Abstract
Fully automated classification algorithms have been successfully applied to diagnose a wide range of neurological and psychiatric diseases. They are sufficiently robust to handle data from different scanners for many applications and in specific cases outperform radiologists. This article provides an overview of current applications taking structural imaging in Alzheimer's Disease and schizophrenia as well as functional imaging to diagnose depression as examples. In this context, we also report studies aiming to predict the future course of the disease and the response to treatment for the individual. This has obvious clinical relevance but is also important for the design of treatment studies that may aim to include a cohort with a predicted fast disease progression to be more sensitive to detect treatment effects.
In the second part, we present our own opinions on i) the role these classification methods can play in the clinical setting; ii) where their limitations are at the moment and iii) how those can be overcome. Specifically, we discuss strategies to deal with disease heterogeneity, diagnostic uncertainties, a probabilistic framework for classification and multi-class classification approaches.
Keywords: Automated diagnosing, MRI, SVM, Dementia, Depression, Schizophrenia
2 Introduction
Multivariate pattern recognition methods are becoming increasingly popular in the field of cognitive neuroscience, specifically their application for the interpretation of brain images. They are hotly debated in the context of mind reading, lie detection and free will (Aharoni et al., 2008; Bles and Haynes, 2008; Soon et al., 2008). In that context, recognition models aim to decode the cognitive states from functional magnetic resonance imaging (fMRI) data (Friston et al., 2008). The second major application has been to provide clinical diagnosis and prognosis in patients with diseases such as Alzheimer's disease (AD). While cognitive scientists are mostly interested in relevant brain regions and potentially their local architecture; high diagnostic accuracy is of greatest importance when using these methods for clinical purposes. These classification methods also rely on recognition models that have been defined by training data. The critical task is the correct labelling of new data while inference on the model parameters is of a lesser interest (Friston et al., 2008).
Recent developments with a clinical context include the application to structural neuroimaging data for the purpose of diagnosing individual patients (Davatzikos et al., 2008a; Davatzikos et al., 2008b; Davatzikos et al., 2009; Duchesnay et al., 2007; Fan et al., 2006; Fan et al., 2007; Klöppel et al., 2008b; Klöppel et al., 2009a; Koutsouleris et al., 2009; Lerch et al., 2008; Teipel et al., 2007a; Teipel et al., 2007b; Vemuri et al., 2008a; Vemuri et al., 2008b). Classifiers are sufficiently sensitive to separate patients with AD or mild cognitive impairment (MCI) (Petersen et al., 2001) from cognitively normal (Davatzikos et al., 2008). In other applications, they identified those cognitively normal subjects who will convert to MCI in the future (Davatzikos et al., 2009). Although most applications have been applied to T1-weighted data, these methods perform well on other imaging modalities such as Positron emission tomography (PET) (Chen et al., 2011; Dukart et al., 2011; Habeck et al., 2008; Walhovd et al., 2010; Zhang et al., 2011) and functional MRI (fMRI) (Costafreda et al., 2009b; Fu et al., 2008; Hahn et al.; Marquand et al., 2008; Nouretdinov et al.). Many of these studies provide evidence that classifiers can be applied clinically to predict the course of disease in individuals and possibly even the combination of symptoms in the individual patient (Klöppel, 2009). This information is not only highly relevant to patient management but also very important for designing treatment trials. Pre-selecting subjects with a predicted rapid decline will allow for shorter and less expensive trials as treatment related changes can be identified more easily.
Since there is a number of review articles on pattern recognition methods (Muller et al., 2001; Rätsch, 2004; Schölkopf and Smola, 2001; Shawe-Taylor and Cristianini, 2004; Vapnik, 1998) and several deal with the specific application to neuroimaging (Bles and Haynes, 2008; Lemm et al., 2011), the main focus of this review will be on the current clinical applications of pattern recognition methods and the steps necessary to integrate them in the clinical routine. To keep this review accessible to readers unfamiliar with classification methods in neuroscience, we begin with a brief outline of the main elements of the analysis pipeline and present an example of a pipeline to discriminate AD from healthy controls. Next, we discuss studies that relied on classification methods for diagnostic purposes and the diagnostic accuracy achievable. In the remainder of this text we discuss current challenges and how these could be overcome.
2.1 Basic elements of a classification pipeline
The analysis pipelines used for generating MRI-based recognition models for an individualised patient diagnosis vary greatly but often include the following four steps:
Training data set: The goal is to acquire a sufficient number of training data sets from individual subjects with well-characterised clinical properties (e.g. clinical diagnosis, pathological measures), which can be used as the gold standard for the classification problem at hand. Since we typically use supervised pattern recognition methods, training data along with the gold standard forms the library for learning disease-specific properties (i.e. patterns) to classify new incoming patients. The training data set has to be large enough to reliably express the disease effect against the “noise” of inter-subject variability.
