Abstract
The ubiquitous EBV causes infectious mononucleosis and is associated with several types of cancers. The EBV genome encodes an early gene product, BARF1, which contributes to pathogenesis, potentially through growth-altering and immune-modulating activities, but the mechanisms for such activities are poorly understood. We have determined the crystal structure of BARF1 in complex with human macrophage-colony stimulating factor (M-CSF), a hematopoietic cytokine with pleiotropic functions in development and immune response. BARF1 and M-CSF form a high-affinity, stable, ring-like complex in both solution and the crystal, with a BARF1 hexameric ring surrounded by three M-CSF dimers in triangular array. The binding of BARF1 to M-CSF dramatically reduces but does not completely abolish M-CSF binding and signaling through its cognate receptor FMS. A three-pronged down-regulation mechanism is proposed to explain the biological effect of BARF1 on M-CSF:FMS signaling. These prongs entail control of the circulating and effective local M-CSF concentration, perturbation of the receptor-binding surface of M-CSF, and imposition of an unfavorable global orientation of the M-CSF dimer. Each prong may reduce M-CSF:FMS signaling to a limited extent but in combination may alter M-CSF:FMS signaling dramatically. The downregulating mechanism of BARF1 underlines a viral modulation strategy, and provides a basis for understanding EBV pathogenesis.
Keywords: crystallography, immune modulation, receptor tyrosine kinase
EBV is a ubiquitous human virus and persists as a latent infection in more than 90% of the world population. It is a gamma-1 herpesvirus that preferentially infects B lymphocytes and epithelial cells, resulting in dysregulation of these cell types in certain hosts. Although best known as the cause of infectious mononucleosis upon primary infection, EBV also is associated with several types of cancer, including Burkitt lymphoma, nasopharyngeal carcinoma (NPC), and gastric carcinoma (1, 2). EBV also may be involved in the pathogenesis of some autoimmune diseases such as multiple sclerosis and systemic lupus erythematosus (1, 2). The role of EBV and the mechanism of host cell dysregulation in cancers and autoimmune diseases remain poorly understood (1, 2).
To understand the viral–host interaction that contributes to the pathogenesis caused by EBV, extensive efforts have been allocated to study proteins encoded by the EBV genome. Among these proteins, an early gene product called “BamHI-A Rightward Frame 1” (BARF1) has been shown to have both immortalizing and transforming activities (3–6) but more recently has been shown to be nonessential for EBV transformation of B cells (7). Interestingly, mutation of the BARF1 gene in the EBV-related rhesus lymphocryptovirus results in a modest reduction in transformation frequency, but the mechanism for this observed reduction is not clear (8). BARF1 is expressed in a high proportion of NPC cases and EBV-positive gastric carcinomas (9–11), and higher levels of BARF1 antibodies are found in NPC patients (12). Recent studies have indicated that BARF1 modulates the host immune response to infection (13). BARF1 has been found to be a functional homolog of the receptor tyrosine kinase colony-stimulating factor receptor (c-FMS or FMS), competing for its ligand, macrophage colony-stimulating factor (M-CSF, or CSF-1) (13). M-CSF is a hematopoietic growth factor involved in the proliferation, differentiation, and survival of monocytes, macrophages, and bone marrow progenitor cells (14). It signals through the cell-surface receptor FMS, which is a class III receptor tyrosine kinase featuring five extracellular Ig-like domains, with the membrane-distal domains responsible for ligand recognition and the membrane-proximal domains involved in homotypic interactions (15, 16). The interaction between BARF1 and M-CSF suggests that BARF1 may work functionally as an immunomodulator. Indeed, BARF1 inhibits IFN-a production by mononuclear cells, likely by binding to M-CSF and reducing the effect of M-CSF on the proliferation of macrophages (7).
Other studies have suggested that BARF1 may work as a survival factor, most likely by inhibiting apoptosis (17), and it also was suggested that the antiproliferative action of BARF1 may be caused by sequestration of the ligand by binding to the receptor-binding site of the ligand (13, 18). In all functional roles, it is suggested that BARF1 binds M-CSF competitively, and in fact it has been demonstrated that BARF1 interacts with all isoforms of M-CSF (soluble or membrane spanning) (13).
