Abstract
The homotypic fusion of endoplasmic reticulum (ER) membranes is mediated by atlastin (ATL), which consists of an N-terminal cytosolic domain containing a GTPase module and a three-helix bundle followed by two transmembrane (TM) segments and a C-terminal tail (CT). Fusion depends on a GTP hydrolysis-induced conformational change in the cytosolic domain. Here, we show that the CT and TM segments also are required for efficient fusion and provide insight into their mechanistic roles. The essential feature of the CT is a conserved amphipathic helix. A synthetic peptide corresponding to the helix, but not to unrelated amphipathic helices, can act in trans to restore the fusion activity of tailless ATL. The CT promotes vesicle fusion by interacting directly with and perturbing the lipid bilayer without causing significant lysis. The TM segments do not serve as mere membrane anchors for the cytosolic domain but rather mediate the formation of ATL oligomers. Point mutations in either the C-terminal helix or the TMs impair ATL’s ability to generate and maintain ER morphology in vivo. Our results suggest that protein–lipid and protein–protein interactions within the membrane cooperate with the conformational change of the cytosolic domain to achieve homotypic ER membrane fusion.
Keywords: membrane perturbation, content mixing, organelle shaping, organelle remodeling, hereditary spastic paraplegia
The fusion of cellular membranes is critical for many biological processes. Homotypic fusion, i.e., the merging of identical membranes, is required for the remodeling of organelles, including the endoplasmic reticulum (ER) and mitochondria. Both organelles contain membrane tubules that are connected into a network by homotypic fusion (1, 2). Much less is known about this process than about heterotypic fusion, which occurs between viral and cellular membranes and between intracellular transport vesicles and target membranes. In viral fusion, the membranes are pulled together by an irreversible conformational change of a single protein (3, 4). In fusion during vesicular transport, three target (t-SNARE) proteins in one membrane and a vesicle (v-SNARE) partner in the other zipper up to form a four-helix bundle in the fused lipid bilayer, a process facilitated by additional proteins (5–8).
The homotypic fusion of ER membranes in metazoans is mediated by the atlastins (ATLs) (9, 10), a class of membrane-bound GTPases that belong to the dynamin family (11). The ATLs contain an N-terminal cytosolic domain consisting of a GTPase module and a three-helix bundle (3HB), as well as two closely spaced transmembrane (TM) segments and a C-terminal tail (CT) (Fig. 1A). A role for the ATLs in ER fusion is suggested by the observation that the depletion of ATLs leads to long, unbranched ER tubules in tissue culture cells (9) and to ER fragmentation in Drosophila melanogaster (10) possibly caused by insufficient fusion between the tubules. Nonbranched ER tubules also are observed upon expression of dominant-negative ATL mutants (9, 12). In addition, antibodies to ATL inhibit ER network formation in Xenopus egg extracts (9). Finally, proteoliposomes containing purified Drosophila ATL undergo GTP-dependent fusion in vitro (10, 13). ATL-mediated homotypic fusion of ER membranes appears to be physiologically important, because mutations in human ATL1, the major isoform in neuronal tissues, can cause hereditary spastic paraplegia (HSP) (14). HSP is a neurodegenerative disease caused by the shortening of the axons in corticospinal motor neurons, leading to progressive spasticity and weakness of the lower limbs.
Fig. 1.
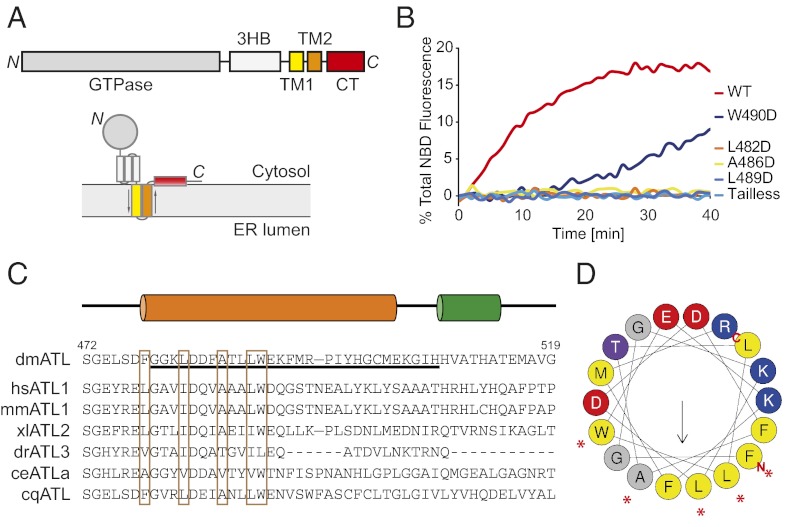
An amphipathic helix in the CT of ATL facilitates fusion. (A) Domain structure and membrane topology of ATL. 3HB, three-helix bundle; TM1 and TM2, transmembrane segments; CT, C-terminal tail. (B) Full-length wild-type Drosophila ATL, ATL lacking the CT (tailless, residues 1–476), and ATL with point mutants in the CT region were reconstituted at equal concentrations into donor and acceptor vesicles. GTP-dependent fusion of donor and acceptor vesicles was monitored by the dequenching of an NBD-labeled lipid present in the donor vesicles. In all cases, fusion was initiated by addition of GTP. (C) Sequence alignment of the CTs from various ATLs. Predicted helices (orange and green cylinders) are indicated, as are the residue numbers for Drosophila ATL. Residues on the hydrophobic face of the first, amphipathic helix are enclosed in brown boxes. The sequence of the synthetic C-terminal peptide used in the following figures is underlined. ce, Caenorhabditis elegans; cq, Culex quinquefasciatus; dm, D. melanogaster; dr, Danio rerio; hs, Homo sapiens; mm, Mus musculus; xl, Xenopus laevis. (D) Helical wheel representation of the first helix (478–495) was generated using program HeliQuest (http://heliquest.ipmc.cnrs.fr/). Hydrophobic, negatively charged, and positively charged residues are shown in yellow, red, and blue, respectively. The termini are indicated, and relatively conserved residues on the hydrophobic face are indicated by asterisks.
Yeast and plant cells do not possess ATLs, but similar GTPases—Sey1p in Saccharomyces cerevisiae and Root Hair Defective 3 (RHD3) in Arabidopsis thaliana—may have an analogous function (9). Mutations in the plant homolog RHD3 cause ER morphology defects (15) similar to those seen after depletion of ATLs. The tubular ER network in yeast is disrupted when the cells lack both Sey1p and one of the tubule-shaping proteins, Rtn1p or Yop1p (9). Normal ER morphology can be reestablished by expression of wild-type Sey1p (9) or human ATL1 (16), supporting the idea that these proteins are functional orthologs. The homotypic fusion of outer mitochondrial membranes also is mediated by membrane-bound GTPases of the dynamin family, the mitofusins in mammals and Fzo1p in yeast (17, 18). Thus, mitochondrial fusion may be achieved by a similar mechanism.