Feature Extraction from Raw Data and Dimensionality Reduction: The input data (e.g. the anatomical or functional characteristics of the disease process) have to be useful for classification or in other words, they need to be meaningful in the context of the disease and comparable (stable) across subjects. The input data can be as coarse as the total intracranial volume (TIV) or as fine as the amount of grey matter in a very small anatomical region, i.e. a voxel. Since neuroimaging data can have 106 or more dimensions depending on resolution and scanned volume, often but not always the numbers of input measures are reduced using various dimensionality reduction methods. Irrespective of the specific type of algorithms, all dimensionality reduction methods aim at generating a compact set of discriminative “features” that can be used for training the classification model instead of the original input data. A popular dimensionality reduction method is principle component analysis (PCA), but many others exist (Guyon and Elisseeff, 2003). This approach is effective in improving signal-to-noise ratio as it extracts potentially relevant information by means of an alternative representation of the data that is based on the covariance matrix. However, PCA is usually applied to the entire brain and therefore it produces features that provide discriminative information only at the whole-brain level. Thus, one major drawback of this dimensionality reduction method is that it misses highly localized pathological information characteristic for certain neurodegenerative disorders (e.g. Huntington's Disease, etc.). A second limitation is that PCA is agnostic with respect to the classification problem at hand, meaning that the algorithm will produce features that e.g. are age-associated if age effects account for most of the variance in the data. In contrast, another strategy that has been employed to decrease dimensionality is the selection of a predefined set of features (e.g. anatomic regions) for classification using some prior knowledge, e.g. medial temporal lobe or hippocampus pathology in AD. Although very powerful, this approach requires an established and validated model describing the functional neuroanatomy of the underlying disease processes. This may be the case for certain neurological conditions (e.g. stroke, Alzheimer's or Huntington's Disease), but in psychiatric disorders, like schizophrenia and depression, the pathophysiological processes are still far less understood. Furthermore, the complex and often heterogeneous clinical phenotypes of these diseases suggest that the underlying neurobiological substrate is also equally complex (see e.g. reviews by Honea et al. (Honea et al., 2005) and Koolschijn et al. (Koolschijn et al., 2009)), potentially spanning multiple brain systems beyond predefined neuroanatomical boundaries. Therefore, the plan to overcome “the curse of dimensionality” by selecting a small set of brain regions may result in a significant loss of discriminative information. Taken together, these two different approaches illustrate that no universal strategy of feature selection might exist, but instead that the optimal approach has to be determined according to the very specific diagnostic / predictive task at hand. A third, recently proposed approach (Gilad-Bachrach et al., 2004; Navot et al., 2006) may provide an intermediate solution to the above-mentioned feature selection problem as it selects patterns of discriminative voxels by evaluating the geometric distance in feature space they induce between different groups in the training data. This margin-based feature selection framework may be capable of providing both global as well as local discriminative information, thus reducing the effect of noisy, unreliable or irrelevant voxels, while avoiding the drawback of the aforementioned dimensionality reduction strategies.
Model Training and Optimization: The classifier uses the training data and the known labels to learn a rule to separate the classes. In this step, a supervised algorithm is selected (e.g. linear discriminant analysis, support vector machine (SVM), neural networks, Gaussian processes etc) and the parameters of the model are optimized to maximally discriminate one group from another on the basis of the training data set. Cross-validation is a widely used approach to tune the parameters of the model. N-fold cross-validation involves randomly dividing the entire data set into N subgroups and then training the algorithm with specific parameters on N-1 subgroups and testing on the left-out subgroup. This process is repeated by leaving each of the sub-groups out one at a time and estimating the average error over all the runs. The model (or the parameters) that gives the best accuracy is picked as the final model. If the number of sub-groups is equal to the number of samples, it is called leave-one out cross-validation since each sample is left out once and used as the test sample. Another important aspect to consider during the model optimization is the operating point. By default, the operating point of any classification algorithm is set to maximize the accuracy but depending upon the problem can be set to maximize sensitivity or specificity. For clinical applications, the operating point can be set to give higher sensitivity if the algorithm results are used for screening a population or high specificity the results are used to identify candidates for high risk therapeutics.
Application to Test Data: The final step is to apply the learned rule to new data that have been pre-processed in the same way as the training data set. Testing the model on a new independent test data set that was not used for training is the best approach to test the generalisability and performance of the developed model. In cases where an independent test set is not available due to limited samples for testing and training one can uses the cross-validation framework described above to estimate the performance of the model
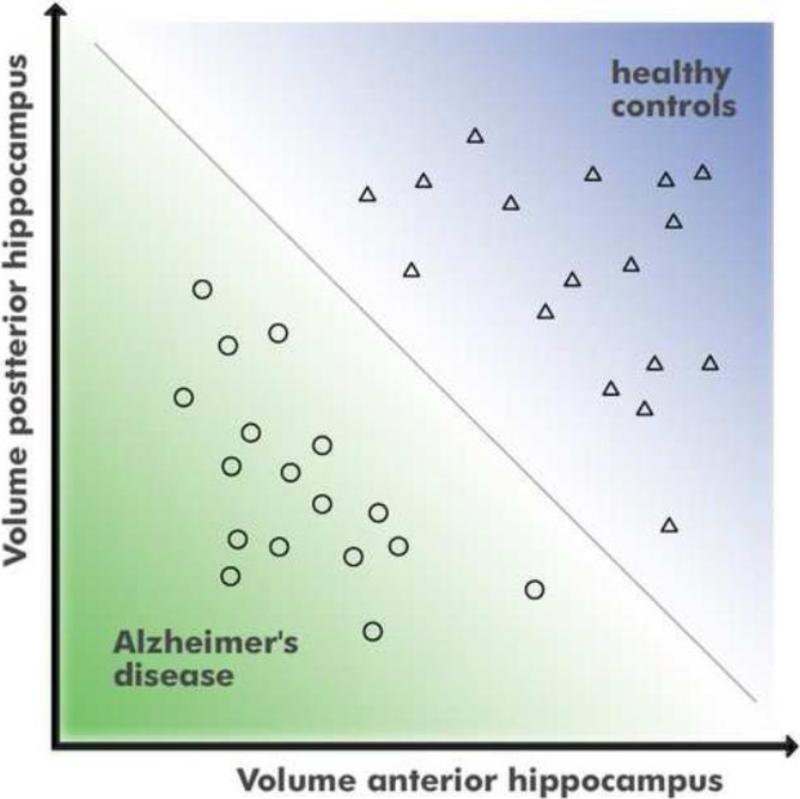
Figure 1 shows artificial data that can be separated into two groups without error by using information from two dimensions. A univariate classifier taking only one dimension (e.g. x-axis in Figure 1) into account could not accurately separate the classes. In an actual application the amount of GM at each anatomical position are the meaningful and comparable features and there could easily be several thousand dimensions instead of just two as shown in Figure 1.