BARF1 is a secreted glycoprotein composed of Ig-like domains. The crystal structure of free BARF1 has been determined and shows that BARF1 oligomerizes into a hexameric ring (18), but how the hexameric BARF1 engages M-CSF, a dimeric cytokine with a four-helix bundle scaffold (16, 19), has been a perplexing question. How the competition between BARF1 and FMS, the cognate M-CSF receptor, is achieved also has been unclear. In this study we show that BARF1 and M-CSF form a high-affinity hexameric complex that dramatically reduces the M-CSF:FMS interaction and signaling, providing further evidence that BARF1 may function as an immune modulator via M-CSF. A high-resolution structure of the BARF1:M-CSF complex, encompassing the hexameric BARF1 ring and three M-CSF dimers in a distant, triangular array, suggests a multipronged approach used by EBV to down-regulate M-CSF:FMS–mediated immune signaling and cell growth.
Results and Discussion
BARF1 and M-CSF Form a High-Affinity, High-Molecular-Weight Complex in Solution.
BARF1 and M-CSF have been shown to bind each other in transfected cells (13), but the binding has not been characterized biochemically. To test if they form a stable complex in vitro, we expressed both proteins in HEK293 cells using the BacMam system (20). The complex was reconstituted by mixing BARF1 with an excess of human M-CSF and was purified with size-exclusion chromatography. In a calibrated size-exclusion column BARF1 and M-CSF form a stoichiomic, stable complex that is clearly separated from and appears much larger than the excess M-CSF protein, which has a dimeric size of ∼30 kDa (Fig. S1A). The apparent size of the complex is ∼250 kDa, corresponding to six or seven copies of BARF1 (monomeric size ∼25 kDa) and M-CSF (monomeric size ∼15 kDa) each. Given the hexameric structure of unliganded BARF1 (18), we interpret the stoichiometry of the complex as 6:6, encompassing the BARF hexamer and six copies of M-CSF monomers.
To quantitate the binding between BARF1 and M-CSF, we used isothermal titration calorimetry (ITC) to measure the thermodynamic profiles of the formation of the complex (Fig. S1B). BARF1 was saturated by the injection of small pulses of M-CSF in a steep sigmoid fashion, indicating an extremely high affinity in the picomolar range.
BARF1 Reduces M-CSF:FMS Binding.
Because BARF1 acts as an M-CSF antagonist (13), we asked if the antagonism was mediated by M-CSF’s cognate receptor FMS. To test if the binding of BARF1 to M-CSF interferes with the binding of M-CSF to FMS, we mixed the three proteins together in gel filtration to analyze whether a stable ternary BARF1:M-CSF:FMS complex formed in solution. We used the entire extracellular segment (ECD) or Ig domains 1–5 (D1–D5) of mouse FMS rather than human FMS, because human FMS–D1–D5 forms aggregates in experimental conditions that mimic the physiological pH and ion strength [Hepes-buffered saline (HBS) with 10 mM Hepes (pH7.5), and 150 mM NaCl]. Because the affinity of mouse M-CSF to mouse FMS–D1–D5 (16) is similar to the affinity of human M-CSF to mouse FMS–D1–D5 (Fig. 1A), the species difference is unlikely an important factor in M-CSF:FMS binding. When both BARF1 and mouse FMS–D1–D5 were mixed with human M-CSF, we observed no ternary complex; only a binary complex between BARF1 and M-CSF was present in solution (Fig. S2). This result suggests that, in the presence of BARF1, the binding between M-CSF and FMS either is prohibited or is weakened to the point that the complex is not held together in gel filtration.