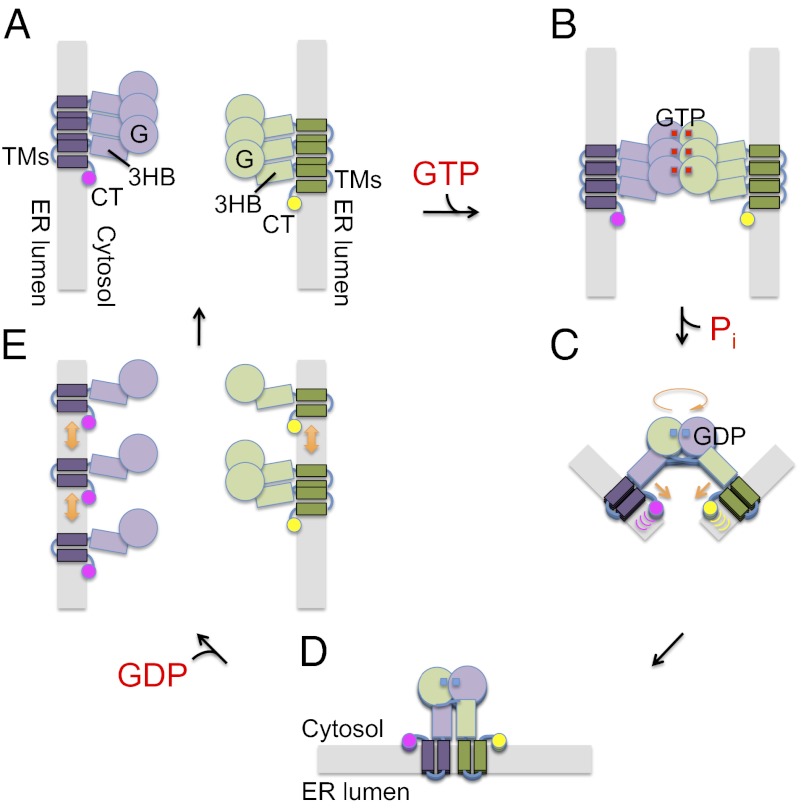
Recent structural and biochemical studies provide significant insight into the mechanism of ATL-mediated homotypic fusion (13, 19). The N-terminal cytosolic domain of human ATL1 forms a nucleotide-dependent dimer in which the GTPase domains face each other (Fig. S1). In one crystal structure, the 3HBs following the GTPase domains point in opposite directions. This structure likely corresponds to a prefusion state in which the full-length proteins still are anchored in different membranes. In another structure, the 3HBs are parallel to one another and have crossed over to dock against the GTPase domain of the partner molecule. In the full-length ATL, the two molecules would sit in the same membrane, likely representing a postfusion state. The conformational change between the pre- and postfusion states, which is triggered by Pi release during the GTP hydrolysis cycle (13), would pull the membranes into close proximity so that they can fuse.
The energy gained from the conformational change in the N-terminal cytosolic domain of ATL appears to be relatively small, because the interaction surface areas are not dramatically different in the pre- and postfusion states. This observation raises the possibility that domains missing in the crystal structures, i.e., the TM segments and the CT, also could play important roles in the fusion process. In fact, deletion of the CT reduces the fusion activity of ATL in vitro (13, 20). Here, we show that an amphipathic helix in the CT interacts with the lipid bilayer and destabilizes it to promote bilayer fusion with little membrane lysis. We also show that the TM segments mediate nucleotide-independent oligomerization of ATL molecules and play an essential role in fusion. Both the CT and TM segments are important for ATL’s ability to generate and maintain ER morphology in vivo.
Results
Amphipathic Helix in the CT Facilitates Fusion.
To test the role of ATL’s CT in membrane fusion, we first used an in vitro lipid-mixing assay. In this assay, purified wild-type or mutant Drosophila ATL is reconstituted into donor and acceptor proteoliposomes at a protein:lipid ratio of 1:2,000. The donor vesicles contain lipids labeled with 7-nitrobenzoxadiazole (NBD) and its FRET acceptor, rhodamine. NBD fluorescence is quenched initially by rhodamine in the proteoliposomes, but subsequent fusion of labeled with unlabeled acceptor proteoliposomes results in dilution of the fluorophores and dequenching of NBD. In agreement with previous results (10, 13), wild-type ATL gave efficient fusion in the presence of GTP and Mg2+, whereas a mutant lacking the entire CT was much less active (Fig. 1B). The CT is not absolutely essential, because the tailless ATL mutant promoted fusion at a fivefold higher concentration (Fig. S2A). To identify the region in the CT that is responsible for enhancement of fusion activity, we compared the sequences of ATLs from different species. The only region of significant sequence conservation immediately follows the second TM segment and is predicted to form an amphipathic helix, which in Drosophila ATL comprises residues 478–502 (Fig. 1 C and D). Several residues on the hydrophobic face of the helix are similar across species (Fig. 1C).
To dissect the function of the amphipathic helix, we tested the effect of point mutations on ATL-mediated membrane fusion. Mutations on the hydrophobic face of the helix, L482D, A486D, and L489D, reduced fusion to a level similar to that seen with the tailless mutant (Fig. 1B). The W490D mutation also reduced fusion, albeit less strongly. The hydrophilic face of the helix appears to be less important, because changing charged residues in the helix (residues D477, D483, E491) to Ala had little effect (Fig. S2C). Thus, the critical part of the CT for ATL-mediated fusion appears to be the hydrophobic residues at the N-terminal part of the helix.
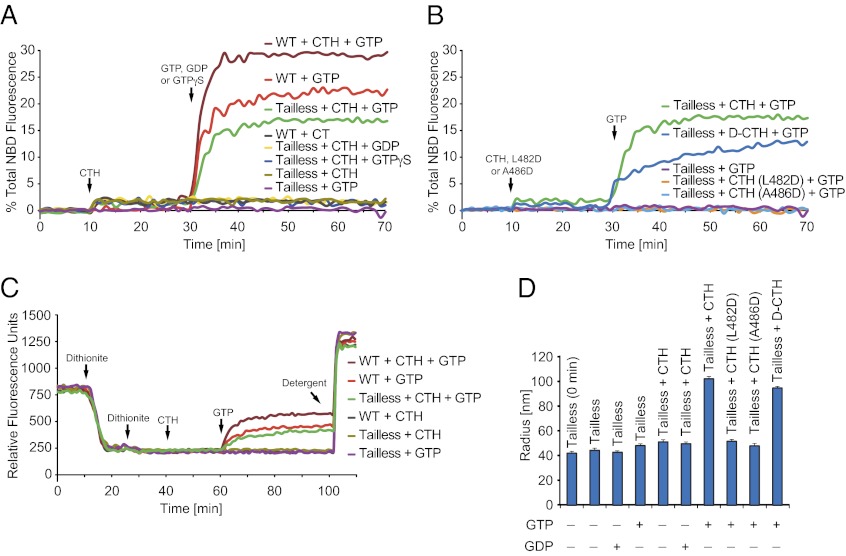
We made the striking observation that a synthetic peptide corresponding to the helix (CTH, residues 479–507 of Drosophila ATL) (Fig. 1C) could act in trans to rescue the fusion activity of the tailless mutant (Fig. 2A). Almost wild-type levels of fusion activity were reached with a peptide concentration of about 15 μM, corresponding to a molar ratio between the peptide and tailless ATL of ∼50:1. The requirement of such a high ratio is expected, because the two partners normally are covalently linked and thus are in close proximity. The CTH also stimulated lipid mixing by full-length wild-type ATL, indicating that CTH does not compete with the endogenous CT in the lipid-mixing reaction (Fig. 2A). Although the peptide alone caused only minor dequenching, significant lipid mixing was absolutely dependent on GTP hydrolysis and was not seen with GDP or GTPγS (Fig. 2A). Similar results were obtained with a slightly shorter peptide (residues 481–507), whereas a significantly shorter peptide (residues 479–494) required higher concentrations for transactivation (Fig. S2D). To test whether transactivation mimics the physiological fusion reaction, we introduced mutations in CTH that impaired fusion activity in full-length ATL. Indeed, peptides containing the L482D and A486D mutations did not stimulate fusion by tailless ATL (Fig. 2B).
Fig. 2.
Transactivation of tailless ATL fusion by a synthetic peptide. (A) The fusion of vesicles containing wild-type or tailless ATL (residues 1–476) was determined by lipid mixing in the absence or presence of a synthetic peptide corresponding to the amphipathic helix (CTH, residues 479–507 of Drosophila ATL; see Fig. 1C). When indicated, the peptide was added at 10 min and the nucleotide was added at 30 min. Controls without nucleotide are shown for comparison. (B) As in A, but with mutant peptides or a peptide consisting of d-amino acids (d-CTH). The slightly lower activity of d-CTH might indicate a small alteration in lipid interaction caused by the chiral nature of the phospholipids. (C) As in A, but with 1 mM sodium dithionite added twice before the reaction and with raw fluorescence data shown. (D) The effective hydrodynamic radii of vesicles containing tailless ATL at a protein:lipid ratio of 1:1,000 were determined by dynamic light scattering with and without the addition of the indicated peptides. The reactions were started at 37 °C by the addition of GTP and terminated after 10 min by the addition of EDTA, before size analysis. One representative experiment is shown, with the radii given as the average of three instrument readings. Error bars indicate the SE.