Figure 1.
Concept of multivariate classification in two dimensions.
It is important to note that there are several parameters that can influence the performance of classifiers. Several studies have studied the effect of pre-processing on the classification performance (Cuingnet et al., 2010; Franke et al., 2010); compared different classification algorithms for structural (Chen and Herskovits, 2010; Plant et al., 2010) and functional (Sato et al., 2009) imaging data as well as compared the performance of anatomical versus statistical regions for separating two classes (Pelaez-Coca et al., 2011). It is unlikely that the same pre-processing pipeline and classification method performs best in all scenarios.
3 Current applications
We start by discussing structural magnetic resonance imaging (sMRI) and AD. This neurodegenerative disease has a clear pathological correlate related to loss of neurons and synapses particularly in the medial temporal lobe. Applications to psychiatric diseases are potentially more challenging since they present with more heterogeneous clinical phenotypes. To this end, we report sMRI-based classification studies in schizophrenia. We conclude this section by summarising fMRI studies useful to diagnose depression and report first evidence for a successful prediction of treatment response using classification methods.
3.1 Dementia
As listed in the introduction, many studies have applied pattern recognition methods to diagnose individual patients with dementia. With only two potential diagnoses available such as AD (the most common form of dementia) and cognitively normal subjects, classifiers have been shown to outperform radiologists (Klöppel et al., 2008a) and to capture the neurodegenerative pathology better than hippocampal volumes which have been traditionally used (Vemuri et al., 2008b). Since there are significant pathological changes before clinical symptoms appear in AD, predicting conversion from MCI to AD (i.e. early diagnosis of AD) is one of the key areas of biomarker research in this field. Visual qualitative estimates of medial temporal atrophy which are performed routinely, predict the progression of MCI to dementia with a sensitivity of 68% and specificity of 69% (DeCarli et al., 2007; Korf et al., 2004). This is outperformed by multivariate classification methods based on sMRI that have accuracy of 80 % (Sensitivity 67; Specificity 93 %) in identifying MCIs who later convert to AD (Teipel et al., 2007b). There are also recent studies showing that pattern recognition method based MRI scores - predict disease progression better when compared to CSF biomarkers (Davatzikos et al., 2010; Vemuri et al., 2009a) and closely correlate with cognitive performance in subjects (Stonnington et al., 2010; Vemuri et al., 2009b). While MRI-based methods might not show best performance for all applications, the above mentioned studies indicate that the relative ease of data acquisition and applying automated diagnosis methods and the non-invasive nature of MRI make it a useful diagnostic measure.
The need for fully automated diagnostic tools in AD has just increased significantly with the release of new diagnostic criteria for AD. Recently both Dubois et al (Dubois et al., 2010) as well as the Alzheimer's Association and National Institute on Aging published (Albert et al., 2011; Jack et al., 2011; McKhann et al., 2011; Sperling et al., 2011) recommendations for updated diagnostic criteria that include AD biomarkers in the diagnostic scheme. Evidence of medial temporal atrophy on structural MRI was one of the major biomarkers included in these recommendations.
3.2 Schizophrenia
In contrast to the promising results in the dementia field, potential MRI-based biomarkers of schizophrenia have been described only by few studies despite the large body of well replicated evidence supporting structural and functional brain alterations in this patient population (Honea et al., 2005). The first study to use advanced pattern recognition methods (SVM) in conjunction with sMRI in this field was published by Davatzikos and colleagues (Davatzikos et al., 2005) based on a population of 69 schizophrenic patients (SCZ) and 79 healthy controls (HC). The authors reported an accuracy over 80.0% for separating either male or female SCZ patients from HC. Similarly, Kawasaki et al. (Kawasaki et al., 2007) observed a classification accuracy of 80% when applying a partial least squares model trained on a group of 30 SCZ patients and 30 HC to an independent test population (n=16 SCZ, n=16 HC). Recently, Ardekani and colleagues (Ardekani et al., 2010) used diffusivity and fractional anisotropy data of 25 SCZ and 25 HC to train a linear discriminant analysis (LDA) classifier. They found that the developed LDA model was capable of correctly classifying 98% of the cases (96% sensitivity, 100% specificity) in a test population of 25 SCZ and 25 HC.
