Cytosine 5′ methylation of CpG dinucleotides within and around genes exerts a major influence on transcription in many plants and animals (1–3). DNA methylation can be causal for transcriptional silencing (4–5) and targets the machinery necessary to assemble specialized chromatin enriched in deacetylated histones (6–8). Once established in somatic cells, CpG methylation patterns within the genome are very stable and provide an attractive mechanism for segregating a large fraction of stably repressed chromatin (9–11). In contrast, DNA methylation is remarkably dynamic during early mammalian development and in certain tumor cells (12, 13). Alterations in the methylation status of the entire genome (14, 15), individual chromosomes (16), and specific genes (17–20) are essential for normal development (21, 22) and can promote tumorigenesis (23, 24). Understanding how these important transitions might be regulated requires the biochemical definition of the enzymatic processes that both methylate and demethylate the genome.
Two mammalian DNA methyltransferases have been functionally defined (25, 26) from a family of related proteins (26, 27). Dnmt1 is essential for inactivation of the X chromosome and genomic imprinting in the mammalian embryo (21). This large enzyme (1,620 amino acids) is targeted to replication foci consistent with the rapid remethylation of DNA in somatic cells (28–30). Like the prokaryotic cytosine-5 methyltransferases with which it shares homology, the enzyme makes use of the Michael addition mechanism to carry out the reaction by first increasing the reactivity of the C-5 position of cytosine (31–33). An enzyme cysteinyl thiolate forms a covalent linkage with C-6 of cytosine, and the carboxyl group of an invariant glutamyl residue protonates the N-3 position to create a cytosine 4, 5 enamine. This reactive moiety attacks the sulfonium linked methyl group of S-adenosyl l-methionine. After transfer of the methyl group, the proton is abstracted from the C-5 of cytosine to reform the 5, 6 double bond and release the enzyme by β-elimination (34, 35). In contrast to the well defined molecular genetics, cell biology, and biochemistry of the Dnmt1 methyltransferase, the enzymatic basis of demethylation of 5-methylcytosine in vivo has been mysterious. Although strategies for demethylating DNA have been proposed (Figs. 1 and 2), none have yet been proven to operate under relevant physiological circumstances in vivo. This issue has now been brought into sharper focus by the recent reports of a mammalian protein with specific demethylase activity for methyl CpG dinucleotides (36), together with a demethylase enzyme complex that acts processively (37) and as published in this issue of Proceedings the finding that this complex converts 5-methylcytosine to cytosine and methanol (38).
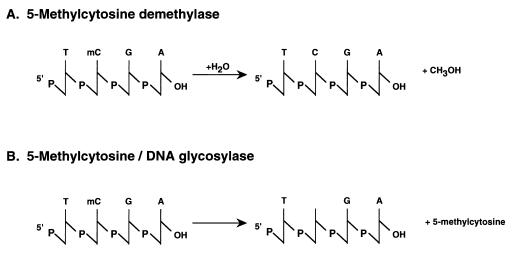
Figure 1.
Active DNA demethylation. Two pathways are illustrated. (A) The 5-methylcytosine demethylase hydrolyzes 5-methylcytosine to cytosine and water (38). (B) The 5-methylcytosine DNA glycosylase abstracts 5-methylcytosine from the phosphodiester backbone, which then is repaired by using endonuclease (43, 44).
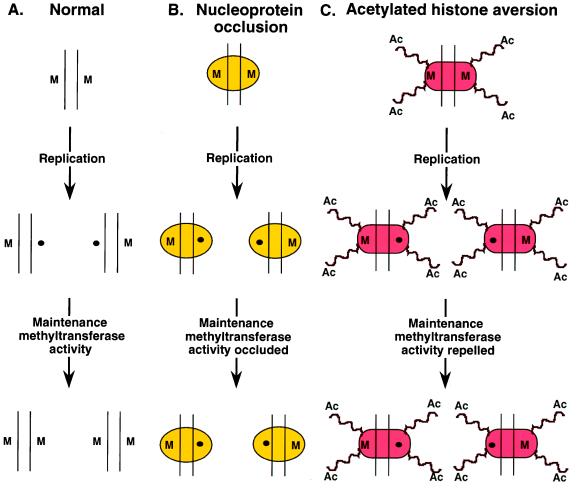
Figure 2.
Replication-coupled DNA demethylation. (A) Normally, each strand of a symmetrically methylated CpG dinucleotide (M) would be segregated after replication daughter chromatids, leaving one strand of DNA methylated (M) and the other not (●). The maintenance methyltransferase activity of Dnmt1 then would restore symmetrical methylation. (B) Regulatory nucleoprotein complexes might occlude Dnmt1 leading to loss of methylation (48, 49). (C) Histone acetylation might repel or inhibit Dnmt1, leading to loss of methylation (47).