Because the lipid mixing we observed may be caused by fusion of only the outer leaflets of the bilayer, we added dithionite before the fusion reaction to test whether the inner leaflets also fuse. Dithionite is poorly membrane permeable and therefore reacts selectively with and abolishes the fluorescence of NBD in the outer leaflet of the bilayer. Thus, any dequenching of NBD after dithionite treatment results from inner leaflet mixing. Wild-type ATL induced inner leaflet mixing, in agreement with previous results (10), whereas tailless ATL alone was inactive but was activated by the addition of CTH in trans (Fig. 2C). As before, CTH also stimulated the fusion activity of wild-type ATL. To confirm CTH-stimulated fusion further, we used dynamic light scattering (DLS) to measure the radius of the proteoliposomes before and after fusion. Fusion was initiated by the addition of GTP and terminated by the addition of EDTA, which chelates the Mg2+ ions required for nucleotide binding and hydrolysis. EDTA also disassembles vesicles that are tethered to one another but not yet fused (10), so that an increase in proteoliposome size observed by DLS would reflect only fusion. As previously reported (10), vesicles containing wild-type ATL underwent a GTP-dependent size increase (Fig. S3). Although tailless ATL induced only a small increase in liposome size, a prominent size increase was observed when CTH was added in trans (Fig. 2D). Together, these results show that CTH stimulates the tailless ATL mutant to catalyze fusion of both the inner and outer leaflets of the lipid bilayer.
We used peptide-stimulated fusion of tailless ATL to analyze the role of the amphipathic helix in greater detail. To test whether the helix functions through protein–protein interactions, we used a synthetic peptide consisting solely of d-amino acids; because of the reversed chirality, the peptide would not be expected to maintain its interactions with tailless ATL, if they existed. The d-amino acid CTH (d-CTH) greatly stimulated lipid mixing of the tailless mutant (Fig. 2B), suggesting that the helix likely does not act through protein–protein interactions. Liposome fusion monitored by DLS also showed that d-CTH has an activity similar to that of the l-amino acid CTH (Fig. 2D). In contrast, only small increases in the average liposome radius were caused by the addition of mutant peptides, L482D and A486D, as expected from the lipid-mixing experiments (Fig. 2B).
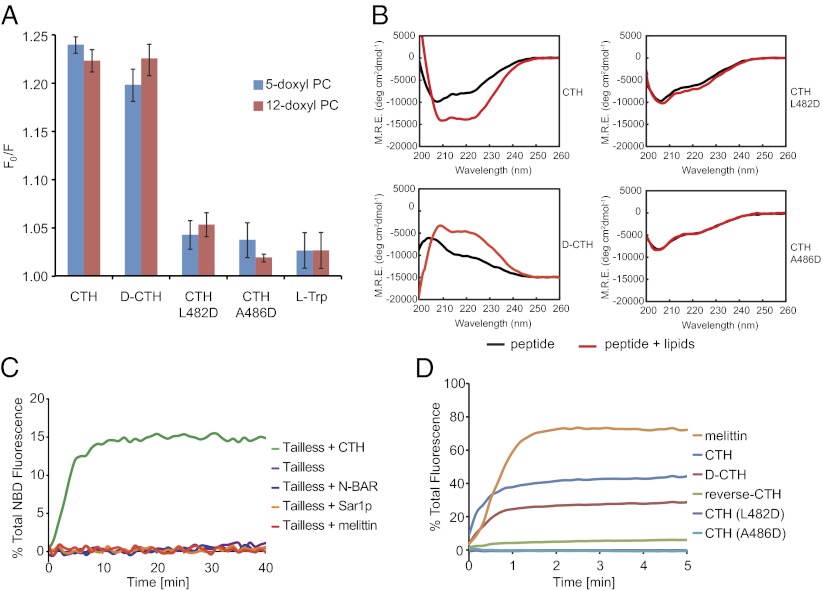
We next tested whether the C-terminal helix of ATL inserts into the lipid bilayer, as has been observed with other amphipathic helices (21). To this end, we monitored the fluorescence of the single Trp residue in the CTH (Trp490 in ATL). Upon addition of liposomes, a slight blue shift was observed, indicating movement of the Trp residue into a more hydrophobic environment (Fig. S4). To test directly whether the Trp residue comes in contact with the hydrophobic tails of lipids, we added the peptide to liposomes containing lipids with doxyl groups in the hydrocarbon chain; the doxyl groups are expected to quench the Trp fluorescence upon contact. Indeed, quenching was observed with doxyl groups located at either position 5 or 12 in the hydrocarbon chain (Fig. 3A). The d-CTH also interacted with the lipid bilayer, whereas an l-amino acid peptide carrying the L482D or A486D mutation showed little binding to the membrane (Fig. 3A).
Fig. 3.
The amphipathic ATL helix interacts with the lipid bilayer. (A) C-terminal peptides [wild type (CTH) or mutants] or a peptide consisting of d-amino acids (d-CTH) were added to liposomes containing or lacking phosphatidylcholine with doxyl groups at position 5 or 12 of the hydrocarbon chain. The quenching of the fluorescence of Trp490 in the peptides was measured and is expressed as F0/F (maximal fluorescence with doxyl-free liposomes divided by maximal fluorescence with doxyl-containing liposomes). A control was done by adding the amino acid Trp instead of a peptide. Shown are the mean and SE of three experiments. (B) Circular dichroism spectra of wild-type, d-amino acid, and mutant CTH peptides were recorded in the absence (black lines) or presence (red lines) of liposomes. (C) The fusion of vesicles by tailless ATL was tested at a protein:lipid ratio of 1:1,000 in the presence of 10 μM of various amphipathic peptides: N-BAR helix (residues 1–24 of Rvs161p), Sar1p helix (residues 1–23 of Sar1p), and melittin. (D) The indicated peptides were added to liposomes loaded with calcein (peptide:lipid ratio 1:50), and the leakage of the dye was monitored by its dequenching.
Circular dichroism measurements provided further evidence for an interaction of the amphipathic helix with lipids. The peptide corresponding to the wild-type sequence became more helical in the presence of liposomes (Fig. 3B). The d-peptide behaved similarly, although the sign of dichroism was reversed, as expected. In contrast, l-peptides containing the L482D or A486D mutations did not become more helical upon addition of liposomes (Fig. 3B). Taken together, these results show that the folding of the amphipathic helix is induced upon interaction with the lipid bilayer and that formation of the helix correlates with its ability to promote fusion.
Surprisingly, other amphipathic peptides that are known to interact with lipid bilayers [including the curvature-inducing N-terminal helices of Sar1p and of the N-BAR domain protein amphiphysin (22–24)] did not promote the fusion activity of the tailless ATL in trans (Fig. 3C). The pore-forming melittin peptide (25) also showed only minimal transactivation, even when added at higher concentrations (Fig. S5). Specificity is supported further by the observation that a synthetic peptide corresponding to the reverse amino acid sequence of CTH (i.e., reading the sequence from the C to the N terminus), which is predicted to have an altered and shortened amphipathic helix (Fig. S6A), showed only low stimulation of fusion by tailless ATL (Fig. S6B). Using the doxyl-quenching assay, we also verified that each of these peptides indeed binds liposomes with composition similar to those used in the lipid-mixing assay (Fig. S7). These results suggest that CTH interacts with the lipid bilayer to stimulate ATL-mediated fusion in a different manner than these other amphipathic peptides.