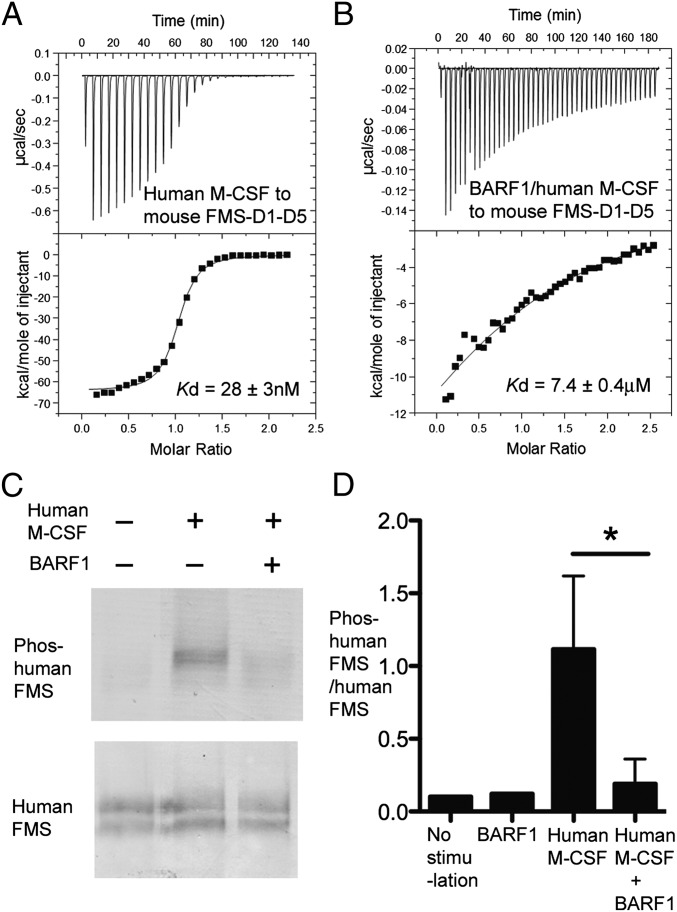
Fig. 1.
BARF1 reduces M-CSF:receptor binding and M-CSF-induced receptor phosphorylation. (A and B) Calorimetric measurements of the binding between human M-CSF (A) and mouse FMS (B) ECD in the absence vs. presence of BARF1. The affinities are shown by the fitted curves. (C) Representative blot of cell-based phosphorylation assays showing that human M-CSF induces the phosphorylation of human FMS Tyr723 much more efficiently in the absence than in the presence of BARF1. COS7 cells expressing human FMS were stimulated with 75 nM of M-CSF or BARF1:M-CSF for 2 min. Cell lysates were analyzed with antibodies against M-CSF receptor and phospho-M-CSF receptor (Tyr723) (D) Quantification of the cell-based phosphorylation assay represented in C. Blots were scanned, and band intensities were quantified using ImageJ 1.45p software. Data are represented as ratios of Tyr723-phosphorylated human FMS to total human FMS levels as challenged by different stimuli, showing that BARF1 efficiently reduces M-CSF signaling but does not reduce signaling to the basal level when no ligand is present. Data for “No stimulation” and “BARF1” represent one experiment; the remaining data represent the mean and SD of three independent experiments. *P < 0.05 using Student’s t test.
To quantify the effect of BARF1 antagonism, we compared the binding of M-CSF to FMS and the binding of BARF1-bound M-CSF (BARF1:M-CSF) to FMS, using ITC (Fig. 1 A and B). In the absence of BARF1, human M-CSF bound mouse FMS–D1–D5 with a high affinity in the nanomolar range, albeit significantly lower than the picomolar affinity between BARF1 and M-CSF. In the presence of BARF1, however, this binding was reduced ∼300-fold to the micromolar affinity range. It should be noted that the BARF1:M-CSF complex does bind to FMS does exist, albeit with a much lower affinity, indicating that the BARF1:M-CSF binding and the FMS:M-CSF binding may be only partially incompatible rather than mutually exclusive.
BARF1 Down-Regulates M-CSF–Induced FMS Phosphorylation.