A Demethylase?
The “amazing demethylase,” as it was recently described by Cedar and Verdine (ref. 39, p. 579), is a member of a conserved family of MBD (methyl CpG-binding domain) proteins (36, 40) related to the MeCP2 transcriptional repressor (41). The demethylase enzyme is identical to MBD2b (36, 40), and a longer MBD2a variant of unknown enzymatic activity also exists. MBD2a is found in vivo whereas there is so far no evidence for naturally occurring MBD2b. The small basic MBD2b protein (262 amino acids, pI 9.1) shows selectivity (10–100 fold) for binding to DNA containing methylated CpG dinucleotides (40). As described, the demethylase has remarkable biochemistry and enzymatic activities. The enzyme was first identified by empirical testing for 5-methylcytosine demethylase activity after the in vitro translation of mRNA derived from an expressed sequence tag cDNA with homology to MeCP2 (36). The polyhistidine-tagged enzyme retained activity after purification on a Ni2+-charged agarose resin, and transfection of human embryonal kidney cells with the MBD2b expressing clones generated detectable demethylase that fractionated on sucrose gradients in the 160–190 K range (36). It is suggested that the MBD2b protein might multimerize (36). Although the molecular identity of a demethylase would appear to be clearly established by these experiments as MBD2b, the demethylase enzyme complex used for mechanistic studies has been fractionated from human cells and is not defined at a molecular level (36–38). Thus, the relationship of this cellular demethylase complex to MBD2b remains unclear. An additional complication is the highly unusual biochemistry of the cellular enzyme fraction compared with the known properties of MBD2b. The initial purification step, gradient elution from the weak anion exchange matrix DEAE-Sephadex A-50, provides 850-fold enrichment with activity reported at 4.9–5.0 M NaCl. Two subsequent chromatographic steps result in a further 75-fold purification by successive elution (also at 4.8–5.0 M NaCl) from both a strong cation exchanger (SP-Sepharose) and a strong anion exchanger (Q-Sepharose). Remarkably, a further 10-fold enrichment is achieved by applying the 4.8–5.0 M NaCl pool from Q-Sepharose, without prior reduction of salt concentration, to DEAE-Sephacel and eluting with 10 mM Tris⋅HCl (pH 7.5) and 10 mM MgCl2. A silver stained SDS polyacrylamide gel describes coelution of activity with three polypeptides of 38–40 kDa, clearly distinct from the known electrophoretic migration of recombinant MBD2b (29 kDa; ref. 38). The repeated elution of demethylase at 4.8–5.0 M NaCl from both strong and weak anion exchangers as well as a strong cation exchanger differ substantially from the chromatographic properties of recombinant MBD2b and from the archetypical MBD family member MeCP2 (40, 41). These discrepancies remain to be resolved; it is conceivable that MBD2b might be post-translationally modified in vivo or that it is associated with a cofactor that substantially alters chromatographic properties. Whatever the precise nature of the demethylase, the 500,000-fold purification of this cellular enzyme allows effective comparison with previously defined “demethylase” activities (36–38).
Distinct Pathways for “Active” Demethylation
The chemistry necessary to directly demethylate 5-methylcytosine is challenging, requiring the disruption of a C-C bond. Earlier work had demonstrated that 5-methylcytosine was replaced by labeled cytosine during the demethylation reaction in erythroleukemia cells, indicative of replacement of the entire nucleotide or base alone (42). One potential mechanism to achieve this aim is through the action of 5-methylcytosine DNA glycosylase, which removes the methylated cytosine from DNA, leaving the deoxyribose intact (ref. 43 and Fig. 1). Local DNA repair then eventually adds back the cytosine in nucleotide form. It has been suggested that targeting of the glycosylase requires an RNA moiety (44). A more active role for RNA as an acceptor of the entire 5-methylcytosine nucleotide in an in vitro demethylation assay has been suggested (45) but later reevaluated (46). The demethylase enzyme complex characterized in the most recent series of papers lacks DNA glycosylase or nuclease activity (36), is resistant to RNase (38), and hydrolyses 5-methylcytosine to cytosine and methanol (38). Therefore, the novel 5-methylcytosine demethylase enzyme complex appears distinct from previously characterized activities (Fig. 1). This confusing situation will only be clarified by the molecular cloning and characterization of the genes encoding the demethylase present in these extracts after their biochemical purification. Knockout experiments should eventually establish whether a particular protein is important for the demethylation of DNA sequences that occurs during development or in tumor cells.
Meanwhile, if MBD2b proves to be the key demethylase and other unidentified activities in the in vitro translation extract or transfected cell are not contributing to demethylation, a whole new chemical problem requires analysis. This hydrolysis of 5-methylcytosine to cytosine and methanol will have a very large activation energy because O-H and C-C bonds have to be broken to allow the formation of C-O and C-H bonds. Cedar and Verdine (39) suggest that demethylation by water should be thermodynamically possible and that comparable molecular events to those occurring during DNA methylation would facilitate the process. This remains to be shown. As noted by the authors (36), their results “identify a new enzyme and biochemical reaction that have not yet been described in any organism” (p. 583).
