 does not co-IP with RECQ1. RECQ1 IP was subjected to Western analysis using anti-XRCC1 or anti-RECQ1 antibodies. In all experiments, input corresponds to 5% of total protein used in IP reactions.
PARP-1 acts as a DNA strand break sensor and is recruited to damage sites where it participates in repair processes [31]. Therefore, we investigated whether the RECQ1-PARP-1 interaction is modulated by H2O2 treatment that primarily induces single-strand breaks (SSBs) and base damage [32]. We immunoprecipitated RECQ1 from nuclear extracts prepared from untreated or H2O2-treated cells and probed for PARP-1 (Fig. 1D). A similar amount of PARP-1 was co-immunoprecipitated with RECQ1 from the extracts of untreated or H2O2 treated cells (Fig. 1D). These results demonstrate that endogenous RECQ1 coexists in a complex with PARP-1, and this interaction is unaffected following cellular exposure to H2O2. Notably, these experiments were performed in the presence of DNAse I to abolish DNA-mediated protein interactions. XRCC1 is a binding partner of PARP-1 and important scaffold protein in the BER pathway [33]. Under the same conditions as above, we did not detect association of RECQ1 with XRCC1 (Fig. 1E), suggesting that PARP-1-RECQ1 interaction is independent of XRCC1. We examined localization of RECQ1 and PARP-1 in HeLa cells before and after treatment with H2O2. Colocalization of RECQ1 and PARP-1 was observed in both cases; RECQ1 and PARP-1 proteins were detected in the nucleus of HeLa cells as reported previously [3, 34], and this localization pattern was not significantly affected by H2O2 (Supplementary Fig. 2).
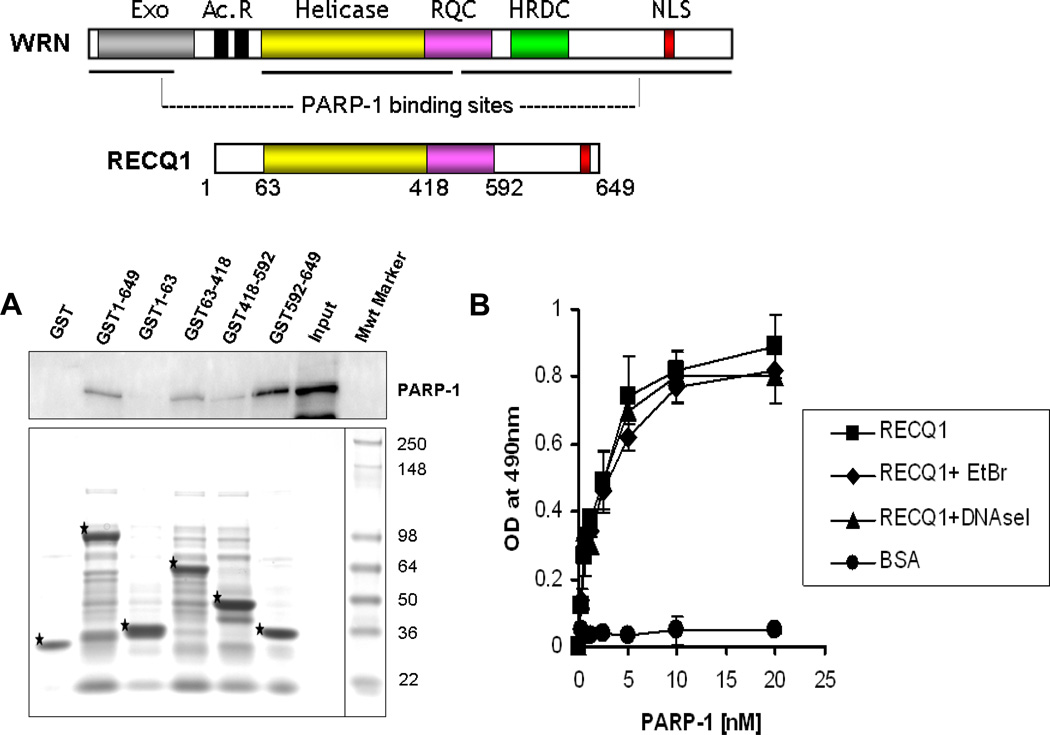
There is prior evidence for interaction of other RecQ helicases with PARP-1 [35–37]. The helicase and RecQ conserved (RQC) domains that mediate interaction between WRN and PARP-1 are conserved among several RecQ homologs including RECQ1 (Fig. 2A). In order to map the PARP-1 interaction domain(s) within RECQ1, GST fusion proteins that encompass truncated versions of human RECQ1 were generated. These fusion proteins were expressed in bacterial cells and GST pull-down experiments were performed using HeLa cell extracts followed by Western blot analysis (Fig. 2A). Cellular PARP-1 efficiently bound full-length RECQ1 and minimally with the polypeptide consisting of the RQC domain. We failed to detect any PARP-1 binding to the N-terminus of RECQ1 (amino acid residues 1–63). However, a polypeptide fragment carrying the C-terminus of RECQ1 (amino acid residues 592–649), which does not have a counterpart in WRN, efficiently bound PARP-1, suggesting that RECQ1 may utilize a distinct domain to interact with PARP-1. Additionally, the polypeptide consisting of the helicase domain of RECQ1 (amino acid residues 63–418) also bound PARP-1. This suggests that PARP-1 has interaction sites in the helicase domain and C-terminal of RECQ1 protein. As these experiments utilized lysates from unstressed cells, it is possible that other potential interacting protein(s) in the cell lysate may compete with PARP-1 for binding to RECQ1.
Figure 2. A direct physical interaction between RECQ1-PARP-1 is mediated via the helicase domain and C-terminal region of RECQ1 in vitro.
A. Top: A schematic diagram of the full-length WRN and RECQ1 proteins showing conserved domains. RQC, RecQ-conserved domain; HRDC, helicase RnaseD conserved domain; NLS, nuclear localization signal. Also shown are Exonuclease (Exo) domain and Acidic repeats (Ac. R) present in WRN protein. Previously reported PARP-1 binding regions of WRN are indicated by horizontal lines. Bottom: Identification of the PARP-1 interacting region of RECQ1. GST alone or GST-RECQ1 fragments (as indicated) bound to glutathione beads were incubated with HeLa extracts (500 µg) overnight at 4°C. After extensive washings, the bound PARP-1 was eluted with SDS sample buffer and analyzed by Western blot using anti-PARP-1 antibody (upper). Coomassie staining of the eluted proteins was done to test expression of various GST-fusion fragments of RECQ1 (lower). GST-RECQ1 proteins are marked by asterisk. Mwt Marker, protein molecular weight marker. B. Recombinant RECQ1 and PARP-1 proteins interact directly as shown by ELISA. Either BSA or purified recombinant RECQ1 was coated onto microtiter plates. Following blocking with 3% BSA, appropriate wells were incubated with the indicated concentrations of recombinant PARP-1 (0–20 nM) for 1 h at 30°C. Parallel wells contained DNaseI (100 U/ml) or EtBr (50 µg/ml) in the binding step to test for DNA-mediated protein interaction. Following washing, RECQ1 bound PARP-1 was detected by ELISA using anti-PARP-1 antibody. The values represent the mean of three independent experiments performed in duplicate with SD indicated by error bars.
A direct interaction between PARP-1 and RECQ1 was confirmed by ELISA using purified recombinant proteins (Fig. 2B; and Supplementary Fig. 3). PARP-1 bound RECQ1 in a protein concentration-dependent manner, whereas a very low OD490 signal was detected in control experiments where BSA (0–76 nM) was substituted for RECQ1. Incubation with either EtBr or DNase I during the binding reaction did not affect the interaction appreciably, indicating that the interaction between RECQ1 and PARP-1 is not DNA-mediated.
Altogether, these results demonstrate that RECQ1 forms a stable complex with PARP-1 in human cells and a direct protein-protein interaction between RECQ1-PARP-1 is primarily mediated via the C-terminus and the helicase domain of RECQ1 in vitro.