Previous studies demonstrated that in bone marrow macrophage nonadherent proliferation assays BARF1 protein was able to neutralize the effects of human M-CSF efficiently in a dose-dependent manner (13). However, it was unclear whether this antagonism had a direct effect on FMS signaling. To test if BARF1 acts on M-CSF:FMS signaling, we compared M-CSF–stimulated FMS tyrosine phosphorylation in the absence and in the presence of BARF1, using a cell-based assay. In human FMS-expressing COS7 cells, M-CSF stimulated FMS Tyr723 phosphorylation in an immediate and direct fashion. In comparison, the same concentration of BARF1:M-CSF complex stimulated FMS Tyr723 phosphorylation much less efficiently (Fig. 1 C and D). Nevertheless, the BARF1:M-CSF complex was able to induce FMS phosphorylation, and the level of receptor phosphorylation was slightly higher than in the control with no M-CSF stimulation (Fig. 1D). Similar results also were observed in human or mouse FMS-expressing HEK293H cells (Fig. S3), a result that is consistent with the results of calorimetry binding showing that BARF1 dramatically weakens but does not abolish the M-CSF:FMS interaction.
Structure of the BARF1:M-CSF Complex.
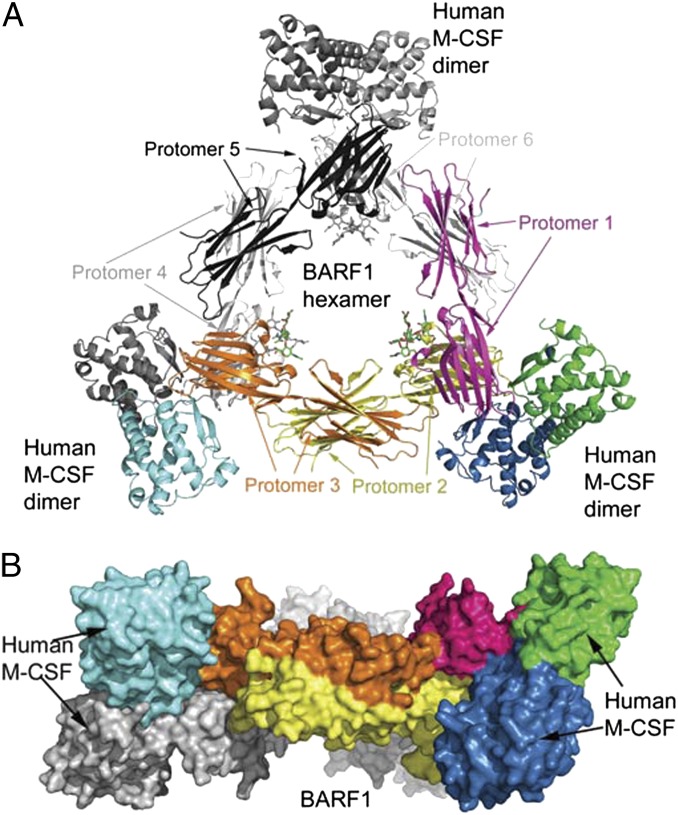
To elucidate how BARF1 interacts with M-CSF, we crystallized the BARF1: human M-CSF complex and determined its structure at 2.2-Å resolution using molecular replacement (Table S1). The structure contained three molecules of BARF1 and three molecules of M-CSF (Fig. 2) in the asymmetric unit, corresponding to half of a hexameric complex. Using the crystallographic twofold symmetry operation, we constructed a complete hexameric assembly, encompassing six BARF1 protomers and six M-CSF protomers (Fig. 2A). The ring-shaped BARF1 hexamer is located in the middle, creating a spacious central channel in the complex that is partially filled by the N-linked glycans extending from the BARF1 Asn95 site. The six M-CSF protomers, divided as three homodimers, surround the BARF hexameric ring at three distant corners, with the longest dimension of each dimer tilted ∼60° to the BARF1 hexameric ring (Fig. 2B).
Fig. 2.
Structure of the 6:6 BARF1:M-CSF complex. (A) Ribbon model of the three M-CSF dimers (corners) bound to the BARF1 hexamer (center). The molecules in the asymmetric unit, corresponding to half the complex, are colored yellow, orange, and magenta for BARF1 and blue, green, and cyan for M-CSF. The other half of the complex, in gray, can be generated with the crystallographic twofold symmetry. N-linked glycans in the complex are depicted as sticks. (B) Surface representation of the complex, in an orientation 90° relative to A.