Although these classification results provide important leads, further open questions related to the clinical applicability of diagnostic MRI-based biomarkers have to be resolved. First, the examined sample sizes are still small and therefore they may not fully represent the broad cross-sectional spectrum of different disease phenotypes subsumed under and possibly coerced into the diagnostic construct of “schizophrenia” (Tsuang et al., 1990). The additional and interacting effects of disease duration and medication (Ho et al., 2011) may further impact on the neurobiological substrate in a complex and still poorly understood way, thus adding a longitudinal dimension to the heterogeneity of this patient population. Therefore, the development of predictive methods in high-risk, prodromal or first-episode populations may surmount these conceptual pitfalls, as demonstrated recently (Koutsouleris et al., 2009; Sun et al., 2009). Nevertheless, the neurobiological boundaries between schizophrenia, schizoaffective and bipolar disorder remain unclear with respect to recent genetic and structural imaging findings, which suggested a considerable degree of pathophysiological overlap between these nosological constructs.
Taken together, it remains unclear whether MRI-based pattern recognition methods trained to dichotomize between HC and SCZ would achieve the level of sensitivity and specificity needed to be integrated into clinical real-world scenarios. In case the cross-sectional and longitudinal clinical heterogeneity of the disease construct is subserved by a similarly complex neurobiological substrate, it is highly likely that such classifiers would fail to provide generalisable diagnostic performance. In this context, recently proposed semi-supervised machine learning algorithms (Filipovych et al., 2010) may provide an alternative to fully supervised machine learning methods in that they are capable of deconstructing the heterogeneity of schizophrenia by modelling the hidden neurobiological clustering within this patient population. In addition, the methodological shift from single predictive models to ensembles of classifiers may produce more robust and generalizable diagnostic biomarkers because ensemble methods have shown to reduce the risk of unfortunate selections of poorly performing single classifiers by averaging the diagnostic decisions of numerous predictive models (Koutsouleris et al.; Koutsouleris et al., 2011; Polikar, 2006).
3.3 Depression
To date the diagnosis of depression is based on behavioural symptoms and course of illness and treatment guidelines are based on clinical empirical evidence and expert consensus. However the interest in neurobiological markers of depression has grown substantially in recent years. Studies have shown that a combination of fMRI and pattern recognition techniques can accurately discriminate depressed subjects from healthy controls (Fu et al., 2008; Hahn et al., 2011; Marquand et al., 2008; Nouretdinov et al.) and predict treatment response (Costafreda et al., 2009a; Costafreda et al., 2009b).
As depression is associated with negative bias and impairments in interpersonal relationships (Fu et al., 2004) it seems advantageous to build up a diagnostic system for depression based on fMRI patterns of brain activation in response to emotional and affective processing. Fu et al (Fu et al., 2008) have shown that pattern of brain activity during sad facial processing when analyzed with SVM correctly classified up to 84% of patients (sensitivity) and 89% of controls subjects (specificity). As expected, Marquand et al (Marquand et al., 2008) obtained lower diagnostic accuracy using pattern of brain activation to verbal working memory. They found that prediction accuracy based on fMRI data during an n-back task was highest (68 % accuracy) in the 2-back condition. Hahn and colleagues (Hahn et al., 2011) showed that integrating predictions based on brain activation associated with emotional and affective processing substantially increased the accuracy to discriminate a heterogeneous group of depressed patients (i.e. patients who were on a variety of medications and with varying degrees of depressive symptoms) from healthy controls. Their best single classifier achieved an accuracy of 72% while the decision tree algorithm integrating the prediction of single classifiers leads to 83% accuracy (80% sensitivity and 87% specificity).
Attempting to predict treatment response, Costafreda et al showed that structural MRI features are predictive of an individual patient's clinical response to antidepressant medication with an accuracy of 89% , while fMRI responses showed the greatest predictive potential for cognitive behaviour therapy with an accuracy of 79% (Costafreda et al., 2009a; Costafreda et al., 2009b).
Mourão-Miranda et al. (2011) applied one-class SVM approach to investigate if patterns of fMRI response to sad facial expressions in depressed patients would be classified as outliers in relation to patterns of healthy control subjects. They found a significant correlation between the OC-SVM predictions and the patients’ Hamilton Rating Scale for Depression (HRSD), i.e. the more depressed the patients were the more of an outlier they were. In addition the OC-SVM split the patient groups into two subgroups whose membership were associated with future response to treatment, i.e. among the patients classified as outliers 70% did not respond to treatment and among those classified as non-outliers 89% responded to treatment.
As a methodological contribution, Nouretdinov et al. (Nouretdinov et al., 2011) applied transductive conformal predictor (TCP) to structural and functional MRI data to investigate diagnostic and prognostic prediction in depression and compute confidence for each prediction. Their approach was as accurate as those obtained with previous results using SVM.
4 Discussion
4.1 Potential clinical applications
The studies presented here show encouraging results and have a number of implications that suggest a general adoption of computer-assisted methods for MRI scan-based diagnosis should be seriously considered. The most important of these implications are that these methods can a) improve diagnosis in places where trained neuroradiologists or cognitive neurologists are scarce; b) increase the speed of diagnosis without compromising accuracy by eschewing lengthy specialist investigations; and c) aid the recruitment of clinically homogeneous patient populations for pharmacological trials, d) reduce subjectivity in diagnostic assessment that is inherent in the traditional mode of radiological practise which is focused on providing a written narrative report based on subjective diagnostic impression. The clinical application of these techniques is largely dependent on the disease under study. For the case of dementia, primary care and local referral play an important role in the diagnosis of a disease as common as AD. In this context, computerised methods may be especially helpful for screening purposes. Identifying those with subthreshold psychotic symptoms who will soon develop schizophrenia is clinically highly relevant to justify medication with often significant side-effects. For all scenarios, including depression, predicting future disease course or response to treatment would be extremely useful to inform patients and relatives but also to avoid unnecessary medication.