3.2. RECQ1-depleted cells display increased PARP activation specifically in response to H2O2 treatment
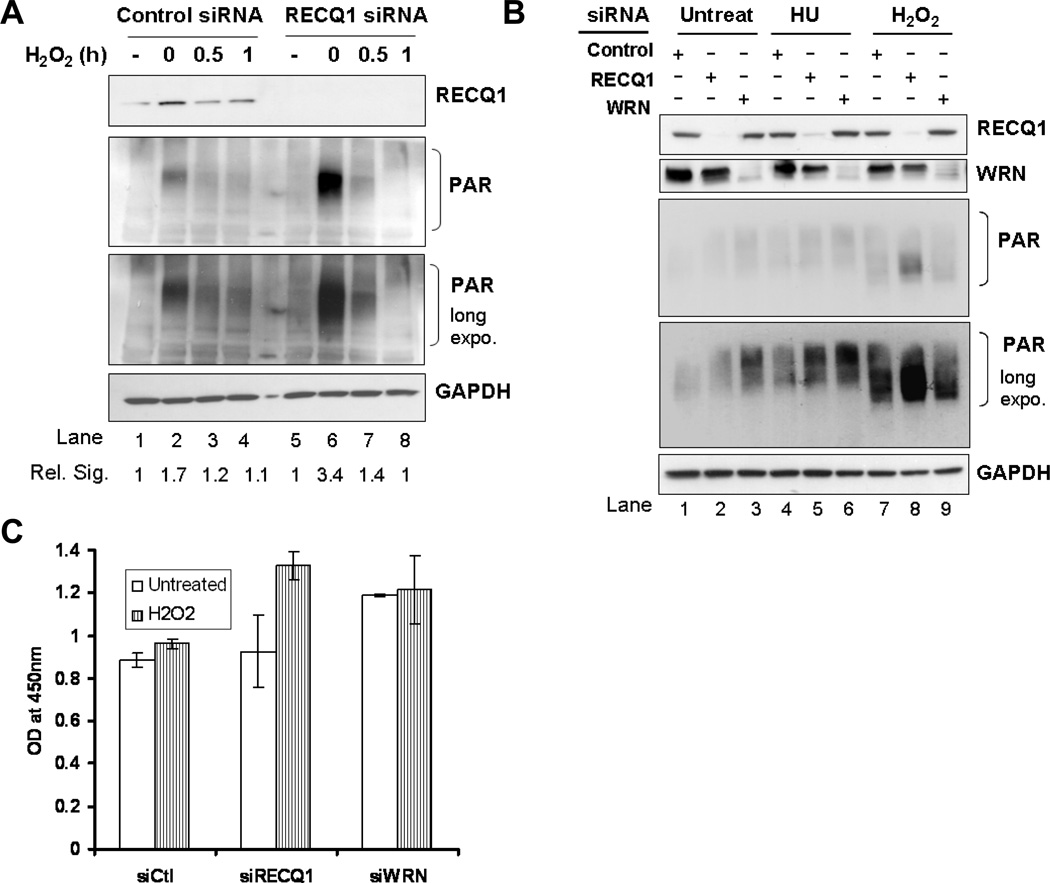
PARP-1 is activated in response to DNA breaks leading to automodification and modification of other receptor proteins with PAR polymers synthesized from NAD+ [38]. Cellular RECQ1 was not found to be poly(ADP)ribosylated, indicating that RECQ1 is not a substrate for PARP (data not shown). To determine if RECQ1 affects PARP activity in untreated or H2O2-treated conditions, we examined PAR levels in whole cells extracts (Fig. 3). Cells transfected with control siRNA showed a little detectable PAR under untreated conditions (Fig. 3A, lane 1) and an increased signal for PAR was observed immediately upon completion of H2O2 treatment (Fig. 3A, lane 2). PAR levels decreased progressively upon recovery from H2O2 treatment and only residual PAR was still observed at 1 h (Fig. 3A, lane 4). We did not detect appreciable PAR levels in lysates of untreated RECQ1-depleted cells (Fig. 3A, lane 5); average total PAR signal was quantitatively comparable in untreated control or RECQ1-depleted cells. Remarkably, following H2O2 treatment, RECQ1-depleted HeLa cells displayed significantly elevated PAR levels as compared to control cells, indicating increased activation of PARP in response to H2O2 (Fig. 3A, lane 6). Total PAR levels decreased with the recovery from H2O2 treatment similarly in RECQ1-depleted cells (Fig. 3A, lane 8). Quantification of total PAR signal on Western blots showed that at 0 min after removal from H2O2, RECQ1-depleted cells displayed approximately 2.7 (± 0.29) fold higher total PAR whereas control siRNA transfected cells displayed an average 1.6 (± 0.08) fold increase in total PAR signal as compared to PAR signal observed in respective untreated conditions.
Figure 3. RECQ1-deficient cells exhibit increased activation of PARP upon H2O2 exposure.
36 h after transfection with control, RECQ1, or WRN siRNA, HeLa cells were treated with 200 µM H2O2 for 20 min and allowed to recover in complete medium. Lysates were prepared at indicated time points following treatment and PARP activation was determined by the intensity of poly-ADP(ribose) (PAR) signal using Western blot analyses (both short and long exposure of Western blot are shown). Depletion of RECQ1 or WRN in the lysates was verified by Western blot and is indicated. GAPDH signal in the same blot serves as loading control. A. RECQ1 depleted cells show increased PAR formation upon H2O2 treatment as compared to control cells (lane 2 vs. lane 6). Control and RECQ1-depleted cells show comparable level of PAR formation in untreated condition. Total PAR signal on Western blot was quantified using ImageJ and signal intensity relative to untreated is indicated in each case. Rel. Sig., relative signal intensity for PAR. B. Depletion of RECQ1 or WRN leads to distinct cellular response with respect to PARP activation upon H2O2 exposure. PAR signal was detected in lysates prepared from control, RECQ1 or WRN-depleted cells that were either untreated or exposed to 2 mM hydroxyurea (HU) for 16h or 200 µM H2O2 for 20 min. C. PARP activity was measured by a quantitative colorimetric assay using lysates from control, RECQ1 or WRN-depleted cells that were either untreated or treated with 200 µM H2O2 for 20 min. PARP activity was measured by taking OD at 450 nm. Values represent mean of three independent experiments with SD indicated by error bars. Ctl, control.
A functional relationship between PARP-1 activation and WRN has been previously reported [36–37, 39]; therefore, we evaluated PAR formation in WRN-depleted cells. Increased PAR levels were observed in untreated cells that were transfected with WRN siRNA as compared to those with control siRNA (Supplementary Fig. 4; and Fig. 3B, lanes 1 and 3); this is in agreement with previous reports suggesting that the absence of WRN results in the generation of DNA breaks and other structures leading to PARP activation in an undamaged state [36–37, 39]. Additionally, PARP is activated upon stalled replication forks induced by HU treatment [40]. We therefore compared the level of PARP activation in control, RECQ1 or WRN-depleted cells that were untreated, treated with HU or H2O2 (Fig. 3B). In untreated condition, approximately 2 (± 0.11)-fold greater signal for total PAR was obtained in WRN-depleted cells as compared to control cells; whereas the total PAR in RECQ1-depleted cells was similar (1.13 (± 0.08)-fold) to control cells. Exposure to HU resulted in PARP activation in each case; control, RECQ1 or WRN-depleted cells displayed 1.4, 1.9 and 1.7 fold higher PAR signal when compared to untreated, respectively suggesting increased PARP activation in RECQ1 or WRN-depleted cells (Fig. 3B, lanes 4, 5, and 6). Exposure to H2O2 induced a much greater PAR signal in RECQ1-depleted cells as compared to control or WRN-depleted cells (Fig. 3B, lanes 7, 8, and 9). In contrast to RECQ1-depleted cells, and consistent with a previous observation in WS cells [41], further PAR formation in WRN-depleted cells upon H2O2 treatment was less than that in control cells. Regardless of the differences in the level of RECQ1 and WRN knockdown, qualitatively similar results were obtained reproducibly.
To ascertain whether increased PAR formation is associated with increased PARP activity, we examined in vitro PARP activity in extracts of control, RECQ1 or WRN-depleted cells (Fig. 3C). As determined by this quantitative assay, H2O2 treatment resulted in about 1.6-fold increase in PARP activity in RECQ1-depleted cells compared to control cells (p < 0.05; Fig. 3C). Untreated WRN-depleted cells exhibited higher PARP activity than control or RECQ1-depleted cells (p < 0.05), but no significant further increase was observed in WRN-depleted cells following H2O2 treatment (p = 0.067; Fig. 3C). These results demonstrate that RECQ1-deficient cells exhibit increased activation of PARP in a specific response to H2O2-induced DNA damage whereas WRN deficiency leads to a constitutive PARP activation.