To test whether the amphipathic ATL helix destabilizes the lipid bilayer, we added the CTH to liposomes loaded with self-quenching concentrations of the fluorescent dye calcein; perturbation of the bilayer would cause the release of calcein, resulting in dilution and a consequent increase in its fluorescence. Indeed, both the l- and d-amino acid CTH induced some calcein leakage from the liposomes at about the same peptide:lipid ratio as used in the fusion assay (Fig. 3D). In contrast, the peptides carrying the L482D or A486D mutations were inactive. The peptide with the reverse sequence also showed reduced activity (Fig. 3D), consistent with its behavior in the fusion assay (Fig. S6B). These results show that the activity of ATL’s C-terminal amphipathic helix is correlated with its ability to destabilize the integrity of the lipid bilayer. However, the helix is less potent than melittin in calcein leakage (Fig. 3D) but is much more active in stimulating the fusion of tailless ATL (Fig. 3C and Fig. S5), indicating it likely does not permeabilize membranes by forming pores.
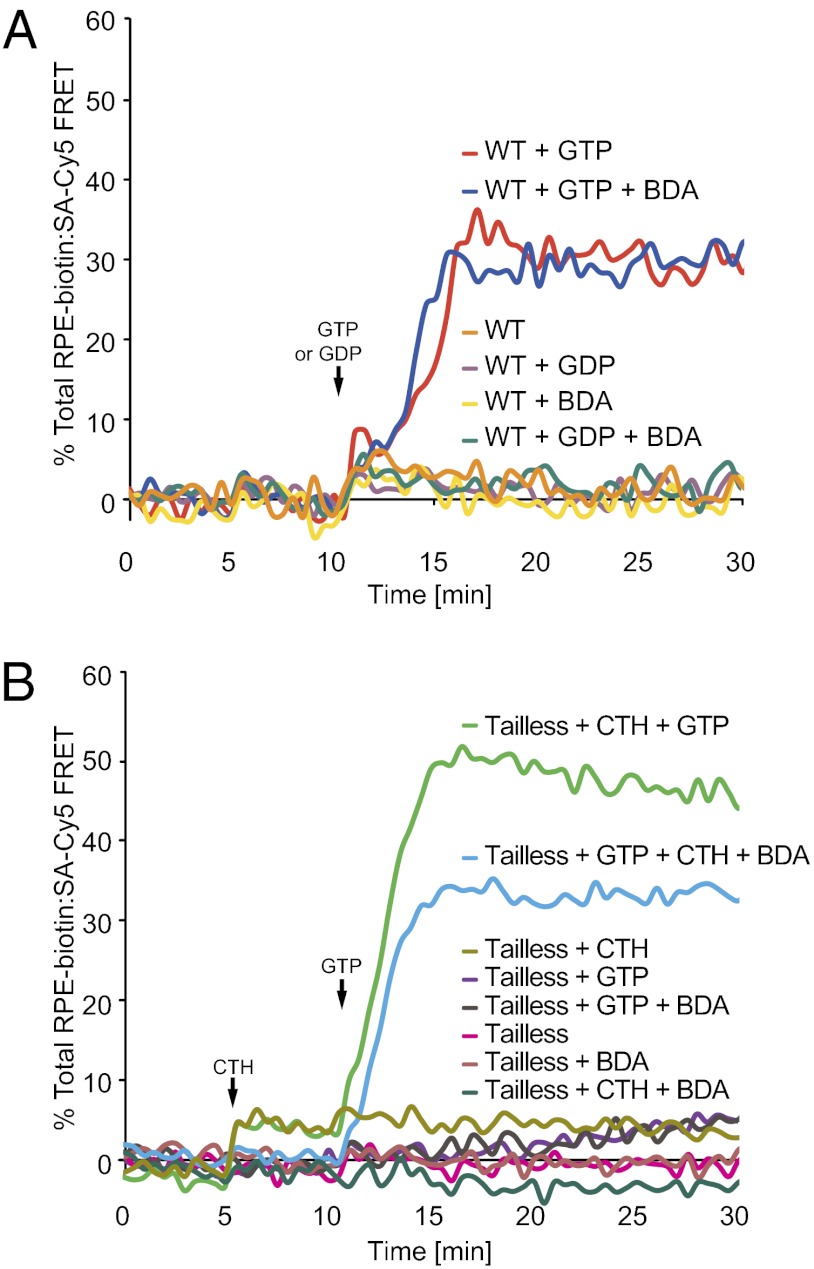
The fact that the CTH increases the permeability of lipid bilayers raises the possibility that it stimulates the fusion activity of tailless ATL by disrupting and reannealing the lipid bilayers rather than by promoting true fusion in which the permeability barrier between the inside and outside of the vesicles is maintained. To test this possibility, we used a content-mixing assay, in which two fluorophores, biotinylated R-phycoerythrin (RPE-biotin) and Cy5-labeled streptavidin (SA-Cy5), are encapsulated into donor and acceptor proteoliposomes, respectively (26). When vesicle contents mix during fusion or lysis, the interaction between the streptavidin and biotin moieties brings R-phycoerythrin and Cy5 close enough for the fluorophores to undergo FRET. Fusion can be distinguished from lysis by adding biotin-dextran (BDA) to the outside of the proteoliposomes, thus preventing FRET between leaked dyes. Maximum FRET fluorescence is determined by adding detergent to a reaction performed in the absence of BDA. Lipid mixing is followed in parallel with content mixing by incorporating Marina Blue- and NBD-labeled lipids into donor vesicles and monitoring the dequenching of Marina Blue (26). We first tested whether wild-type ATL could induce content mixing during liposome fusion, because it has not yet been demonstrated that ATL mediates true, nonleaky fusion rather than just lipid mixing. Proper encapsulation of each dye into the proteoliposomes first was confirmed by adding one FRET partner outside proteoliposomes containing the other FRET partner; FRET occurred only when detergent was added to lyse the vesicles (Fig. S8 A and B). In the actual fusion reaction, wild-type ATL catalyzed GTP-dependent content mixing with nearly no lysis (Fig. 4A). The kinetics of content mixing corresponded to that of lipid mixing (Fig. S8C). Addition of the CTH to proteoliposomes containing tailless ATL induced significant nonleaky fusion as well, whereas almost no content mixing was observed in the absence of peptide or GTP (Fig. 4B). Again, GTP-dependent content mixing proceeded in parallel with lipid mixing (Fig. S8D). A small increase in FRET was observed in the content-mixing assay when CTH was added in the absence of GTP (Fig. 4B), indicating that the lipid binding of the peptide causes some leakage. Nevertheless, it is clear that the CTH does not induce massive lysis and primarily promotes nonleaky membrane fusion by tailless ATL, similar to the reaction with wild-type ATL.
Fig. 4.
CTH-stimulated fusion of tailless ATL involves mixing of vesicle contents. (A) Content mixing and leakage mediated by wild-type ATL was measured by FRET between RPE-biotin and SA-Cy5 encapsulated in donor and acceptor vesicles, respectively. GTP or GDP was added, as indicated. BDA with a molecular mass of 70 kDa was added where indicated to block the FRET contribution from content dyes released by membrane lysis. The data are presented as the increase in Cy5 fluorescence, expressed as a percentage of the total fluorescence determined after the addition of detergent in the absence of BDA. Nucleotide or buffer was added at 10 min. (B) As in A, but using vesicles reconstituted with tailless ATL. The CTH was added at 5 min, and GTP or buffer was added at 10 min.