The basic implementation of how these methods can be applied clinically is shown in Fig. 2. It includes a training dataset acquired at a highly specialized imaging centre (shown by the top row in Fig. 2) which has access to both a large training data set as well as a gold standard measure for classification. This information will be used to develop diagnostic algorithms and be made available at primary referral centres. When the radiologist sees a new patient's scan there, he/she would be able to use the library/diagnostic algorithm made available to make an informed decision about underlying disease in the patient (shown in the bottom panel of figure 2).
Figure 2.
Potential setting for a clinical application.
4.2 Necessary future developments and directions
Multi-class classification methods for clinical applications
While two-class classification problems have typically been developed, one of the key issues in the clinic is the differential diagnosis of patients across several disease subtypes. For example in neurodegenerative dementia, there are at least three common pathologies underlying dementia subtypes – AD, Lewy body disease and Frontotemporal lobar degeneration.
Such multi-class problems can be solved either by applying pair-wise classifiers where there is a two-class algorithm to separate pairs of disease subtypes or by applying a one-against all classifier where a two-class algorithm is used to separate each subtype from all others. While the approach described above works well in most multi-class problems, combining information from several independent two-class classifiers represents ambiguity when providing a single diagnosis for a patient especially in the presence of mixed diseases. A suggested alternative approach for this problem is to apply simultaneous clustering of subjects solely based on their image features and using a simplified mixture model to label subjects as belonging to specific disease subtype clusters (Vemuri et al., 2011). More methods need to be developed and/or applied for the problem of differential diagnosis of disease subtypes.
Development of robust classification methods
A general application to the clinical setting will lead to suboptimal conditions in terms of data acquisition. This could start with somewhat outdated hardware and scanning sequences as well as limited equipment for stimulus presentation and response recording for fMRI activation studies. It also has time constraints, partly because subjects are often unable to remain still for prolonged intervals and partly because longer scan time is more expensive. A related problem is the presence of additional brain pathology unrelated to the clinical question. This will be particularly relevant when classifiers are applied to the elderly where small stroke lesions or vascular pathology pose a challenge to the classifiers. So far, studies have excluded such subjects from the analyses (also see section on outlier rejection) and we are not aware of any studies that have formally addressed this problem.
Addressing disease heterogeneity
Another important problem comes from a substantial heterogeneity that exists between patients. While studies in cognitive neuroimaging recruit subjects similar in age or education and usually include those with any type of brain pathology, this is in stark contrast to the clinical setting where subjects may have several types of pathology and differ in disease stage and demographic variables. An example for this is dementia where mixed dementia cases constitute about 40 % of all dementia cases.
Large training data sets increase classification accuracy (Franke et al., 2010; Klöppel et al., 2009b) and may be able to reduce problems with disease heterogeneity as they can integrate the whole spectrum of clinical and pathological manifestations. Standardized data sharing has started for structural imaging (Marcus et al., 2007; Mueller et al., 2005) but it is still rare for fMRI data. This is partly due to the higher heterogeneity of stimuli and the dependence on e.g. instructions for research on higher cognitive functions.
Outlier detection approaches
This approach can be applied to situations where one is interested in accessing deviations from a specific class or population, e.g. treating patient classification as an outlier detection problem (Mourão-Miranda et al., 2011). This option may be suitable when the current case to be classified differs substantially from those in the training set. Classification of such cases will be unreliable and they could be rejected already in the pre-processing stage by comparing them to each training group. Another interesting possibility may also be the rejection of outliers in the training set which could become more homogeneous and representative of each class.
Dealing with diagnostic uncertainties
The need for large datasets necessarily often entails the need to include cases with lower diagnostic certainty. Besides, the definition of a gold standard may often be a matter of debate. Consequently, this uncertainty should be returned to the clinician who is making the actual diagnoses. What is needed is therefore a framework that integrates uncertainties that start with the training data and continue through the classification process.
Probabilistic classification approaches, such as Gaussian Process Classifiers (Rasmussen and Williams, 2006), have the advantage of furnishing predictive probabilities for the test examples (i.e. they encode a measure of predictive confidence, which can separate examples that are confidently classified from the more ambiguous ones). This approach has been recently been applied to classify whole-brain pattern of brain activity in response to thermal pain (Marquand et al., 2010) and to classify depressed patients vs. healthy controls (Hahn et al., 2010). In addition approaches such as the Sparse Multinomial Logistic Regression (Krishnapuram and Carin, 2005; Ryali et al., 2010) can be used to furnish predictive probabilities in multi-class problems. Alternatively, measures of predictive confidence can also be obtained from classification algorithms that do not readily produce probability outputs (e.g. SVM) using ensemble-based learning approaches. These ensemble strategies generate probability outputs for new unseen subjects by combining the outputs of numerous classification models according to e.g. majority voting, boosting or mixture of experts schemes (Polikar, 2006).
A complementary strategy to deal with diagnostic uncertainties is the use of semi-supervised classification methods. The assumption so far has been that the clinical or pathological diagnosis which is considered as the gold standard for developing the classifier is available for all subjects. This is far from reality in the clinic where mild forms of the disease are difficult to classify into disease or normal groups. Therefore there may be two groups of patients available – labelled where the gold standard is available and unlabelled where the gold standard is unavailable or unreliable.
One might consider using only the labelled datasets for training the classifier but this will not perform well in the cases where there are a small number of labelled data sets or the actual distribution of the disease features is more complex that the distribution of the labelled features. In such cases, semi-supervised methods are often used (Vapnik, 1998). These methods have recently been shown to be efficient in using all the unlabelled as well as labelled data sets to develop methods with high generalisability and accuracy (Filipovych et al., 2010).