3.3. RECQ1-depletion does not affect homologous recombination repair of I-SceI-induced double strand breaks or results in sensitivity to PARP inhibitor
Oxidative stress by H2O2 treatment has not been shown to directly affect DSB-induced homologous recombination (HR); however, defects in HR are exacerbated in the presence of oxidative stress [42]. Additionally, inhibition of PARP-1 is synthetic lethal with defects in HR [43–44]. Both WRN and BLM-depleted cells demonstrated decreased efficiency in HR, which was linked with constitutively hyperactivated PARP and correlated with their significantly decreased survival after treatment with PARP inhibitor [39]. In contrast, our results show that depletion of RECQ1 does not result in constitutive activation of PARP in untreated cells. Given the contribution of WRN and BLM in repairing DSBs by HR and the biochemical activities of RECQ1, we wanted to determine whether RECQ1 directly affects DSB-induced HR frequencies.
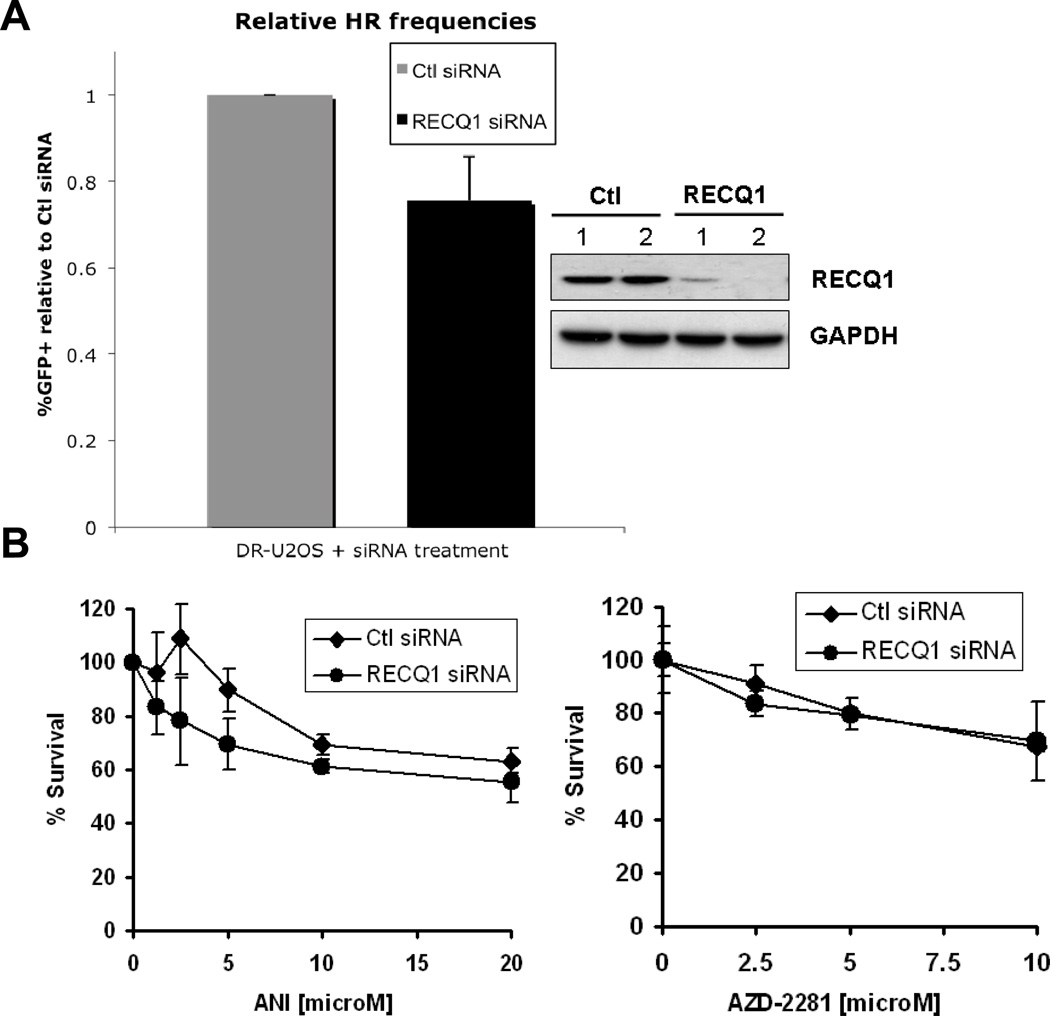
We measured homology-directed repair of I-SceI-induced DSBs in RECQ1-depleted human U2OS cells containing an integrated DR-GFP reporter substrate (DR-U2OS) [25]. In these experiments, only a minor reduction of HR was detected in RECQ1-depleted cells, which was not statistically significant compared to control siRNA-treated cells (p = 0.09; Fig. 4A). In contrast, BLM-depletion resulted in decreased HR compared to the control siRNA transfected cells (p < 0.001; Supplementary Fig. 5A), consistent with previous results [39, 45]. In agreement with their relative HR-deficiency, BLM-depleted U2OS cells displayed increased sensitivity to a PARP inhibitor ANI over the entire dose range that was tested (p < 0.01; Supplementary Fig. 5B) while RECQ1-depleted U2OS cells were not sensitive to ANI (p = 0.102; Fig. 4B). Control or RECQ1-depleted cells were similarly sensitive to AZD2281 (Olaparib) (Fig. 4B), another potent PARP inhibitor [46]. Our results demonstrate that while RecQ helicases such as BLM plays a prominent role in HR including repair of I-SceI-induced DSB, RECQ1 may play a role in HR mediated repair of other more physiologically relevant DNA lesions such as those induced by oxidative stress.
Figure 4. RECQ1 does not play a direct role in homologous recombination repair of DNA double-strand breaks.
A. DR-U2OS cells were transfected with control or RECQ1 siRNA twice. Upon second siRNA transfection (at I-SceI TF), cells were either harvested for Western blot analysis or additionally transfected with pCAGGS (empty vector) or pCABSce (I-SceI expression vector). 48 h after second siRNA transfection (at FACs), cells were harvested for Western blot analysis or analyzed for GFP fluorescence. Restoration of a functional GFP gene through complete homologous recombination (HR) was used to determine HR proficiency. %GFP positive cells were quantitated and relative frequencies were determined by normalizing to control siRNA transfected cells. Mean of relative %GFP positive cells and S.E.M. from four independent experiments are shown. Depletion of RECQ1 at I-SceI TF and at FACs, indicated by 1 and 2, respectively, was verified by Western blotting and is shown. GAPDH is used as loading control. B. RECQ1-depleted cells are not sensitive to PARP inhibitors. Cells transfected with control or RECQ1 siRNA were subjected to continuous exposure to increasing dose of ANI or AZD2281 in complete medium and their survival was measured 72 h later by MTS assay. Percentage of control growth was plotted for each data point, representing the mean ± SD of three independent experiments performed in quadruplicate. Ctl, control.