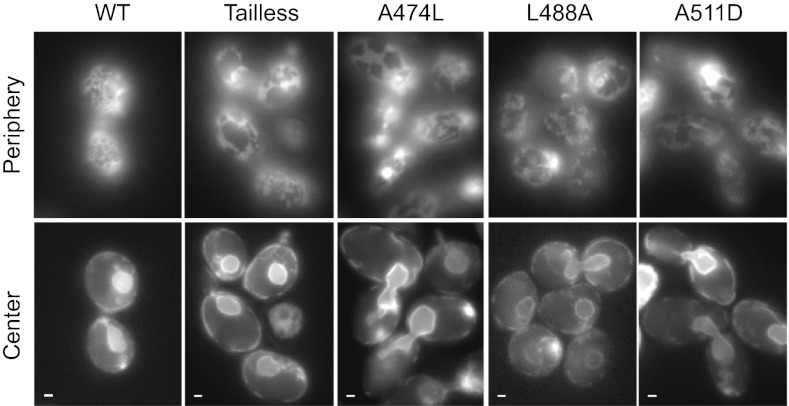
We next tested the role of the C-terminal amphipathic helix in vivo, taking advantage of the fact that human ATL1 can replace the functional ortholog Sey1p in S. cerevisiae; the abnormal cortical ER morphology in cells lacking Sey1p and the tubule-shaping protein Yop1p can be restored by expression of human ATL1 from a CEN plasmid under the constitutive MET25 promoter (16). Restoration of ER morphology also was observed when ATL1 was expressed at lower levels from the weaker endogenous SEY1 promoter (Fig. 5). The tailless ATL1 mutant or the A511D mutant (equivalent to A486D in Drosophila ATL) did not rescue the morphology defects (Fig. 5; for expression levels, see Fig. S9), indicating that the amphipathic helix is required for efficient ER network formation in vivo. As expected from the in vitro experiments (Fig. S2A), at higher expression levels (under the MET25 promoter), the tailless mutant was able to restore the tubular ER network (Fig. S2B).
Fig. 5.
ATL function tested in vivo in yeast cells. Wild-type human ATL1 or the indicated mutants were expressed under the endogenous SEY1 promoter in S. cerevisiae cells lacking Sey1p and Yop1p (sey1Δ yop1Δ cells). See Fig. S9 for expression levels. The ER was visualized by expressing Sec63-GFP, focusing the microscope on either the periphery or the center of the cells. (Scale bars, 1 μm.)
Analysis of the Role of TM Segments in Fusion.
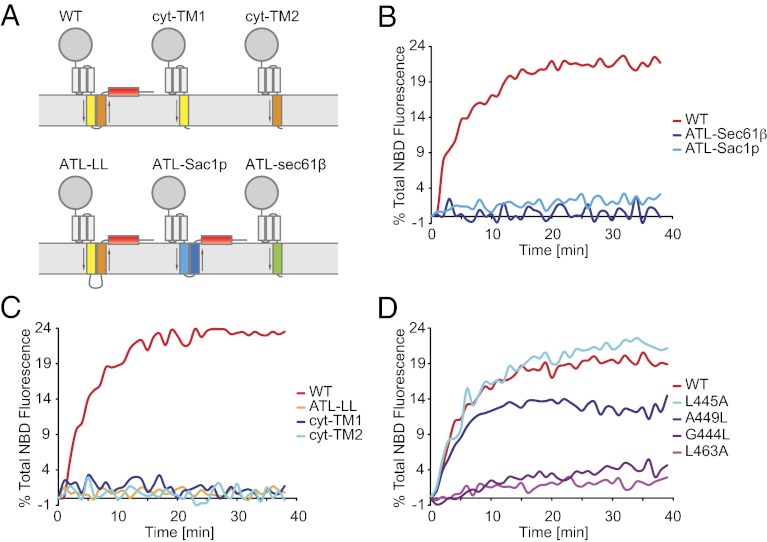
We next analyzed the role of the two closely spaced TM segments in promoting fusion, again using Drosophila ATL. To investigate whether the TM segments serve merely as membrane anchors, we replaced them with unrelated TMs and tested these mutants in the lipid-mixing assay. Replacement with the TM of the tail-anchored human Sec61β protein (ATL-Sec61β) resulted in a mutant with no detectable fusion activity (Fig. 6 A and B), although it was reconstituted into liposomes efficiently (Fig. S10D). A similar result was obtained when the TMs were replaced with the two TM segments of yeast Sac1p (ATL-Sac1p), which are separated by a loop of only ∼10 amino acids and thus form a hairpin structure similar to that in ATL. Mutant proteins lacking either TM1 and CT (cyt-TM2) or TM2 and CT (cyt-TM1) also were inactive (Fig. 6 A and C). Fusion of mutants lacking the CT (cyt-TM2, cyt-TM1, and ATL-Sec61β) was not restored when the CTH peptide was added in trans (Fig. S11A). Even when these mutants were reconstituted at a fivefold-higher ratio of protein:lipid, only cyt-TM2 showed some low fusion activity in the presence of the peptide (Fig. S11B). Taken together, these results indicate that the TM segments are not serving just as membrane anchors for the cytosolic domain during ATL-mediated membrane fusion. Furthermore, the distance between the two TM segments is important, because the insertion of seven residues (AAEEEEA) into the intervening loop (ATL-LL) rendered the protein inactive (Fig. 6C), even though membrane integration was not affected (Fig. S10D).
Fig. 6.
Testing TM mutants of ATL in the fusion assay. (A) Schematic representation of the ATL constructs used. TM1 and TM2 are colored in yellow and orange, respectively, and the CT is colored in red. The membrane orientation of the TM segments is indicated by arrows. The TMs of Sac1p are shown in cyan and blue, and the TM of Sec61β is shown in green. (B) Fusion assays with wild-type ATL or TM replacement mutants. (C) As in B, but with TM deletion or insertion mutants. (D) As in B, but with point mutants in the TM segments.
To identify residues that are important for the function of the TM segments in membrane fusion, we mutated both conserved and less conserved amino acids (Fig. S12). Substitutions were made to nonpolar residues to avoid the possibility that the mutant proteins simply would be compromised in membrane integration. Although many mutants behaved like the wild-type protein or had moderately reduced fusion activity (e.g., L445A or A449L, respectively), the G444L and L463A mutants were almost inactive (Fig. 6D; see Fig. S12 for the effect of all mutations tested). Both the A449L and L463A mutants also were defective in vivo, because the corresponding human ATL1 mutants (A474L and L488A) did not restore normal ER morphology in yeast cells lacking Sey1p and Yop1p (Fig. 5). Taken together, these results indicate that the specific amino acid sequence of the TM segments is important for their function in membrane fusion.
One potential function for the TMs is to mediate the oligomerization of ATL molecules, a possibility suggested by the observation that the overexpression of a fragment containing the TMs and CT of ATL causes a dominant-negative effect on ER morphology (9). Indeed, ATL molecules form oligomers in digitonin, but they dissociate in SDS or Triton X-100 (Fig. S13). This oligomerization is not mediated by the GTPase domains, because it occurs even when nucleotide binding is prevented by the addition of EDTA.
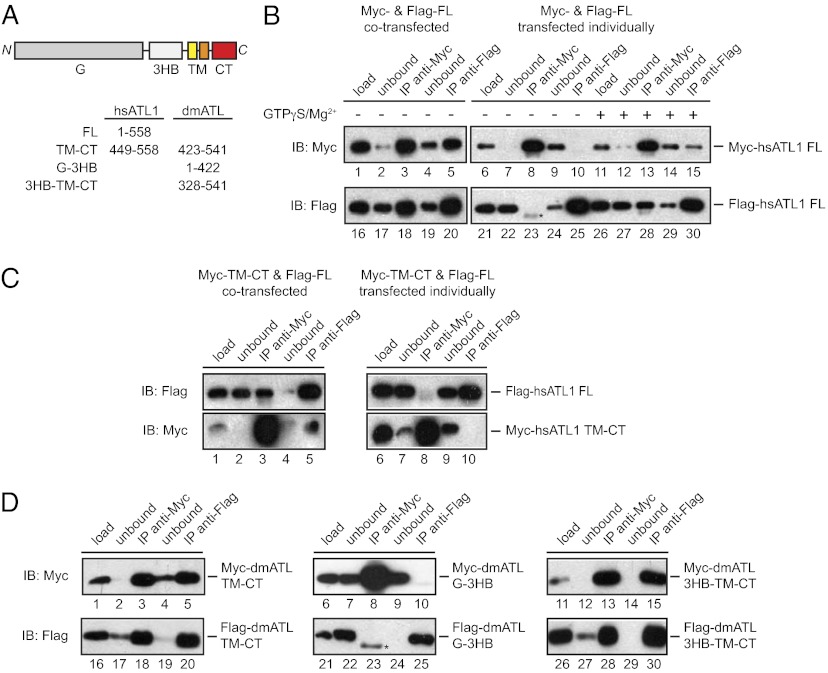
To test directly whether the TM segments of ATL interact within the membrane, we performed coimmunoprecipitation experiments. As described previously (12, 27), when Myc- and Flag-tagged full-length human ATL1 (Myc-/Flag-hsATL1 FL) was cotransfected into COS-7 cells, Myc antibodies were able to precipitate Flag-hsATL1 (Fig. 7 A and B, lane 18), and, conversely, Flag antibodies could precipitate Myc-hsATL1 (Fig. 7B, lane 5). EDTA was included during the incubation to prevent nucleotide binding and thus coimmunoprecipitation caused by dimerization through the GTPase domains. In contrast, when Myc- and Flag-tagged proteins were transfected individually into COS-7 cells, and the extracts were mixed, no coimmunoprecipitation was observed (Fig. 7B, lanes 10 and 23). The interaction between ATL molecules was restored when GTPγS and MgCl2 were added to allow GTP-dependent dimerization of ATL molecules (Fig. 7B, lanes 15 and 28). These results indicate that nucleotide-independent oligomerization requires that both partners reside in the same membrane. An Myc-tagged construct containing only the TMs and CT of human ATL1 (hsATL1 TM-CT) (Fig. 7A) also interacted with Flag-hsATL1 FL when the proteins were expressed in the same cells but not in different cells (Fig. 7C, lanes 3 and 5 versus lanes 8 and 10). Taken together, these results suggest that the TMs mediate the nucleotide-independent association of ATL molecules.