Replication-Coupled Demethylation
Aside from the evidence for active demethylation by dedicated demethylase enzymes and use of the DNA repair pathway, it is probable that DNA replication also will contribute to eliminating 5-methylcytosine from DNA. We have described the targeting of the Dmnt1 methyltransferase to the replication fork and the rapid maintenance methylation of CpG dinucleotides (28–30). Chromatin structures and nucleoprotein complexes associated with transcriptional activity might interfere with the maintenance methyltransferase activity of Dnmt1, thereby progressively reducing the level of DNA methylation with each cell division event. Examples of this phenomenon include the selective loss of DNA methylation in Neurospora after growth of cells in Trichostatin A, an inhibitor of histone deacetylase (47), and the requirements for protein association during DNA replication to demethylate specific DNA sequences (48, 49). As illustrated in Fig. 2, during replication, both strands of DNA making up a symmetrically methylated CpG dinucleotide in the parental chromosome will be segregated to daughter chromatids. This leads both daughter chromatids to contain hemimethylated DNA. Under normal circumstances, the maintenance methyltransferase activity of Dnmt1 will restore symmetric methylation. It is possible that sequence-specific transacting factors bound over the sites of DNA methylation might prevent access of the methyltransferase enzyme to these sites, leading to the progressive demethylation of DNA, potentially in a sequence specific manner. With respect to more global genome demethylation, particular histone modifications such as acetylation might exert a “teflon effect,” preventing efficient access of the methyltransferase enzyme. These models predict that DNA replication will be an integral component of demethylation.
Significance
DNA methylation is a major force in mammalian development and tumorigenesis. It provides a defense mechanism against the expression of exogenous DNA in plants and animals that constrains the expression of transgenes (50–52) and, when misdirected, contributes to tumorigenesis (53). Understanding the biochemistry of DNA methylation dynamics is a problem of tremendous biomedical importance. To achieve this goal, the characterization of the molecular mechanisms that target both DNA methylation and demethylation is essential. Such knowledge should enable the manipulation of these events effectively in the hope of achieving the most effective expression of exogenous DNA. DNA methylation dynamics present a mystery that needs to be solved.
Acknowledgments
We thank Adrian Bird, Howard Cedar, Brian Hendrich, Marylin Monk, and Susan Wallace for useful discussions and Thuy Vo for manuscript preparation.
Footnotes
A commentary on this article begins on page 6107.
References
- 1.Razin A. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng H-H, Bird A. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 3.Raina R, Schlappi M, Fedoroff N. Novartis Symp. 1997;214:133–143. doi: 10.1002/9780470515501.ch8. [DOI] [PubMed] [Google Scholar]
- 4.Buschhausen G, Wittig B, Graessmann M, Graessmann A. Proc Natl Acad Sci USA. 1987;84:1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kass S U, Landsberger N, Wolffe A P. Curr Biol. 1997;7:157–167. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 6.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Nat Genet. 1998;19:187–189. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 7.Nan X, Ng H-H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 8.Eden S, Hashimshony T, Keshet I, Cedar H, Thorne A W. Nature (London) 1998;394:842. doi: 10.1038/29680. [DOI] [PubMed] [Google Scholar]
- 9.Riggs A D, Pfeifer G P. Trends Genet. 1992;8:169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- 10.Yoder J A, Walsh C P, Bestor T H. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 11.Bird A, Tweedie S. Philos Trans R Soc London B. 1995;349:249–253. doi: 10.1098/rstb.1995.0109. [DOI] [PubMed] [Google Scholar]
- 12.Shemer R, Razin A. In: Epigenetics. Russo V E A, Martienssen R A, Riggs A D, editors. Plainview, New York: Cold Spring Harbor Lab. Press; 1996. pp. 215–230. [Google Scholar]
- 13.Schmutte C, Jones P A. Biol Chem. 1998;379:377–388. doi: 10.1515/bchm.1998.379.4-5.377. [DOI] [PubMed] [Google Scholar]
- 14.Monk M, Boubelik M, Lehnert S. Development (Cambridge, UK) 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 15.Kafri T, Gao X, Razin A. Proc Natl Acad Sci USA. 1993;90:10558–10562. doi: 10.1073/pnas.90.22.10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant S G, Chapman V M. Annu Rev Genet. 1988;22:199–233. doi: 10.1146/annurev.ge.22.120188.001215. [DOI] [PubMed] [Google Scholar]