3.4. PARP-1-dependent repair of H2O2-induced DNA damage in RECQ1-depleted cells
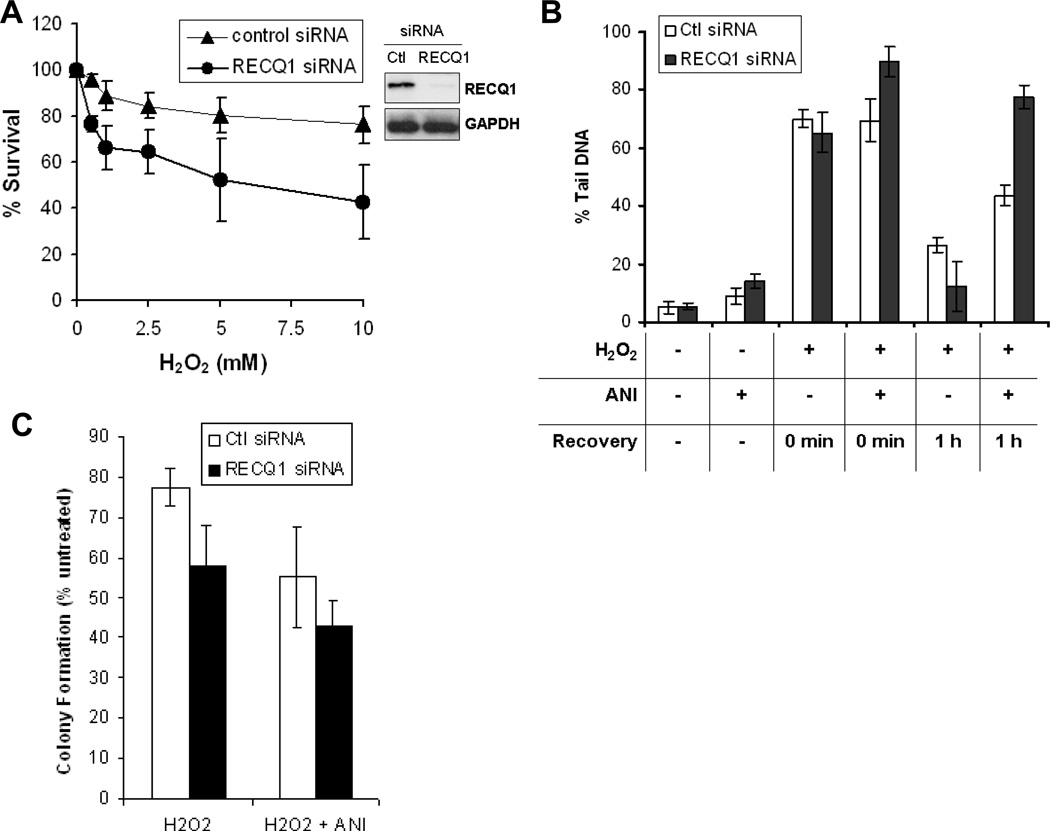
PARP-1 plays important roles in repair pathways for DNA damage caused by oxidizing agents [22]. Thus we sought to determine the sensitivity to oxidative DNA damage and repair efficiency of RECQ1-depleted cells (Fig. 5). Control or RECQ1 siRNA-transfected cells were exposed to increasing concentrations of H2O2 for 10 min at 4°C and their survival was measured 72 h later by MTS assay. In multiple experiments, RECQ1-depleted cells exhibited moderately increased sensitivity to H2O2 when compared to control siRNA-transfected cells (Fig. 5A).
Figure 5. RECQ1-depleted cells repair H2O2-induced DNA damage in PARP activity-dependent manner.
A. RECQ1-depleted cells are sensitive to H2O2 exposure. Cells transfected with control or RECQ1 siRNA were exposed in quadruplicate to increasing doses of H2O2 for 10 min followed by growth in complete medium and their survival was measured 72 h later by MTS assay. Percentage of control growth was plotted for each data point, representing the mean ± SD of three independent experiments. Depletion of RECQ1 is shown by Western blot. GAPDH serves as loading control. B. Total DNA strand breaks in control or RECQ1-depleted cells. Alkaline Comet Assay was used to measure the total DNA strand breaks with or without prior exposure to PARP inhibitor ANI (100 µM, 16 h) in control or RECQ1-depleted cells that were either untreated or allowed to recover for indicated time following treatment with H2O2 (10 µM, 5 min). The total DNA strand breaks were quantitated as % tail DNA representing the mean of three independent experiments with SD indicated by error bars. At least 100 cells were analyzed in each experiment. C. Colony formation of H2O2-treated control or RECQ1-depleted cells grown in the presence or absence of ANI. Cells were incubated with or without ANI (10 µM, 16 h) before treatment with H2O2 (1 mM, 10 min) or left untreated. Following removal of H2O2, cells were grown in the presence or absence of ANI (10 µM) in complete medium and colonies were stained and counted after 14 days. As reported previously, depletion of RECQ1 resulted in reduced colony formation [3]. Results are expressed as the relative percentage of colonies as compared to untreated controls for each siRNA following adjustment for plating efficiency using untreated plates and are the means of duplicate experiments, each performed in triplicate. Ctl, control.
We next measured the total amount of DNA strand breaks in untreated and H2O2-treated cells using an alkaline comet assay (Fig. 5B). No remarkable difference in total DNA damage was observed between control and RECQ1-depeleted cells that were untreated or treated with H2O2 (Fig. 5B). Given the physical interaction between RECQ1 and PARP-1 and increased H2O2-induced PARP activation in RECQ1-depleted cells, we examined whether PARP-1 activity is important for the repair of H2O2-induced DNA damage in RECQ1-depleted cells. Control or RECQ1-depleted cells were pretreated with ANI to specifically inhibit PARP activity, and total strand breaks were analyzed under untreated conditions or after treatment with H2O2. ANI treatment did not alter the overall level of strand breaks in control cells that were untreated or H2O2-treated. Inhibition of PARP activity by ANI did not affect spontaneous damage (p = 0.072) in untreated RECQ1-depleted cells; however, presence of ANI resulted in increased strands breaks in RECQ1-depleted cells upon H2O2 exposure (p < 0.05; Fig. 5B). This suggests that H2O2-induced DNA strand breaks are partly repaired in PARP activity-dependent manner in HeLa cells depleted of RECQ1. We compared the total DNA damage in control or RECQ1-depleted cells following 1 h recovery from H2O2 treatment. In the absence of ANI, the DNA damage in control or RECQ1-depleted cells was similarly reduced at 1 h following recovery from H2O2 (Fig. 5B), similar to repair kinetics observed in HeLa cells [47].
PARP inhibition has been shown to reduce the rate of strand break repair [48–49]. Inhibition of PARP activity by ANI resulted in reduced repair in both control and RECQ1-depleted cells (Fig. 5B); at 1 h following recovery from H2O2 treatment, the total percentage of damage was greater in RECQ1-depleted cells as compared to the control cells (p < 0.05). Thus, RECQ1 may play a role in repair of H2O2-induced DNA damage left unrepaired when PARP is inhibited. In colony formation assay, RECQ1-depleted cells showed reduced survival as compared to control siRNA transfected cells following H2O2 treatment (p < 0.05) (Fig. 5C). Colony formation of H2O2-treated control or RECQ1-depleted cells in the continued presence of ANI revealed that PARP inhibition reduced cell survival in each case (p > 0.05) and the sensitivity of RECQ1-depleted cells to H2O2 was not exacerbated in the presence of ANI (Fig. 5C), suggesting alternative processing of DNA damage intermediates in combined deficiency of RECQ1 and PARP activity.
3.5. RECQ1 is rapidly recruited to chromatin following induction of oxidative stress
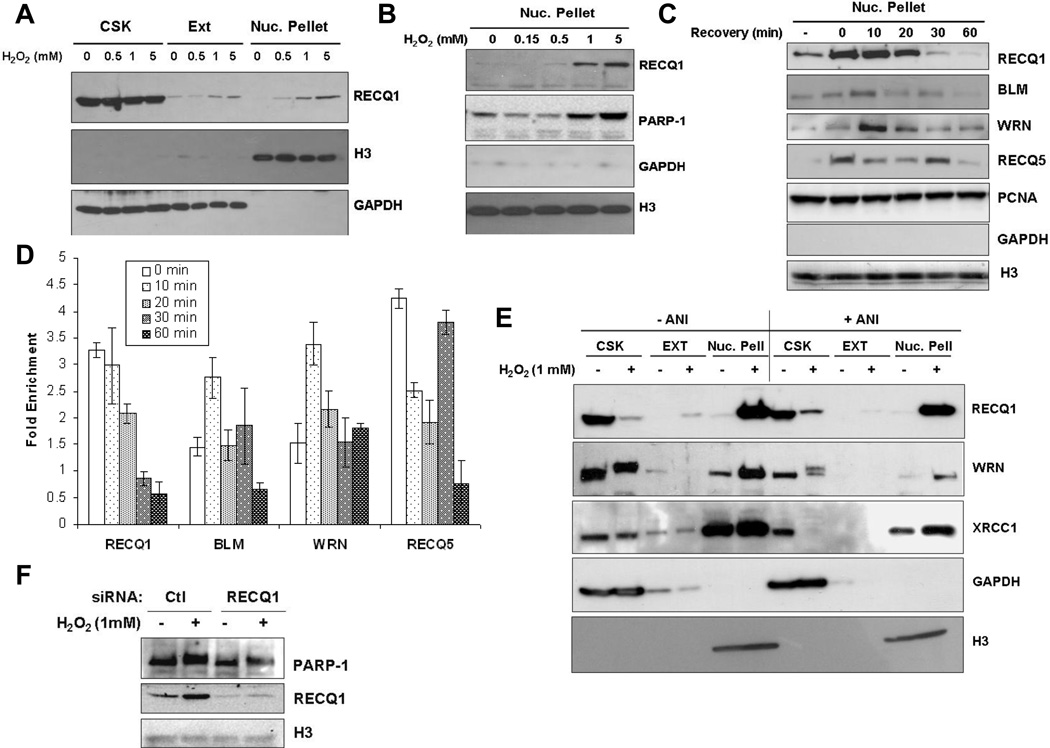
To explore the roles of RECQ1 in cellular mechanisms required for the repair of oxidative DNA damage, we evaluated the distribution of endogenous RECQ1 in various subcellular compartments following treatment with H2O2 (Fig. 6) using a previously reported sequential fractionation [27]. HeLa cells either untreated or treated for 20 min with increasing concentrations of H2O2 (0–5 mM) were subjected to successive detergent extraction to isolate cytoskeleton, cytoplasmic and soluble nuclear proteins, and insoluble nuclear pellet containing proteins that were tightly bound to chromatin and/or nuclear matrix. RECQ1 protein in untreated cells predominantly fractionated with soluble proteins, with only a very minor fraction associated with chromatin in the insoluble nuclear pellet (Fig. 6A). Following H2O2 treatment, a greater amount of RECQ1 was present in the insoluble fraction that also contained histones (Fig. 6A). Moreover, the intensity of RECQ1 signal in the insoluble nuclear pellet correlated with the dose of H2O2 treatment (Fig. 6A and 6B). Consistent with a previous observation [50], H2O2 treatment also enriched PARP-1 in chromatin (Fig. 6B). These results indicate that following H2O2 exposure, a portion of RECQ1 is recruited to damaged chromatin, perhaps to participate in DNA repair.