Fig. 7.
The TMs of ATL mediate nucleotide-independent oligomerization. (A) Domain structure of ATL. The residue numbers of constructs used in coimmunoprecipitation are listed. dmATL, Drosophila melanogaster ATL; hsATL1, Homo sapiens ATL1. (B) Myc- and Flag-tagged hsATL1 were cotransfected into COS-7 cells and solubilized in digitonin or were transfected individually followed by mixing of the digitonin-solubilized cell extracts. Immunoprecipitation (IP) was performed with anti-Myc or anti-Flag antibodies. When indicated, 1 mM GTPγS and 5 mM MgCl2 were added. The samples were analyzed by SDS/PAGE and immunoblotting (IB) with anti-Myc or anti-Flag antibodies. Ten percent of the starting material (load) and of the material not bound to the antibodies (unbound) was analyzed also. (C) As in B, but with COS-7 cells expressing Myc-hsATL1 TM-CT and Flag-hsATL1. (D) As in B, but with COS-7 cells expressing Myc- and Flag-tagged dmATL constructs. A contaminating Ig band from the antibody-conjugated matrix used in the IP is indicated by an asterisk.
A recent report suggested that the 3HB, and not the TMs, are involved in oligomerization of Drosophila ATL (28). In agreement with this report, we found that Myc- and Flag-tagged constructs containing the 3HB, TMs, and CT associated with one another (Fig. 7D, lanes 28 and 30). However, in contrast to Pendin et al. (28), we found robust self-association with a construct containing only the TMs and CT (Fig. 7D, lanes 18 and 20). This self-association, together with evidence that the CT does not interact with other domains of ATL, suggests that the TMs indeed can mediate oligomerization. Furthermore, although Pendin et al. (28) found some self-association with a fragment containing the GTPase (G) domain and the 3HB, the interaction was very weak in their hands and was negligible in ours (Fig. 7D, lanes 23 and 25), suggesting only a minor contribution by the 3HB. Thus, our data indicate that the TMs are the major requirement for nucleotide-independent oligomerization of ATL molecules.
Discussion
Our results provide important insight into the mechanism of ATL-mediated homotypic ER fusion. We show that the previously identified GTPase-induced conformational change of the cytosolic domain is insufficient to achieve efficient fusion. Rather, both the CT and the TM segments play important and specific roles in membrane fusion.
The crucial segment of the CT is a conserved amphipathic helix that follows immediately after the second TM segment. Although the amphipathic helix of the CT is not absolutely essential for fusion, it is physiologically important, because ATL mutants lacking the CT cause HSP (29–31), and tailless human ATL and a point mutant in the C-terminal helix are defective in maintaining ER morphology in S. cerevisiae. Our results show that mutations in the helix dramatically reduce fusion activity and that the fusion defect of an ATL mutant lacking the C terminus is rescued when a synthetic peptide of the C-terminal amphipathic helix is added in trans. The peptide stimulates genuine GTP-dependent fusion of tailless ATL, as shown by lipid mixing in both leaflets of the bilayer, by a size increase of the proteoliposomes that was not caused by mere tethering of the vesicles, and, most importantly, by mixing of vesicle contents. The C-terminal helix likely does not function by interacting with other domains of the ATL molecule, because a CTH composed of d-amino acids also stimulates fusion in trans. In addition, the CTH stimulates the fusion of wild-type ATL, whereas an inhibitory effect might have been expected if it blocked the endogenous sequence from interacting with another domain of ATL. Using various biophysical assays, we show that instead the CTH interacts with lipids and becomes more helical upon contact with the lipid bilayer. The relevance of lipid interaction is supported by the observation that mutations in the CT of full-length ATL that reduce fusion activity also affect lipid binding of the CTH. Given that both the N and C termini of the helix are on the cytosolic side of the membrane (12, 27), and that the most conserved part of the helix comprises only ∼15 amino acids, it seems likely that the helix sits in the cytosolic leaflet, with its hydrophobic residues inserted into the bilayer.
The C-terminal helix probably does not promote fusion by forming a lipidic pore like melittin or by generating membrane curvature like the amphipathic helices in Sar1p and N-BAR, because these peptides do not mimic the activity of the CTH in the lipid-mixing assay. The ATL helix may function by destabilizing the lipid bilayer, because the CTH releases calcein from the vesicle interior but fusion-defective mutants do not. Although the exact mechanism by which the ATL helix disturbs the integrity of the lipid bilayer is unclear, we speculate that it displaces negatively charged phospholipid head groups and exposes the hydrocarbon chains of lipid molecules at the membrane surface, a perturbation that would lower the energy barrier for fusion between the approaching bilayers.
The fact that the C-terminal helix destabilizes the lipid bilayer raised the question of whether ATL mediates genuine fusion or merely rupture and reannealing of the lipid bilayer. Here, we show that, indeed, wild-type ATL causes GTP-dependent mixing of vesicle contents without significant lysis. Even the combination of tailless ATL mutant and CTH mediates true fusion, although some leakiness was observed, consistent with the calcein release experiments. The increased lysis is likely caused by the high concentration of the CTH in this reaction; in wild-type ATL, the helix would be linked covalently to the rest of the protein and would act only locally at the site of fusion. The content-mixing data provide strong evidence that ATL is capable of mediating complete membrane fusion without significantly compromising the permeability barrier of the lipid bilayer. Maintaining the barrier during fusion would be important to retain luminal ER proteins and maintain stores of calcium ions in the ER lumen.
Our results indicate that the TM segments are even more important than the CT for ATL’s function. The TMs are more than just membrane anchors, because they cannot be replaced by unrelated TMs. In addition, even fairly conservative point mutations compromise fusion, highlighting their sequence-specific function. Several of these mutants also are defective in maintaining ER morphology in S. cerevisiae. One role for the TMs is to mediate nucleotide-independent oligomerization of ATL molecules, as shown by coimmunoprecipitation and sucrose gradient centrifugation experiments. We propose that the association between the TM segments clusters ATL molecules in the membrane before fusion.
Our results suggest a refined model for ATL-mediated membrane fusion in which the CT and TMs of ATL cooperate with the N-terminal cytosolic domain. First, several ATL molecules in a membrane would associate with each other through their TM segments. Second, these complexes would interact with similarly assembled ATL molecules in another membrane; the interaction of ATL molecules across the two membranes would require GTP binding. It also is conceivable that the first and second steps are coordinated rather than occurring in a strictly consecutive manner. Third, GTP hydrolysis and phosphate release would trigger a conformational change that would pull the membranes toward each other for fusion. Because the energetic gain from the conformational change of a single pair of opposing ATL molecules may be relatively small, we speculate that nucleotide-independent oligomerization of ATL molecules would increase the efficiency of fusion by allowing several ATL molecules in each membrane to undergo synchronously the conformational changes leading to fusion. Local perturbation of the membrane bilayer by the CT also could contribute to the process by lowering the energy barrier for the approach and eventual merging of the membranes. Finally, once fusion has been completed and the postfusion conformation is reached, GDP would be released, allowing the nucleotide-dependent ATL dimers to dissociate and to start a new round of fusion.