Klöppel et al.: Research highlights.
Overview of clinical applications of classification methods
Description of current limitations
Outlook and future directions
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aharoni E, et al. Can neurological evidence help courts assess criminal responsibility? Lessons from law and neuroscience. Ann N Y Acad Sci. 2008;1124:145–160. doi: 10.1196/annals.1440.007. [DOI] [PubMed] [Google Scholar]
- Albert M, et al. The diagnosis of mild cognitive impairment due to Alzheimer's disease: Recommendations from the National Institute on Aging and Alzheimer's Association Workgroup. Alzheimer Dem. 2011 doi: 10.1016/j.jalz.2011.03.008. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardekani BA, et al. Diffusion tensor imaging reliably differentiates patients with schizophrenia from healthy volunteers. Hum Brain Mapp. 2010;32:1–9. doi: 10.1002/hbm.20995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bles M, Haynes JD. Detecting concealed information using brain-imaging technology. Neurocase. 2008;14:82–92. doi: 10.1080/13554790801992784. [DOI] [PubMed] [Google Scholar]
- Chen K, et al. Characterizing Alzheimer's disease using a hypometabolic convergence index. Neuroimage. 2011 doi: 10.1016/j.neuroimage.2011.01.049. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Herskovits EH. Machine-learning techniques for building a diagnostic model for very mild dementia. Neuroimage. 2010;52:234–244. doi: 10.1016/j.neuroimage.2010.03.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costafreda SG, et al. Prognostic and diagnostic potential of the structural neuroanatomy of depression. PloS One. 2009a;4:e6353. doi: 10.1371/journal.pone.0006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costafreda SG, et al. Neural correlates of sad faces predict clinical remission to cognitive behavioural therapy in depression. Neuroreport. 2009b;20:637–641. doi: 10.1097/WNR.0b013e3283294159. [DOI] [PubMed] [Google Scholar]
- Cuingnet R, et al. Automatic classification of patients with Alzheimer's disease from structural MRI: A comparison of ten methods using the ADNI database. Neuroimage. 2010 doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, et al. Whole-brain morphometric study of schizophrenia revealing a spatially complex set of focal abnormalities. Arch Gen Psychiatry. 2005;62:1218–1227. doi: 10.1001/archpsyc.62.11.1218. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, et al. Detection of prodromal Alzheimer's disease via pattern classification of magnetic resonance imaging. Neurobiol Aging. 2008a;29:514–523. doi: 10.1016/j.neurobiolaging.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, et al. Individual patient diagnosis of AD and FTD via high-dimensional pattern classification of MRI. Neuroimage. 2008b;41:1220–1227. doi: 10.1016/j.neuroimage.2008.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, et al. Longitudinal progression of Alzheimer's-like patterns of atrophy in normal older adults: the SPARE-AD index. Brain. 2009;132:2026–2035. doi: 10.1093/brain/awp091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, et al. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCarli C, et al. Qualitative estimates of medial temporal atrophy as a predictor of progression from mild cognitive impairment to dementia. Arch Neurol. 2007;64:108–115. doi: 10.1001/archneur.64.1.108. [DOI] [PubMed] [Google Scholar]
- Dubois B, et al. Revising the definition of Alzheimer's disease: a new lexicon. Lancet Neurol. 2010;9:1118–1127. doi: 10.1016/S1474-4422(10)70223-4. [DOI] [PubMed] [Google Scholar]
- Duchesnay E, et al. Classification based on cortical folding patterns. IEEE Trans Med Imaging. 2007;26:553–565. doi: 10.1109/TMI.2007.892501. [DOI] [PubMed] [Google Scholar]
- Dukart J, et al. Combined Evaluation of FDG-PET and MRI Improves Detection and Differentiation of Dementia. PloS One. 2011;6:e18111. doi: 10.1371/journal.pone.0018111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, et al. Diagnosis of Brain Abnormality Using both Structural and Functional MR Images. Conf Proc IEEE Eng Med Biol Soc. 2006;1:1044–1047. doi: 10.1109/IEMBS.2006.259260. [DOI] [PubMed] [Google Scholar]
- Fan Y, et al. Multivariate examination of brain abnormality using both structural and functional MRI. Neuroimage. 2007;36:1189–1199. doi: 10.1016/j.neuroimage.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Filipovych R, et al. Semi-supervised cluster analysis of imaging data. Neuroimage. 2010;54:2185–2197. doi: 10.1016/j.neuroimage.2010.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K, et al. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage. 2010;50:883–892. doi: 10.1016/j.neuroimage.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Friston K, et al. Bayesian decoding of brain images. Neuroimage. 2008;39:181–205. doi: 10.1016/j.neuroimage.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Fu CH, et al. Attenuation of the neural response to sad faces in major depression by antidepressant treatment: a prospective, event-related functional magnetic resonance imaging study. Arch Gen Psychiatry. 2004;61:877–889. doi: 10.1001/archpsyc.61.9.877. [DOI] [PubMed] [Google Scholar]
- Fu CH, et al. Pattern classification of sad facial processing: toward the development of neurobiological markers in depression. Biol Psychiatry. 2008;63:656–662. doi: 10.1016/j.biopsych.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Gilad-Bachrach R, et al. Margin based feature selection-theory and algorithms. ACM; 2004. p. 43. [Google Scholar]