Figure 6. RECQ1 associates with damaged chromatin in PARP activity-independent manner.
Sequential extraction of HeLa cells without and with H2O2 treatment. CSK and EXT fractions contain cytoplasmic, cytoskeletal, and soluble nuclear proteins. Insoluble nuclear pellet fractions contain proteins that are tightly bound to chromatin and/or the nuclear matrix. GAPDH and histone H3 serve as loading controls and markers for the fractions containing soluble proteins and chromatin, respectively. Results were documented using different exposures of the Western blots. A. RECQ1 associates with chromatin in a H2O2 dose-dependent manner. HeLa cells were treated with the indicated concentrations of H2O2 for 20 min at 37°C followed by sequential fractionation. B. H2O2 dose-dependent chromatin association of RECQ1 is accompanied by enrichment of PARP-1 on damaged chromatin. Nuclear pellet fractions containing chromatin bound proteins are shown. C. Rapid and reversible association of RECQ1 with damaged chromatin. HeLa cells were treated with 1 mM H2O2 for 20 min at 37° followed by incubation in H2O2-free medium for recovery, and subjected to sequential fractionation at indicated time points. Nuclear pellet fractions show relocalization of RECQ1, WRN, BLM and RecQ5β to damaged chromatin at different time during recovery. Levels of PCNA remain unchanged and serve as loading control. Untr., Untreated. D. Quantitation of chromatin enrichment of RecQ proteins at different time points following recovery from H2O2 (1 mM, 20 min) treatment. Western blots were quantitated using ImageJ. Signals were normalized using Histone (H3) and protein signal in the untreated was used to calculate fold enrichment following H2O2 treatment. Mean of relative fold enrichment and S.E.M. from three independent experiments are shown. E. H2O2-induced chromatin binding of RECQ1 and WRN is independent of PARP activity. HeLa cells grown in the presence or absence of ANI (100 µM) for 24 h, were mock-treated or treated with 1 mM H2O2 for 20 min. Cells were then sequentially fractionated and proteins from each fraction analyzed by Western blotting. The apparent decrease of RECQ1, WRN and XRCC1 proteins in the CSK fraction reflects the decrease of soluble nuclear proteins, which now associate tightly with the chromatin-enriched nuclear pellet. F. RECQ1-depleted cells show H2O2 -induced chromatin enrichment of PARP-1. PARP-1 was detected by Western blot in the chromatin fractions prepared from control or RECQ1-depleted cells that were either untreated or treated with 1 mM H2O2 for 20 min.
As other human RecQ proteins have been implicated in the repair of oxidative DNA damage [51–53], we assessed their chromatin association following recovery from H2O2 treatment (Fig. 6C). Similar to RECQ1, H2O2 exposure also induced enrichment of WRN, BLM and RecQ5β on chromatin (Fig. 6C and 6D). Poor specificity of the commercially available antibodies in our hands prevented us from including RECQL4 in these analyses (data not shown). The level of PCNA, a protein that is constitutively present on chromatin, showed no change after H2O2 treatment. Maximum RECQ1 recruitment to chromatin was first obtained immediately after treatment (0 min) and stayed at a similar level until 10 min after recovery; chromatin-associated RECQ1 then progressively decreased over time following recovery from H2O2 (Fig. 6C and 6D). At 30 and 60 min after recovery from H2O2, the RECQ1 level in chromatin fractions were lower than untreated cells (Fig. 6C and 6D); suggesting reorganization of cellular pool of RECQ1 following DNA damage response. In contrast, maximum recruitment of WRN and BLM were reproducibly observed at 10 min after recovery. We note that the recruitment of RECQ5β was most robust among these RecQ helicases; RECQ5β accumulated immediately after treatment (0 min), decreased thereafter, but the signal spiked again at 30 min suggesting it’s participation in early as well as late stages of repair (Fig. 6C and 6D). Collective results implicate these RecQ helicases in repair of H2O2-induced DNA damage. Our observation that RECQ1 is rapidly and reversibly recruited to the oxidatively-damaged chromatin suggests an early role of RECQ1 in recognition and/or repair of H2O2-induced DNA damage.
3.6. H2O2-induced chromatin association of RECQ1 is independent of PARP activity
Given the role of PARP-1 in recruiting DNA repair proteins to the site of damage [31, 47], we examined whether H2O2-induced chromatin recruitment of RECQ1 is also dependent on PARP activity. We treated HeLa cells with PARP inhibitor ANI prior to H2O2 exposure and evaluated the amount of chromatin associated RECQ1 following H2O2 exposure. ANI treatment did not alter the total cellular RECQ1 protein level (Supplementary Fig. 6). Using the sequential extraction protocol as described above, quantitatively comparable enrichment of RECQ1 was obtained in chromatin fractions of H2O2-exposed cells in the presence or absence of ANI (Fig. 6E). The relative enrichment of WRN following H2O2 was similar in the presence or absence of ANI although PARP inhibition decreased the amount of WRN with and without H2O2 induced stress (Fig. 6E). Nonetheless, PARP inhibition did not impede H2O2-induced chromatin association of WRN or RECQ1. It has been suggested that PAR synthesis is required for the recruitment of XRCC1 to sites of oxidative DNA damage [31]; however, we found H2O2 -dependent increase in chromatin accumulation of XRCC1 in the presence of ANI in HeLa cells (Fig. 6E). This is consistent with the observation that a deficiency of PARP-1 did not affect chromatin recruitment of XRCC1 and several other BER proteins [47]. Analyses of subcellular fractions from control or RECQ1-depleted cells demonstrated that PARP-1 is similarly enriched in the chromatin containing fractions following H2O2 treatment (Fig. 6F).
4. Discussion
Despite the critical importance of RecQ helicases in genome maintenance, biological functions of the most abundant human RecQ protein have remained unclear. Our results demonstrate that RECQ1 exhibits a rapid response to oxidative DNA damage through its interaction with chromatin. Both RECQ1 and WRN are recruited to damaged chromatin; however, depletion of RECQ1, but not of WRN, results in increased activation of PARP when cells are exposed to H2O2. In contrast to WRN-deficient cells which are resistant to H2O2 [41], RECQ1-depleted cells are moderately sensitive to H2O2 and show dependence upon PARP for the repair of H2O2-induced DNA damage. Similar to WRN, RECQ1 associates with PARP-1 in vivo; however, distinct domains within these two RecQ proteins might mediate physical interaction with PARP-1 in vitro. Collectively, these results identify novel functions of RECQ1 and further emphasize functional specialization among human RecQ homologs.