- 17.Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Genes Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 18.Li E, Beard C, Jaenisch R. Nature (London) 1993;366:2282–2292. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 19.Norris D P, Patel D, Kay G F, Penny G D, Brockdorff N, Sheardown S A, Rastan S. Cell. 1994;77:41–51. doi: 10.1016/0092-8674(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 20.Jones P A, Laird P W. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 21.Li E, Bestor T H, Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 22.Neumann B, Barlow D P. Curr Opin Genet Dev. 1996;6:159–163. doi: 10.1016/s0959-437x(96)80045-1. [DOI] [PubMed] [Google Scholar]
- 23.Issa J P, Baylin S B. Nat Med. 1996;2:281–282. doi: 10.1038/nm0396-281. [DOI] [PubMed] [Google Scholar]
- 24.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J P. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 25.Bestor T H, Laudano A P, Mattaliano R, Ingram V M. J Mol Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 26.Okano M, Xie S, Li E. Nat Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 27.Yoder J A, Bestor T H. Hum Mol Genet. 1998;7:279–284. doi: 10.1093/hmg/7.2.279. [DOI] [PubMed] [Google Scholar]
- 28.Leonhardt H, Page A W, Weier H U, Bestor T H. Cell. 1992;71:865–874. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 29.Chuang L S, Ian H I, Koh T W, Ng H H, Xu G, Li B F. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 30.Gruenbaum Y, Szyf M, Cedar H, Razin A. Proc Natl Acad Sci USA. 1983;80:4919–4921. doi: 10.1073/pnas.80.16.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santi D V, Garrett C E, Barr P J. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- 32.Randerath K, Tseng W C, Harris J S, Lu L J. Recent Res Cancer Res. 1983;84:283–297. doi: 10.1007/978-3-642-81947-6_22. [DOI] [PubMed] [Google Scholar]
- 33.Chen L, MacMillan A M, Chang W, Ezez-Nikpay K, Lane W S, Verdine G L. Biochemistry. 1991;30:11018–11026. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- 34.Chen L, MacMillan A M, Verdine G L. J Am Chem Soc. 1993;115:5318–5319. [Google Scholar]
- 35.Yoder J A, Soman N S, Verdine G L, Bestor T H. J Mol Biol. 1997;270:385–395. doi: 10.1006/jmbi.1997.1125. [DOI] [PubMed] [Google Scholar]
- 36.Bhattacharya S K, Ramchanctani S, Cervoni N, Szyf M. Nature (London) 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 37.Cervoni N, Bhattacharya S, Szyf M. J Biol Chem. 1999;274:8363–8366. doi: 10.1074/jbc.274.13.8363. [DOI] [PubMed] [Google Scholar]
- 38.Ramchandani S, Bhattacharya S K, Cervoni N, Szyf M. Proc Natl Acad Sci USA. 1999;96:6107–6112. doi: 10.1073/pnas.96.11.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cedar H, Verdine G L. Nature (London) 1999;397:579–580. doi: 10.1038/17492. [DOI] [PubMed] [Google Scholar]
- 40.Hendrich B, Bird A. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis J D, Meehan R R, Henzel W J, Maurer-Foy I, Jeppesen P, Klein F, Bird A. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 42.Razin A, Szyf M, Kafri T, Roll M, Giloh H, Scarpa S, Carotti D, Cantoni G L. Proc Natl Acad Sci USA. 1986;83:2827–2831. doi: 10.1073/pnas.83.9.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jost J-P, Siegmann M, Sun L, Leung R. J Biol Chem. 1995;270:9734–9739. doi: 10.1074/jbc.270.17.9734. [DOI] [PubMed] [Google Scholar]
- 44.Jost J-P, Fremont M, Siegmann M, Hofsteenge J. Nucleic Acids Res. 1997;25:4545–4550. doi: 10.1093/nar/25.22.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss A, Keshet I, Razin A, Cedar H. Cell. 1996;86:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- 46.Swisher J F A, Rand E, Cedar H, Pyle A M. Nucleic Acids Res. 1998;26:5573–5580. doi: 10.1093/nar/26.24.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selker E U. Proc Natl Acad Sci USA. 1998;95:9430–9435. doi: 10.1073/pnas.95.16.9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuo K, Silke J, Georgiev O, Marti P, Giovannini N, Rungger D. EMBO J. 1998;16:1446–1453. doi: 10.1093/emboj/17.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsieh C L. Mol Cell Biol. 1999;19:46–56. doi: 10.1128/mcb.19.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matzke M A, Matzke A J M, Eggleston W B. Trends Plant Sci. 1996;1:382–388. [Google Scholar]
- 51.Pikaart M J, Recillas-Targa F, Felsenfeld G. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garrick D, Fiering S, Martin D I, Whitelaw E. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 53.Cameron E E, Bachman K E, Myohanen S, Herman J G, Baylin S B. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]