- Guyon I, Elisseeff A. An introduction to variable and feature selection. The Journal of Machine Learning Research. 2003;3:1157–1182. [Google Scholar]
- Habeck C, et al. Multivariate and univariate neuroimaging biomarkers of Alzheimer's disease. Neuroimage. 2008;40:1503–1515. doi: 10.1016/j.neuroimage.2008.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn T, et al. Integrating neurobiological markers of depression. Arch Gen Psychiatry. 2011;68:361–368. doi: 10.1001/archgenpsychiatry.2010.178. [DOI] [PubMed] [Google Scholar]
- Ho BC, et al. Long-term antipsychotic treatment and brain volumes: a longitudinal study of first-episode schizophrenia. Arch Gen Psychiatry. 2011;68:128–137. doi: 10.1001/archgenpsychiatry.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honea R, et al. Regional deficits in brain volume in schizophrenia: a meta-analysis of voxel-based morphometry studies. Am J Psychiatry. 2005;162:2233–2245. doi: 10.1176/appi.ajp.162.12.2233. [DOI] [PubMed] [Google Scholar]
- Jack C, et al. Introduction to the recommendations from the National Institute on Aging and the Alzheimer Association workgroup on diagnostic guidelines for Alzheimer's disease. Alzheimer Dem. 2011 doi: 10.1016/j.jalz.2011.03.004. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki Y, et al. Multivariate voxel-based morphometry successfully differentiates schizophrenia patients from healthy controls. Neuroimage. 2007;34:235–242. doi: 10.1016/j.neuroimage.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Klöppel S, et al. Accuracy of dementia diagnosis: a direct comparison between radiologists and a computerized method. Brain. 2008a;131:2969–2974. doi: 10.1093/brain/awn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel S, et al. Automatic classification of MR scans in Alzheimer's disease. Brain. 2008b;131:681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel S. Brain morphometry and functional imaging techniques in dementia: methods, findings and relevance in forensic neurology. Curr Opin Neurol. 2009;22:612–616. doi: 10.1097/WCO.0b013e328332ba0f. [DOI] [PubMed] [Google Scholar]
- Klöppel S, et al. Automatic detection of preclinical neurodegeneration: presymptomatic Huntington disease. Neurology. 2009a;72:426–431. doi: 10.1212/01.wnl.0000341768.28646.b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel S, et al. A plea for confidence intervals and consideration of generalizability in diagnostic studies. Brain. 2009b;132:e102. doi: 10.1093/brain/awn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolschijn PC, et al. Brain volume abnormalities in major depressive disorder: a meta-analysis of magnetic resonance imaging studies. Hum Brain Mapp. 2009;30:3719–3735. doi: 10.1002/hbm.20801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf ES, et al. Medial temporal lobe atrophy on MRI predicts dementia in patients with mild cognitive impairment. Neurology. 2004;63:94–100. doi: 10.1212/01.wnl.0000133114.92694.93. [DOI] [PubMed] [Google Scholar]
- Koutsouleris N, et al. Use of neuroanatomical pattern classification to identify subjects in at-risk mental states of psychosis and predict disease transition. Arch Gen Psychiatry. 2009;66:700–712. doi: 10.1001/archgenpsychiatry.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsouleris N, et al. Use of neuroanatomical pattern regression to predict the structural brain dynamics of vulnerability and transition to psychosis. Schizophr Res. 2010;123:175–187. doi: 10.1016/j.schres.2010.08.032. [DOI] [PubMed] [Google Scholar]
- Koutsouleris N, et al. Early recognition and disease prediction in the at-risk mental states for psychosis using neurocognitive pattern classification. Schizophr Bull. 2011 doi: 10.1093/schbul/sbr037. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnapuram B, Carin L. Sparse multinomial logistic regression: Fast algorithms and generalization bounds. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2005:957–968. doi: 10.1109/TPAMI.2005.127. [DOI] [PubMed] [Google Scholar]
- Lemm S, et al. Introduction to machine learning for brain imaging. Neuroimage. 2011 doi: 10.1016/j.neuroimage.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Lerch JP, et al. Automated cortical thickness measurements from MRI can accurately separate Alzheimer's patients from normal elderly controls. Neurobiol Aging. 2008;29:23–30. doi: 10.1016/j.neurobiolaging.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Marcus DS, et al. Open Access Series of Imaging Studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J Cogn Neurosci. 2007;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- Marquand A, et al. Quantitative prediction of subjective pain intensity from whole-brain fMRI data using Gaussian processes. Neuroimage. 2010;49:2178–2189. doi: 10.1016/j.neuroimage.2009.10.072. [DOI] [PubMed] [Google Scholar]
- Marquand AF, et al. Neuroanatomy of verbal working memory as a diagnostic biomarker for depression. Neuroreport. 2008;19:1507–1511. doi: 10.1097/WNR.0b013e328310425e. [DOI] [PubMed] [Google Scholar]
- McKhann G, et al. The diagnosis of dementia due to Alzheimer's disease: Recommendations from the National Institute on Aging and the Alzheimer's Assocation Workgroup. Alzheimer Dem. 2011 doi: 10.1016/j.jalz.2011.03.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, et al. The Alzheimer's disease neuroimaging initiative. Neuroimaging Clin N Am. 2005;15:869–877. xi–xii. doi: 10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K, et al. An introduction to kernel-based learning algorithms. IEEE transactions on neural networks. 2001;12:181–201. doi: 10.1109/72.914517. [DOI] [PubMed] [Google Scholar]