Human RecQ proteins localize to DNA repair foci after a variety of DNA damage [54–62]; however, there are limited data regarding their localization in response to oxidative stress. Endogenous RECQL4 was reported to accumulate in the nucleoli of cells in response to oxidative stress [35], whereas another study demonstrated that BLM and RECQL4 form nuclear foci following H2O2 treatment [63]. This suggests differences in the subnuclear distribution of human RecQ helicases before and after oxidative damage. We discovered that RECQ1 rapidly accumulates to chromatin in response to oxidative damage; moreover, WRN, BLM and RECQ5β are also enriched in chromatin containing fractions although the extent and timing of their recruitment to damaged chromatin varied. Oxidative damage-dependent chromatin association of these RecQ proteins is independent of ATM kinase (data not shown) and PARP, which are important sensors of oxidative stress in human cells [64]. This observation raises the question of what could be the signal for triggering the relocalization of RECQ1 and other RecQ proteins after oxidative damage. A possible signal could be a post-translational modification of RECQ1 that allows its increased interaction with chromatin. Indeed chromatin-associated RECQ1 was found to be phosphorylated in the cells that had been exposed to IR or UV [3]. Preliminary analysis of the RECQ1 in oxidatively stressed cells failed to detect any phosphorylation associated with the chromatin fractions (S.S., unpublished results); however, other modifications such as acetylation or oxidation cannot be excluded. Alternatively, modification of RECQ1-interacting proteins involved in assembly of repair complexes could also lead to its recruitment to the damaged chromatin in response to oxidative stress.
After treatment with H2O2, HeLa cells repair most strand breaks within 1 h [65]. Our results suggest that when cells are exposed to H2O2, RECQ1 is among the first RecQ proteins to arrive on chromatin; it remains associated for the period of time required for the repair of strand breaks and then diffuses away. RECQ1 and its protein partners may be part of the DNA damage response by localizing to sites of oxidative lesions where RECQ1 utilizes its catalytic activities to process genomic DNA structures. Remarkably, RECQ1 catalyzes unwinding of duplex DNA containing oxidative base lesions [66]. Despite some overlap, the observed differences in the recruitment kinetics of RecQ proteins might reflect their unique requirement at distinct steps of damage response. It is possible that the activities of certain RecQ proteins are assigned to dedicated pathways or subpathways of oxidative DNA damage repair [67–68].
Interaction with PARP-1 also supports a putative role of RECQ1 in oxidative DNA damage response. Our data indicates that RECQ1 binding to PARP-1 involves its helicase domain and C-terminus. WRN and RECQL4 are the other known PARP-1 interacting RecQ proteins in humans [35–36]. In addition to a centrally located helicase domain, many RecQ helicases contain additional conserved RQC and HRDC (helicase and RNaseD C-terminal) domains which are implicated in protein interactions and DNA binding [1]. Furthermore, RecQ helicases have N- and C-terminal extensions that are postulated to lend unique functional characteristics to each helicase [1]. PARP-1 was reported to be major interacting protein with the WRN RQC domain [36], however, further analyses identified additional PARP-1 binding sites spanning the helicase and HRDC domains and extending to the C-terminus of WRN [37] (Fig. 2A). RECQL4, which lacks RQC and HRDC domains, interacts with PARP-1 via its C-terminal portion [35]. RECQ1 contains a RQC domain but lacks HRDC domain [69]; nonetheless, the last 14 amino acids of the RECQ1 C-terminus (amino acid residues 592–649) which mediates in vitro interaction with PARP-1 (this study) contain a motif that is partly homologous to WRN HRDC domain (Supplementary Fig. 7). The C-terminus of RECQ1 is likely to be critical for mediating protein-protein interactions as well as regulating sub-cellular localization through its nuclear localization signal motif [28].
Human RecQ helicases have an intriguing ability to unwind duplex DNA as well as mediate strand annealing of complementary ssDNA molecules in vitro [19]. Biochemical activities of RECQ1 are modulated by RPA and ATP [11]. DNA-dependent ATP hydrolysis is essential for the helicase activity of RECQ1 [14] whereas ATP binding to RECQ1 inhibits its strand annealing activity [16]. RPA physically interacts with RECQ1 (and other RecQ helicases) and stimulates its helicase activity [14] but strongly inhibits the ability of RECQ1 to efficiently mediate annealing between complementary ssDNA [16]. Conceivably, a mode of regulating the opposing activities of RECQ1 in biological setting may be through its interaction with cellular protein partners. We have demonstrated that RECQ1-PARP-1-RPA associate in a common protein complex. Interaction with PARP-1 may be important in providing a microenvironment to regulate dual activities of RECQ1 as appropriate for the cellular context since PARP-1 also modulates cellular ATP pools [70].
Recently it was reported that PARP is hyperactivated in HR-defective cells that are also sensitive to PARP inhibitors [39]. Notably, this also included BLM and WRN shRNA-depleted cells [39]. Our findings show that in contrast, depletion of RECQ1 by itself does not lead to PARP hyperactivation, and RECQ1-depleted cells are not sensitive to PARP inhibitor. We also determined that RECQ1-depleted cells are not compromised in their ability to repair I-SceI-induced DSBs by HR. However, given the biochemical and cellular characteristics of RECQ1 [11, 71], we cannot rule out a role of RECQ1 in suppressing recombination between diverged sequences, similar to mismatch repair-deficient cells [72]. Interestingly, PARP-1 was recently shown to enhance the mismatch-dependence of 5’-directed excision in human mismatch repair in vitro [30]. Although it is unknown whether RECQ1 and PARP-1 are involved in common HR regulatory mechanisms, it is relevant to note that in addition to interacting with each other, RECQ1 and PARP-1 are capable of interacting with the common components of mismatch repair system which are involved in the suppression of recombination between diverged sequences [24, 30].
A significant difference between RECQ1 and WRN-depleted cells was found to be in their ability to activate PARP in response to oxidative damage. We find that RECQ1-depleted cells depend on PARP activity for the repair of H2O2-induced DNA damage. This indicates that a fraction of H2O2-induced DNA lesions require RECQ1 for their repair. When RECQ1 is absent, these lesions are possibly repaired by an alternative mechanism that involves increased activation of PARP. Compensatory mechanisms of DNA damage processing likely contribute to survival in the combined absence of RECQ1 and PARP activity.
Elevated levels of oxidative DNA lesions have been detected in BS, WS and RTS patient cells [51–53], and human WRN, BLM, RECQL4 and RecQ5β have been shown to interact with and modulate activities of several proteins involved in BER [53, 73–75]. When exposed to oxidative damage by H2O2, WS cells fail to activate PARP-1 and bypass cell proliferation arrest [41]. One interpretation of these results may be that the elevated level of endogenous damage in WS cells prevents further induction of DNA damage response following subsequent H2O2 exposure. This, however, does not rule out contribution from additional helicases in BER as suggested by several studies [76–77]. It is also plausible that temporarily and biochemically distinct helicase functions are utilized for the repair of endogenous and exogenously induced damage. A potential candidate may be RECQ1, as recent work implicates RECQ1 in a variety of repair pathways [11, 71]. Data presented here associate RECQ1 with oxidative DNA damage response but a direct role of RECQ1 in specific repair pathway remains to be elucidated.
Supplementary Material
Highlights.
RECQ1 interacts with PARP-1 in vivo and in vitro
Hydrogen peroxide induces PARP activity independent chromatin binding of RECQ1
RECQ1-depleted cells overactivate PARP specifically in response to H2O2 treatment
RECQ1-depleted cells show modest HR deficiency and mild PARP inhibitor sensitivity
Acknowledgement
This work is supported by NIH/NIGMS grant 7SC1GM093999-02 (to SS) and NCI U54CA137788/U54CA132378 Pilot Project (to SS and MJ). We acknowledge technical help from Ms. Ying Guo. We thank Drs. Sergei Nekhai and Yuri Obukhov for their help with mass spectrometry analyses. Mass spectrometry was performed at Howard University College of Medicine RCMI Proteomic Core Facility supported by NIH/NCRR grant 2 G12RR003048 from the Research Centers in Minority Institutions (RCMI) Program (Division of Research Infrastructure, National Center for Research Resources, NIH).