The BARF1 hexamer in the complex is similar to the free BARF1 structure [Protein Data Bank (PDB) ID code 2CH8], in which each BARF1 protomer contains two Ig-like domains (Ig1 and Ig2) joined together tightly with a hydrophobic boundary (18). The ring is formed by the N-terminal Ig1 docking face-to-face on the Ig1 of a neighboring BARF1 protomer and the C-terminal Ig2 docking side-to-side on the Ig2 of another neighboring BARF1 protomer. The BARF1 hexamer is a rigid entity, reinforced by a two-layer ring structure, with each layer consisting of three molecules of BARF1 and the two layers stacking on top of each other. Consequently, there are only a few structural differences between the unbound and bound BARF1, the largest change being the conformation of a loop that is involved in BARF1:M-CSF interaction (discussed below) (Fig. S4). The M-CSF molecules in the complex are grouped into disulfide-linked, head-to-head dimers similar to the free and the FMS-bound M-CSF structures (PDB ID codes 1HMC and 3EJJ) (16, 19), and the core of each protomer is composed of four helices (αA, αB, αC, and αD) and two β-strands (β1 and β2). In the BARF bound form, only helix αB shows a significant conformational change (discussed below).
Each M-CSF dimer uses its top, membrane-distal face (assuming a signaling orientation when it dimerizes FMS; in this orientation, the two β-strands are at the top), around its dimerization seam, to associate with one of the three tips of the BARF hexameric ring, each tip consisting of the dimerization seam of two neighboring BARF1 Ig1 domains. The overall shape of the hexameric complex therefore is a triangle, placing the three M-CSF dimers at the corners and remote from one another. Although the BARF1 hexamer can rotate freely in solution when it is loaded with circulating M-CSF, it must lie flat, with the ring parallel to the membrane of the M-CSF–expressing cells, to capture the membrane-anchored splice isoforms of M-CSF with all three M-CSF–binding sites loaded.
BARF1:M-CSF Interaction.
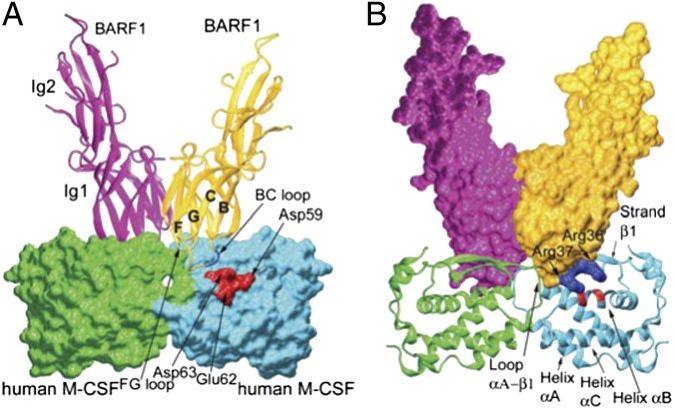
What is the basis for the high affinity between BARF1 and M-CSF? Analysis of the BARF1:M-CSF interface suggests a complex binding arrangement on a largely rigid scaffold. Because both BARF1 and M-CSF use dimerization seams to bind each other, the BARF1:M-CSF interaction is composite: Each BARF1 protomer interacts with two M-CSF protomers, and each M-CSF protomer interacts with two BARF1 protomers. On the BARF1 side, the interface is provided by two protruding ends of the Ig1 domains, with a deep groove separating the two protruding areas, and the M-CSF interface is provided by a relatively flat and continuous face (Fig. 3 and Fig. S5) that fits into the BARF1 groove at the dimerization seam. The total composite interface of BARF1:M-CSF buries ∼1,500 Å2 of solvent-accessible area. For clarification, we describe the interaction of each BARF1 protruding end with two M-CSF protomers, corresponding to about half of each 2:2 composite interface.
Fig. 3.
Interaction between each M-CSF dimer and two BARF1 protomers in the complex. (A) The BARF1 ribbon model on the M-CSF surface model, showing that the Ig1 domains of the BARF-1 molecules (ribbons) clamp the M-CSF dimer (surface) at the mutual dimeric seams. The strands leading to the BC and GF loops are marked as “B,” “C,” “F,” and “G”. (B) The BARF1 surface model on the M-CSF ribbon model, showing the M-CSF dimer fitting into the BARF1 groove. In both panels the clustered charge–charge interactions are shown in blue for positive charges and red for negative charges.