It has been suggested that fusion involves the oligomerization of ATL molecules in different membranes through their 3HBs (20, 28). Although it is possible that an interaction between the 3HBs of opposing ATL molecules occurs during the nucleotide-dependent conformational change, our results show that the 3HB plays a minor or negligible role in the nucleotide-independent oligomerization of ATL molecules. Rather, the TMs mediate this association of ATL molecules in the same membrane before nucleotide binding.
Our results on the mechanism of ATL-mediated fusion may be relevant to other fusion reactions as well. As in the ATLs, the assembly of the soluble domains of t- and v-SNAREs results in a relatively modest energy gain (32), indicating that other mechanisms, such as those described here for ATL, also may be necessary for efficient fusion. In fact, the v-SNARE protein synaptobrevin possesses a short helix adjacent to the TM segment that is believed to interact with the lipid bilayer before fusion (33, 34), similar to the C-terminal amphipathic helix of ATL. In flavivirus fusion proteins, a conserved amphipathic helix immediately preceding the TM segments also is postulated to interact with lipids at early phases of the fusion process (35, 36). The ability to use a synthetic peptide to activate fusion in trans allowed us to provide the best evidence yet that amphipathic helices affect membrane fusion by inserting into the lipid phase.
In other fusion reactions, the TM segments also may be more than simply membrane anchors. The TM of the SNARE protein synaptobrevin appears to sit in the membrane at an angle and is believed to destabilize the lipid bilayer (34, 37), a result that is supported by experiments with a synthetic peptide (38). In addition, point mutations in the TMs of both syntaxin and synaptobrevin compromise fusion (34, 39), although perhaps not as strongly as mutations in ATL. Oligomerization of SNARE proteins through their TM regions also has been reported, similar to our findings for ATL (for review, see ref. 40). In the case of viral fusion proteins, specific TM requirements vary (40). Replacing the TM of the HIV-1 gp41 protein with some unrelated TMs caused a reduction in fusion efficiency (41), whereas the replacement with another TM had no effect (42). In the case of vesicular stomatitis virus G protein (43) or influenza virus hemagglutinin (44), other TMs worked just as well as the native TM. On the other hand, point mutations in the TMs of these fusion proteins can reduce their activity (45, 46). Thus, it seems that often the TMs in other fusion reactions also may play a specific role, although perhaps they are not as crucial as in ATL.
Finally, we propose that the mechanism of ATL-mediated fusion is shared by related GTPases involved in homotypic fusion. The functional orthologs of ATL in yeast and plants (Sey1p in S. cerevisiae and RHD3 in A. thaliana) have similar GTPase domains, a helical domain, and two closely spaced TM segments (9). They also contain a conserved amphipathic helix immediately following the second TM segment. The mitofusin/Fzo1p involved in the fusion of the outer mitochondrial membrane also has a related GTPase domain, a coiled-coil domain, and two closely spaced TM segments. Although the overall mechanism therefore may be the same for all these GTPases, mitofusin/Fzo1p contains C-terminal heptad repeats instead of an amphipathic helix, and these repeats have been proposed to form antiparallel interactions with a mitofusin/Fzo1p molecule in the apposed membrane (47). This difference indicates that there may be variations in the common theme of homotypic membrane fusion.
Materials and Methods
Lipid-Mixing Assay.
Full-length, codon-optimized Drosophila ATL was expressed in Escherichia coli as a GST fusion and was purified on glutathione agarose. The GST moiety was cleaved off by thrombin and removed with glutathione agarose. Detergent-mediated reconstitution was used to integrate ATL into preformed donor liposomes [82:15:1.5:1.5 mole percent of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC):1,2-dioleoyl-sn-glycero-3-phosphoserine (DOPS):NBD-1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE):rhodamine-DPPE] and acceptor liposomes (83.5:15:1.5 mole percent of POPC:DOPS:dansyl-DPPE) as described (10). The fusion assays were performed as described previously (13). For dithionite quenching, 1 μL 100 mM dithionite in buffer [25 mM Hepes (pH 10), 100 mM KCl, 10% (vol/vol) glycerol] was added during the lipid-mixing assay when indicated. The initial background fluorescence was subtracted from the raw fluorescence readings, and these values are expressed as percentages of the maximum fluorescence after detergent addition.
Content-Mixing Assay.
Fusion reactions were carried out as for lipid-mixing experiments, except that RPE-biotin was incorporated into the lumen of donor vesicles, and SA-Cy5 was incorporated into the lumen of acceptor vesicles (26). Content mixing and leakage were determined by FRET between RPE-biotin and SA-Cy5, and content mixing only was determined after the addition of BDA with a molecular mass of 70,000 Da to the outside of the vesicles, as previously described (26). Lipid mixing was determined in parallel by dequenching of the fluorescence of Marina Blue-PE (26). Dansyl-DPPE (excitation at 336 nm and emission at 517 nm) was included in the lipid mix of the acceptor vesicles to determine total lipid concentration, but it did not interfere with the fusion assays. All measurements were made using a SpectraMax M5 Microplate Reader (Molecular Devices, LLC). Thesit (C12E9) was added to reactions lacking BDA to determine the maximum fluorescence. The initial background fluorescence was subtracted from the raw fluorescence readings, and these values are expressed as percentages of the maximum fluorescence after detergent addition.
Dynamic Light Scattering.
Fusion reactions using proteoliposomes containing tailless or wild-type ATL were carried out using the procedures and conditions used in the lipid-mixing experiments. Peptide (15 μM) was added 10 min before GTP addition, where indicated. EDTA-containing buffer was added to stop vesicle fusion before size analysis. The mean effective hydrodynamic radii of proteoliposomes were determined using a DynaPro Nanostar instrument (Wyatt).
Doxyl-Quenching Assay.
Peptide was mixed with liposomes (84.5:15:0.5 mole percent POPC:DOPS:NBD-DPPE) at a peptide:lipid molar ratio of 1:40, and the fluorescence of Trp was measured. Emission intensity at the peak maximum in the presence of liposomes with or without doxyl-PC was used to determine the extent of quenching. Experiments were performed at room temperature on the SpectraMax M5 Microplate Reader (Molecular Devices).
Circular Dichroism.
Circular dichroism experiments were performed on a Jasco J-815 instrument at 25 °C. Where indicated, 30 μM peptide in 10 mM potassium phosphate (pH 7.5), 100 mM KCl, and liposomes (85:15 mole percent POPC:DOPS, final concentration of 1 mM lipids) were included. Spectra were collected from 200–260 nm at a bandwidth of 1 nm and a scan speed of 100 nm/min.
Calcein-Leakage Assay.
Liposomes (84.5:15:0.5 mole percent POPC:DOPS:Texas Red-DPPE) were prepared as described previously (10), except that calcein was encapsulated in the liposomes at a concentration of 100 mM. Then 2 μM peptide was added to 0.1 mM lipids in a 96-well plate, and the fluorescence (excitation at 490 nm, emission at 520 nm) was monitored at room temperature using a SpectraMax M5 Microplate Reader (Molecular Devices).
Fluorescence Microscopy in Yeast.
Yeast cells lacking SEY1 and YOP1 were cultured and visualized as previously described (9).
Mammalian Tissue Culture, Transfection and Coimmunoprecipitation.
COS7 Cells were maintained at 37 °C with 5% CO2 in DMEM containing 10% (vol/vol) FBS and were transfected using FuGENE HD (Roche). Coimmunoprecipitation was performed as described previously (9).