Abbreviations
- SSB
single strand breaks
- DSB
double strand breaks
- BER
base excision repair
- HR
homologous recombination
- PAR
poly(ADP)ribose
- PARP
poly(ADP)ribose polymerase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interests
The authors declare that there are no conflicts of interest
Contributor Information
Sudha Sharma, Email: sudha.sharma@howard.edu.
Pornima Phatak, Email: pornima.phatak@howard.edu.
Alexei Stortchevoi, Email: alexei.stortchevoi@howard.edu.
Maria Jasin, Email: m-jasin@ski.mskcc.org.
Jeannine R. LaRocque, Email: jlk99@georgetown.edu.
References
- 1.Sharma S, Doherty KM, Brosh RM., Jr Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawabe T, Tsuyama N, Kitao S, Nishikawa K, Shimamoto A, Shiratori M, Matsumoto T, Anno K, Sato T, Mitsui Y, Seki M, Enomoto T, Goto M, Ellis NA, Ide T, Furuichi Y, Sugimoto M. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene. 2000;19:4764–4772. doi: 10.1038/sj.onc.1203841. [DOI] [PubMed] [Google Scholar]
- 3.Sharma S, Brosh RM., Jr Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS ONE. 2007;2:e1297. doi: 10.1371/journal.pone.0001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Futami K, Kumagai E, Makino H, Goto H, Takagi M, Shimamoto A, Furuichi Y. Induction of mitotic cell death in cancer cells by small interference RNA suppressing the expression of RecQL1 helicase. Cancer Sci. 2008;99:71–80. doi: 10.1111/j.1349-7006.2007.00647.x. [DOI] [PubMed] [Google Scholar]
- 5.Futami K, Kumagai E, Makino H, Sato A, Takagi M, Shimamoto A, Furuichi Y. Anticancer activity of RecQL1 helicase siRNA in mouse xenograft models. Cancer Sci. 2008;99:1227–1236. doi: 10.1111/j.1349-7006.2008.00794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li D, Frazier M, Evans DB, Hess KR, Crane CH, Jiao L, Abbruzzese JL. Single nucleotide polymorphisms of RecQ1, RAD54L, and ATM genes are associated with reduced survival of pancreatic cancer. J Clin Oncol. 2006;24:1720–1728. doi: 10.1200/JCO.2005.04.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D, Liu H, Jiao L, Chang DZ, Beinart G, Wolff RA, Evans DB, Hassan MM, Abbruzzese JL. Significant effect of homologous recombination DNA repair gene polymorphisms on pancreatic cancer survival. Cancer Res. 2006;66:3323–3330. doi: 10.1158/0008-5472.CAN-05-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puranam KL, Blackshear PJ. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- 9.Seki M, Miyazawa H, Tada S, Yanagisawa J, Yamaoka T, Hoshino S, Ozawa K, Eki T, Nogami M, Okumura K, et al. Molecular cloning of cDNA encoding human DNA helicase Q1 which has homology to Escherichia coli Rec Q helicase and localization of the gene at chromosome 12p12. Nucleic Acids Res. 1994;22:4566–4573. doi: 10.1093/nar/22.22.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puranam KL, Kennington E, Sait SN, Shows TB, Rochelle JM, Seldin MF, Blackshear PJ. Chromosomal localization of the gene encoding the human DNA helicase RECQL and its mouse homologue. Genomics. 1995;26:595–598. doi: 10.1016/0888-7543(95)80181-k. [DOI] [PubMed] [Google Scholar]
- 11.Sharma S, Brosh RM., Jr Unique and important consequences of RECQ1 deficiency in mammalian cells. Cell Cycle. 2008;7:989–1000. doi: 10.4161/cc.7.8.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeRoy G, Carroll R, Kyin S, Seki M, Cole MD. Identification of RecQL1 as a Holliday junction processing enzyme in human cell lines. Nucleic Acids Res. 2005;33:6251–6257. doi: 10.1093/nar/gki929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma S, Stumpo DJ, Balajee AS, Bock CB, Lansdorp PM, Brosh RM, Jr, Blackshear PJ. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol Cell Biol. 2007;27:1784–1794. doi: 10.1128/MCB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui S, Klima R, Ochem A, Arosio D, Falaschi A, Vindigni A. Characterization of the DNA-unwinding activity of human RECQ1, a helicase specifically stimulated by human replication protein A. J Biol Chem. 2003;278:1424–1432. doi: 10.1074/jbc.M209407200. [DOI] [PubMed] [Google Scholar]
- 15.Cui S, Arosio D, Doherty KM, Brosh RM, Jr, Falaschi A, Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004;32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM., Jr Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J Biol Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 17.Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, Hickson ID, Vindigni A. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem. 2008;283:17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 18.Bugreev DV, Brosh RM, Jr, Mazin AV. RECQ1 possesses DNA branch migration activity. J Biol Chem. 2008;283:20231–20242. doi: 10.1074/jbc.M801582200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brosh RM, Jr, Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35:7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riley PA. Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int J Radiat Biol. 1994;65:27–33. doi: 10.1080/09553009414550041. [DOI] [PubMed] [Google Scholar]
- 21.Lesko SA, Drocourt JL, Yang SU. Deoxyribonucleic acid-protein and deoxyribonucleic acid interstrand cross-links induced in isolated chromatin by hydrogen peroxide and ferrous ethylenediaminetetraacetate chelates. Biochemistry. 1982;21:5010–5015. doi: 10.1021/bi00263a026. [DOI] [PubMed] [Google Scholar]
- 22.Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001;477:7–21. doi: 10.1016/s0027-5107(01)00091-4. [DOI] [PubMed] [Google Scholar]
- 24.Doherty KM, Sharma S, Uzdilla LA, Wilson TM, Cui S, Vindigni A, Brosh RM., Jr RECQ1 helicase interacts with human mismatch repair factors that regulate genetic recombination. J Biol Chem. 2005;280:28085–28094. doi: 10.1074/jbc.M500265200. [DOI] [PubMed] [Google Scholar]
- 25.Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andegeko Y, Moyal L, Mittelman L, Tsarfaty I, Shiloh Y, Rotman G. Nuclear retention of ATM at sites of DNA double strand breaks. J Biol Chem. 2001;276:38224–38230. doi: 10.1074/jbc.M102986200. [DOI] [PubMed] [Google Scholar]
- 28.Seki T, Tada S, Katada T, Enomoto T. Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem Biophys Res Commun. 1997;234:48–53. doi: 10.1006/bbrc.1997.6535. [DOI] [PubMed] [Google Scholar]
- 29.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Kadyrov FA, Modrich P. PARP-1 enhances the mismatch-dependence of 5'-directed excision in human mismatch repair in vitro. DNA Repair (Amst) 2011;10:1145–1153. doi: 10.1016/j.dnarep.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaruga P, Dizdaroglu M. Repair of products of oxidative DNA base damage in human cells. Nucleic Acids Res. 1996;24:1389–1394. doi: 10.1093/nar/24.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meder VS, Boeglin M, de Murcia G, Schreiber V. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J Cell Sci. 2005;118:211–222. doi: 10.1242/jcs.01606. [DOI] [PubMed] [Google Scholar]
- 35.Woo LL, Futami K, Shimamoto A, Furuichi Y, Frank KM. The Rothmund-Thomson gene product RECQL4 localizes to the nucleolus in response to oxidative stress. Exp Cell Res. 2006;312:3443–3457. doi: 10.1016/j.yexcr.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 36.von Kobbe C, Harrigan JA, May A, Opresko PL, Dawut L, Cheng WH, Bohr VA. Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol Cell Biol. 2003;23:8601–8613. doi: 10.1128/MCB.23.23.8601-8613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Kobbe C, Harrigan JA, Schreiber V, Stiegler P, Piotrowski J, Dawut L, Bohr VA. Poly(ADP-ribose) polymerase 1 regulates both the exonuclease and helicase activities of the Werner syndrome protein. Nucleic Acids Res. 2004;32:4003–4014. doi: 10.1093/nar/gkh721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindahl T, Satoh MS, Poirier GG, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20:405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 39.Gottipati P, Vischioni B, Schultz N, Solomons J, Bryant HE, Djureinovic T, Issaeva N, Sleeth K, Sharma RA, Helleday T. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res. 2010;70:5389–5398. doi: 10.1158/0008-5472.CAN-09-4716. [DOI] [PubMed] [Google Scholar]