At the interface between each BARF1 and an M-CSF dimer, the BC loop (linking the B- and C-strands) and FG loop (linking the F- and G-strands) of the BARF1 Ig1 domain fit into a slightly concave area formed by helix αB and strand β1 of one M-CSF protomer and the αA–β1 loop of the other M-CSF protomer (Fig. 3). On the BARF1 side, the BC loop is larger and more protruding than the FG loop, providing the majority of the interface area. Notably, the greatest structural difference between free BARF1 and the M-CSF–bound BARF1 is in the BC loop (Figs. S4 and S6). To bind M-CSF, this loop needs to make a conformational change, breaking the hydrophobic interaction between Val38 and Trp35/Leu40/Leu108, and this remodeling of the main-chain conformation (Fig. S6) flips the side chain of Val38 from being mostly buried to being completely exposed, forming hydrophobic interactions with Ile33, Thr29 (atom Cγ2), and Thr64 (Cγ2) of M-CSF. A main-chain hydrogen bond (BARF1 Val38 N–M-CSF Asp63 O) also is formed because of this conformational change, which pulls the end of the M-CSF αB helix toward the center of the dimer (discussed below). Remarkably, the residues near Val38 were predicted successfully to be a putative ligand-binding site for BARF1 by structural comparison alone (18). However, the utilization of the flipped Val38, rather than the exposed Trp35/Val38/Leu40/Leu108 hydrophobic area, as an insert in ligand binding was unexpected.
Except for the small hydrophobic contact surrounding BARF1 Val38 at the center of the interface, the BARF1:M-CSF interaction is predominantly hydrophilic, with a large number of hydrogen bonds and salt bridges (Fig. S5 and Tables S2–S4). The BARF1 FG loop (Ser83-Ala84-Asn85-Thr85) consists of mostly neutral polar residues. The BC loop (Arg36-Arg37-Val38-Ser39) also is mostly hydrophilic. The tip of this loop, BARF1 Arg36-Arg37, is paired with a patch of negatively charged residues on M-CSF (Glu36, Asp59, Glu63, and Asp63), most of which are from M-CSF helix αB, forming a large number of salt bridges. These salt bridges are interdigitated, i.e., M-CSF Glu63 is sandwiched by BARF1 Arg36 and Arg37, and BARF Arg36 is surrounded by M-CSF Glu36, Asp59, and Glu63, greatly increasing the intimacy of this group of charge interactions.
Interestingly, the intermolecular disulfide bridge linking the two–M-CSF monomer, Cys31-Cys31, is located at the geometric center of the 2:2 composite interface and contacts BARF1 Ala84. By associating with the interface, this disulfide bond, serves as a pivot point where the two–M-CSF monomers can bend and rotate as a result of ligand binding, molding into a specific BARF1-induced conformation (discussed below).
Indirect Interference of BARF1 with MCSF:FMS Binding and Signaling Architecture.