Further details on methods are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank A. Stein for helpful suggestions and R. King and A. Stein for critical reading of the manuscript. T.Y.L. is supported by a fellowship from the National Science Foundation, and R.W.K is supported by a European Molecular Biology Organization Long-Term Fellowship. W.A.P. is supported by the National Institute of Diabetes and Digestive and Kidney Diseases intramural program. T.A.R. is a Howard Hughes Medical Institute investigator. J.H. is supported by National Basic Research Program of China 973 Program, Grant 2010CB83370; National Science Foundation of China Grant 3097144; and an International Early Career Scientist grant from the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 12850 (volume 109, number 32).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208385109/-/DCSupplemental.
References
- 1.Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol. 2001;205:149–214. doi: 10.1016/s0074-7696(01)05004-5. [DOI] [PubMed] [Google Scholar]
- 2.Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu Rev Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 3.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sapir A, Avinoam O, Podbilewicz B, Chernomordik LV. Viral and developmental cell fusion mechanisms: Conservation and divergence. Dev Cell. 2008;14:11–21. doi: 10.1016/j.devcel.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 6.Martens S, McMahon HT. Mechanisms of membrane fusion: Disparate players and common principles. Nat Rev Mol Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 7.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 11.Zhao X, et al. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet. 2001;29:326–331. doi: 10.1038/ng758. [DOI] [PubMed] [Google Scholar]
- 12.Rismanchi N, Soderblom C, Stadler J, Zhu PP, Blackstone C. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum Mol Genet. 2008;17:1591–1604. doi: 10.1093/hmg/ddn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bian X, et al. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc Natl Acad Sci USA. 2011;108:3976–3981. doi: 10.1073/pnas.1101643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salinas S, Proukakis C, Crosby A, Warner TT. Hereditary spastic paraplegia: Clinical features and pathogenetic mechanisms. Lancet Neurol. 2008;7:1127–1138. doi: 10.1016/S1474-4422(08)70258-8. [DOI] [PubMed] [Google Scholar]
- 15.Zheng H, Kunst L, Hawes C, Moore I. A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J. 2004;37:398–414. doi: 10.1046/j.1365-313x.2003.01969.x. [DOI] [PubMed] [Google Scholar]
- 16.Anwar K, et al. The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J Cell Biol. 2012;197:209–217. doi: 10.1083/jcb.201111115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermann GJ, et al. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byrnes LJ, Sondermann H. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc Natl Acad Sci USA. 2011;108:2216–2221. doi: 10.1073/pnas.1012792108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moss TJ, Andreazza C, Verma A, Daga A, McNew JA. Membrane fusion by the GTPase atlastin requires a conserved C-terminal cytoplasmic tail and dimerization through the middle domain. Proc Natl Acad Sci USA. 2011;108:11133–11138. doi: 10.1073/pnas.1105056108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 22.Gallop JL, et al. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee MC, et al. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 24.Peter BJ, et al. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 25.Raghuraman H, Chattopadhyay A. Melittin: A membrane-active peptide with diverse functions. Biosci Rep. 2007;27:189–223. doi: 10.1007/s10540-006-9030-z. [DOI] [PubMed] [Google Scholar]
- 26.Zucchi PC, Zick M. Membrane fusion catalyzed by a Rab, SNAREs, and SNARE chaperones is accompanied by enhanced permeability to small molecules and by lysis. Mol Biol Cell. 2011;22:4635–4646. doi: 10.1091/mbc.E11-08-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu PP, et al. Cellular localization, oligomerization, and membrane association of the hereditary spastic paraplegia 3A (SPG3A) protein atlastin. J Biol Chem. 2003;278:49063–49071. doi: 10.1074/jbc.M306702200. [DOI] [PubMed] [Google Scholar]
- 28.Pendin D, et al. GTP-dependent packing of a three-helix bundle is required for atlastin-mediated fusion. Proc Natl Acad Sci USA. 2011;108:16283–16288. doi: 10.1073/pnas.1106421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tessa A, et al. SPG3A: An additional family carrying a new atlastin mutation. Neurology. 2002;59:2002–2005. doi: 10.1212/01.wnl.0000036902.21438.98. [DOI] [PubMed] [Google Scholar]
- 30.Ivanova N, et al. Hereditary spastic paraplegia 3A associated with axonal neuropathy. Arch Neurol. 2007;64:706–713. doi: 10.1001/archneur.64.5.706. [DOI] [PubMed] [Google Scholar]
- 31.Loureiro JL, et al. Novel SPG3A and SPG4 mutations in dominant spastic paraplegia families. Acta Neurol Scand. 2009;119:113–118. doi: 10.1111/j.1600-0404.2008.01074.x. [DOI] [PubMed] [Google Scholar]
- 32.Wiederhold K, Fasshauer D. Is assembly of the SNARE complex enough to fuel membrane fusion? J Biol Chem. 2009;284:13143–13152. doi: 10.1074/jbc.M900703200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kweon DH, Kim CS, Shin YK. Regulation of neuronal SNARE assembly by the membrane. Nat Struct Biol. 2003;10:440–447. doi: 10.1038/nsb928. [DOI] [PubMed] [Google Scholar]
- 34.Xu Y, Zhang F, Su Z, McNew JA, Shin YK. Hemifusion in SNARE-mediated membrane fusion. Nat Struct Mol Biol. 2005;12:417–422. doi: 10.1038/nsmb921. [DOI] [PubMed] [Google Scholar]
- 35.Allison SL, Stiasny K, Stadler K, Mandl CW, Heinz FX. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J Virol. 1999;73:5605–5612. doi: 10.1128/jvi.73.7.5605-5612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang W, et al. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol. 2003;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowen M, Brunger AT. Conformation of the synaptobrevin transmembrane domain. Proc Natl Acad Sci USA. 2006;103:8378–8383. doi: 10.1073/pnas.0602644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langosch D, et al. Peptide mimics of SNARE transmembrane segments drive membrane fusion depending on their conformational plasticity. J Mol Biol. 2001;311:709–721. doi: 10.1006/jmbi.2001.4889. [DOI] [PubMed] [Google Scholar]
- 39.Han X, Wang CT, Bai J, Chapman ER, Jackson MB. Transmembrane segments of syntaxin line the fusion pore of Ca2+-triggered exocytosis. Science. 2004;304:289–292. doi: 10.1126/science.1095801. [DOI] [PubMed] [Google Scholar]
- 40.Langosch D, Hofmann M, Ungermann C. The role of transmembrane domains in membrane fusion. Cell Mol Life Sci. 2007;64:850–864. doi: 10.1007/s00018-007-6439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyauchi K, et al. Role of the specific amino acid sequence of the membrane-spanning domain of human immunodeficiency virus type 1 in membrane fusion. J Virol. 2005;79:4720–4729. doi: 10.1128/JVI.79.8.4720-4729.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilk T, Pfeiffer T, Bukovsky A, Moldenhauer G, Bosch V. Glycoprotein incorporation and HIV-1 infectivity despite exchange of the gp160 membrane-spanning domain. Virology. 1996;218:269–274. doi: 10.1006/viro.1996.0190. [DOI] [PubMed] [Google Scholar]
- 43.Odell D, Wanas E, Yan J, Ghosh HP. Influence of membrane anchoring and cytoplasmic domains on the fusogenic activity of vesicular stomatitis virus glycoprotein G. J Virol. 1997;71:7996–8000. doi: 10.1128/jvi.71.10.7996-8000.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozerski C, Ponimaskin E, Schroth-Diez B, Schmidt MF, Herrmann A. Modification of the cytoplasmic domain of influenza virus hemagglutinin affects enlargement of the fusion pore. J Virol. 2000;74:7529–7537. doi: 10.1128/jvi.74.16.7529-7537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melikyan GB, Lin S, Roth MG, Cohen FS. Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion. Mol Biol Cell. 1999;10:1821–1836. doi: 10.1091/mbc.10.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cleverley DZ, Lenard J. The transmembrane domain in viral fusion: Essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc Natl Acad Sci USA. 1998;95:3425–3430. doi: 10.1073/pnas.95.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koshiba T, et al. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]