- 40.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Von Kobbe C, May A, Grandori C, Bohr VA. Werner syndrome cells escape hydrogen peroxide-induced cell proliferation arrest. FASEB J. 2004;18:1970–1972. doi: 10.1096/fj.04-1895fje. [DOI] [PubMed] [Google Scholar]
- 42.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 44.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 45.LaRocque JR, Jasin M. Mechanisms of recombination between diverged sequences in wild-type and BLM-deficient mouse and human cells. Mol Cell Biol. 2010;30:1887–1897. doi: 10.1128/MCB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Menear KA, Adcock C, Boulter R, Cockcroft XL, Copsey L, Cranston A, Dillon KJ, Drzewiecki J, Garman S, Gomez S, Javaid H, Kerrigan F, Knights C, Lau A, Loh VM, Jr, Matthews IT, Moore S, O'Connor MJ, Smith GC, Martin NM. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin- 1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J Med Chem. 2008;51:6581–6591. doi: 10.1021/jm8001263. [DOI] [PubMed] [Google Scholar]
- 47.Woodhouse BC, Dianova, Parsons JL, Dianov GL. Poly(ADP-ribose) polymerase-1 modulates DNA repair capacity and prevents formation of DNA double strand breaks. DNA Repair (Amst) 2008;7:932–940. doi: 10.1016/j.dnarep.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 48.Fisher AE, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strom CE, Johansson F, Uhlen M, Szigyarto CA, Erixon K, Helleday T. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 2011;39:3166–3175. doi: 10.1093/nar/gkq1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heale JT, Ball AR, Jr, Schmiesing JA, Kim J-S, Kong X, Zhou S, Hudson DF, Earnshaw WC, Yokomori K. Condensin I Interacts with the PARP-1-XRCC1 Complex and Functions in DNA Single-Strand Break Repair. Molecular Cell. 2006;21:837–848. doi: 10.1016/j.molcel.2006.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pagano G, Zatterale A, Degan P, d'Ischia M, Kelly FJ, Pallardo FV, Kodama S. Multiple involvement of oxidative stress in Werner syndrome phenotype. Biogerontology. 2005;6:233–243. doi: 10.1007/s10522-005-2624-1. [DOI] [PubMed] [Google Scholar]
- 52.Nicotera TM, Notaro J, Notaro S, Schumer J, Sandberg AA. Elevated superoxide dismutase in Bloom's syndrome: a genetic condition of oxidative stress. Cancer Res. 1989;49:5239–5243. [PubMed] [Google Scholar]
- 53.Schurman SH, Hedayati M, Wang Z, Singh DK, Speina E, Zhang Y, Becker K, Macris M, Sung P, Wilson DM, 3rd, Croteau DL, Bohr VA. Direct and indirect roles of RECQL4 in modulating base excision repair capacity. Hum Mol Genet. 2009;18:3470–3483. doi: 10.1093/hmg/ddp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J Cell Biol. 2001;153:367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gray MD, Wang L, Youssoufian H, Martin GM, Oshima J. Werner helicase is localized to transcriptionally active nucleoli of cycling cells. Exp Cell Res. 1998;242:487–494. doi: 10.1006/excr.1998.4124. [DOI] [PubMed] [Google Scholar]
- 56.Marciniak RA, Lombard DB, Johnson FB, Guarente L. Nucleolar localization of the Werner syndrome protein in human cells. Proc Natl Acad Sci U S A. 1998;95:6887–6892. doi: 10.1073/pnas.95.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakamoto S, Nishikawa K, Heo SJ, Goto M, Furuichi Y, Shimamoto A. Werner helicase relocates into nuclear foci in response to DNA damaging agents and co-localizes with RPA and Rad51. Genes Cells. 2001;6:421–430. doi: 10.1046/j.1365-2443.2001.00433.x. [DOI] [PubMed] [Google Scholar]
- 58.Lan L, Nakajima S, Komatsu K, Nussenzweig A, Shimamoto A, Oshima J, Yasui A. Accumulation of Werner protein at DNA double-strand breaks in human cells. J Cell Sci. 2005;118:4153–4162. doi: 10.1242/jcs.02544. [DOI] [PubMed] [Google Scholar]
- 59.Franchitto A, Pichierri P. Bloom's syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J Cell Biol. 2002;157:19–30. doi: 10.1083/jcb.200110009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng L, Kanagaraj R, Mihaljevic B, Schwendener S, Sartori AA, Gerrits B, Shevelev I, Janscak P. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 2009;37:2645–2657. doi: 10.1093/nar/gkp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fan W, Luo J. RecQ4 facilitates UV light-induced DNA damage repair through interaction with nucleotide excision repair factor xeroderma pigmentosum group A (XPA) J Biol Chem. 2008;283:29037–29044. doi: 10.1074/jbc.M801928200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- 63.Werner SR, Prahalad AK, Yang J, Hock JM. RECQL4-deficient cells are hypersensitive to oxidative stress/damage: Insights for osteosarcoma prevalence and heterogeneity in Rothmund-Thomson syndrome. Biochem Biophys Res Commun. 2006;345:403–409. doi: 10.1016/j.bbrc.2006.04.093. [DOI] [PubMed] [Google Scholar]
- 64.Barzilai A, Yamamoto K. DNA damage responses to oxidative stress. DNA Repair (Amst) 2004;3:1109–1115. doi: 10.1016/j.dnarep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 65.Collins AR, Ma AG, Duthie SJ. The kinetics of repair of oxidative DNA damage (strand breaks and oxidised pyrimidines) in human cells. Mutat Res. 1995;336:69–77. doi: 10.1016/0921-8777(94)00043-6. [DOI] [PubMed] [Google Scholar]
- 66.Suhasini AN, Sommers JA, Mason AC, Voloshin ON, Camerini-Otero RD, Wold MS, Brosh RM., Jr FANCJ helicase uniquely senses oxidative base damage in either strand of duplex DNA and is stimulated by RPA to unwind the damaged DNA substrate in a strand-specific manner. J Biol Chem. 2009 doi: 10.1074/jbc.M109.012229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Svilar D, Goellner EM, Almeida KH, Sobol RW. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid Redox Signal. 2011;14:2491–2507. doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Asagoshi K, Tano K, Chastain PD, 2nd, Adachi N, Sonoda E, Kikuchi K, Koyama H, Nagata K, Kaufman DG, Takeda S, Wilson SH, Watanabe M, Swenberg JA, Nakamura J. FEN1 functions in long patch base excision repair under conditions of oxidative stress in vertebrate cells. Mol Cancer Res. 2010;8:204–215. doi: 10.1158/1541-7786.MCR-09-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu L, Chan KL, Ralf C, Bernstein DA, Garcia PL, Bohr VA, Vindigni A, Janscak P, Keck JL, Hickson ID. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin DS, Bertino JR, Koutcher JA. ATP depletion + pyrimidine depletion can markedly enhance cancer therapy: fresh insight for a new approach. Cancer Res. 2000;60:6776–6783. [PubMed] [Google Scholar]
- 71.Wu Y, Brosh RM., Jr Distinct roles of RECQ1 in the maintenance of genomic stability. DNA Repair (Amst) 2010;9:315–324. doi: 10.1016/j.dnarep.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elliott B, Jasin M. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol Cell Biol. 2001;21:2671–2682. doi: 10.1128/MCB.21.8.2671-2682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rossi ML, Ghosh AK, Bohr VA. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst) 2010;9:331–344. doi: 10.1016/j.dnarep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Speina E, Dawut L, Hedayati M, Wang Z, May A, Schwendener S, Janscak P, Croteau DL, Bohr VA. Human RECQL5beta stimulates flap endonuclease 1. Nucleic Acids Res. 2010;38:2904–2916. doi: 10.1093/nar/gkp1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gupta PK, Sirover MA. Altered temporal expression of DNA repair in hypermutable Bloom's syndrome cells. Proc Natl Acad Sci U S A. 1984;81:757–761. doi: 10.1073/pnas.81.3.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tuo J, Muftuoglu M, Chen C, Jaruga P, Selzer RR, Brosh RM, Jr, Rodriguez H, Dizdaroglu M, Bohr VA. The Cockayne Syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J Biol Chem. 2001;276:45772–45779. doi: 10.1074/jbc.M107888200. [DOI] [PubMed] [Google Scholar]
- 77.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, Shen B. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell. 2008;32:325–336. doi: 10.1016/j.molcel.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.