- Mourão-Miranda J, Hardoon DR, Hahn T, Marquand AF, Williams SC, Shawe-Taylor J, Brammer M. Patient classification as an outlier detection problem: an application of the one-class support vector machine. NeuroImage. 2011;58(3):793–804. doi: 10.1016/j.neuroimage.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navot A, et al. Nearest neighbor based feature selection for regression and its application to neural activity. Advances in Neural Information Processing Systems. 2006;18:995. [Google Scholar]
- Nouretdinov I, et al. Machine learning classification with confidence: Application of transductive conformal predictors to MRI-based diagnostic and prognostic markers in depression. Neuroimage. 2011 doi: 10.1016/j.neuroimage.2010.05.023. In press. [DOI] [PubMed] [Google Scholar]
- Pelaez-Coca M, et al. Discrimination of AD and normal subjects from MRI: anatomical versus statistical regions. Neurosci Lett. 2011;487:113–117. doi: 10.1016/j.neulet.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Petersen RC, et al. Current concepts in mild cognitive impairment. Arch Neurol. 2001;58:1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- Plant C, et al. Automated detection of brain atrophy patterns based on MRI for the prediction of Alzheimer's disease. Neuroimage. 2010;50:162–174. doi: 10.1016/j.neuroimage.2009.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikar R. Ensemble based systems in decision making. Circuits and Systems Magazine, IEEE. 2006;6:21–45. [Google Scholar]
- Rasmussen CE, Williams CKI. Gaussian Processes for Machine Learning. MIT Press; 2006. [Google Scholar]
- Rätsch G. A Brief Introduction into Machine Learning. Citeseer. 2004 [Google Scholar]
- Ryali S, et al. Sparse logistic regression for whole-brain classification of fMRI data. Neuroimage. 2010;51:752–764. doi: 10.1016/j.neuroimage.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato JR, et al. Evaluating SVM and MLDA in the extraction of discriminant regions for mental state prediction. Neuroimage. 2009;46:105–114. doi: 10.1016/j.neuroimage.2009.01.032. [DOI] [PubMed] [Google Scholar]
- Schölkopf B, Smola A. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond (Adaptive Computation and Machine Learning) The MIT Press; Cambridge, MA: 2001. [Google Scholar]
- Shawe-Taylor J, Cristianini N. Kernel methods for pattern analysis. Cambridge Univ Pr.; 2004. [Google Scholar]
- Soon CS, et al. Unconscious determinants of free decisions in the human brain. Nat Neurosci. 2008;11:543–545. doi: 10.1038/nn.2112. [DOI] [PubMed] [Google Scholar]
- Sperling RA, et al. Toward defining the preclinical stages of Alzheimer's disease: Recommendations from the National Institute on Aging and the Alzheimer Assocation Workgroup. Alzheimer Dem. 2011 doi: 10.1016/j.jalz.2011.03.003. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonnington CM, et al. Predicting clinical scores from magnetic resonance scans in Alzheimer's disease. Neuroimage. 2010 doi: 10.1016/j.neuroimage.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, et al. Elucidating a magnetic resonance imaging-based neuroanatomic biomarker for psychosis: classification analysis using probabilistic brain atlas and machine learning algorithms. Biol Psychiatry. 2009;66:1055–1060. doi: 10.1016/j.biopsych.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teipel SJ, et al. Morphological substrate of face matching in healthy ageing and mild cognitive impairment: a combined MRI-fMRI study. Brain. 2007a;130:1745–1758. doi: 10.1093/brain/awm117. [DOI] [PubMed] [Google Scholar]
- Teipel SJ, et al. Multivariate deformation-based analysis of brain atrophy to predict Alzheimer's disease in mild cognitive impairment. Neuroimage. 2007b;38:13–24. doi: 10.1016/j.neuroimage.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, et al. Heterogeneity of schizophrenia. Conceptual models and analytic strategies. Br J Psychiatry. 1990;156:17–26. doi: 10.1192/bjp.156.1.17. [DOI] [PubMed] [Google Scholar]
- Vapnik V. Statistical Learning Theory. Wiley Interscience; New York: 1998. [Google Scholar]
- Vemuri P, et al. Alzheimer's disease diagnosis in individual subjects using structural MR images: Validation studies. Neuroimage. 2008a;39:1186–1197. doi: 10.1016/j.neuroimage.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P, et al. Antemortem MRI based STructural Abnormality iNDex (STAND)-scores correlate with postmortem Braak neurofibrillary tangle stage. Neuroimage. 2008b;42:559–567. doi: 10.1016/j.neuroimage.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P, et al. MRI and CSF biomarkers in normal, MCI, and AD subjects: predicting future clinical change. Neurology. 2009a;73:294–301. doi: 10.1212/WNL.0b013e3181af79fb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P, et al. MRI and CSF biomarkers in normal, MCI, and AD subjects: diagnostic discrimination and cognitive correlations. Neurology. 2009b;73:287–293. doi: 10.1212/WNL.0b013e3181af79e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P, et al. Antemortem differential diagnosis of dementia pathology using structural MRI: Differential-STAND. Neuroimage. 2011;55:522–531. doi: 10.1016/j.neuroimage.2010.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd KB, et al. Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. AJNR Am J Neuroradiol. 2010;31:347–354. doi: 10.3174/ajnr.A1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, et al. Multimodal classification of Alzheimer's disease and mild cognitive impairment. Neuroimage. 2011 doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]