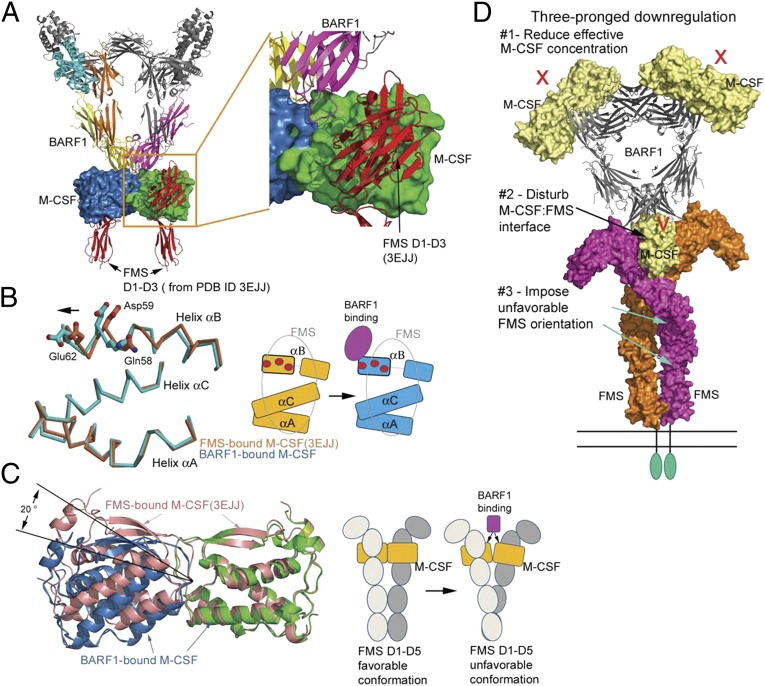
Given that BARF1 reduces the binding between M-CSF and FMS and M-CSF–induced FMS phosphorylation (Fig. 1 and Fig. S3), the question arises how BARF1 structurally disrupts M-CSF:FMS interactions. Intriguingly, when FMS is docked onto the BARF1:M-CSF complex, no overlap between BARF1 and FMS is observed when the M-CSF protomers in the BARF1:M-CSF and FMS:M-CSF complexes are overlaid, although the shortest distance between BARF1 and FMS is only ∼6 Å (Fig. 4A). How does BARF1 reduce FMS-binding to M-CSF without contacting FMS? Comparing the BARF1:M-CSF and FMS:M-CSF structures suggests two significant BARF1-induced conformational changes in M-CSF that can mediate such indirect inhibition (Fig. 4 B and C).
Fig. 4.
Mechanism of BARF1 down-regulation of M-CSF:FMS signaling. (A) Docking FMS to the BARF1:M-CSF complex, based on the previous M-CSF:FMS complex structure (PDB ID code 3EJJ), revealing that there is no overlap between BARF1 and FMS. (B) The conformational change of the M-CSF helix αB upon BARF1 binding. (Left) The helices of BARF1-bound M-CSF (cyan) are superimposed on the helices of FMS-bound M-CSF (salmon), revealing half of the helix αB shifting toward incoming BARF1. The M-CSF residues important for FMS binding are shown as balls and sticks. (Right) Of the three FMS-binding helices of M-CSF, only half of helix αB is shifted by BARF1, creating a partial mismatch with FMS (gray oval). The same set of FMS-binding residues is shown as red circles in the cartoon. (C) The bending of M-CSF dimer by BARF1 and the effect of the bending on FMS conformation. (Left) The BARF1-bound M-CSF dimer (blue and green) is overlaid on the FMS-bound M-CSF dimer (salmon) by superimposing one of their M-CSF protomers. This superimposition reveals that BARF1 forces M-CSF to bend by 20° relative to the center of the dimer, compared with the FMS-bound state. (Right) The cartoons demonstrate the possible effect of such bending on the FMS conformation. (D) A model summarizing the three-pronged approach that BARF-1 may use to down-regulate M-CSF signaling mediated by FMS.
The first conformational change is the shifting of a portion of the FMS-binding area on M-CSF toward BARF1. The shifted area is around the αB helix of M-CSF. This helix is an integral part of the large M-CSF:FMS interface, which includes the αA, αB, and αC helices of M-CSF. αB is the major contact for the second Ig-like domain of FMS, which has proved to be the most important domain for ligand binding in both class III (including KIT, FMS, FLT3, and PDGFRs) (15, 21–23) and the related class V (VEGFRs) receptor tyrosine kinases (24, 25). The BC loop of BARF1 interacts extensively with the M-CSF αB helix via the intimate, interdigitated salt-bridge network (discussed above) and a main-chain hydrogen bond near the end of the helix (BARF1 Val-38 N–M-CSF Asp63 O), and these interactions pull the αB helix, resulting in a shift of the helix at the C-terminal half (residues 58–66) and a kink in the middle of the helix (around residue 55). This change is likely to be detrimental for M-CSF:FMS interaction, because the M-CSF Gln58, Asp59, and Glu62 residues are all FMS-contacting residues (16). Importantly, Asp59 and Glu62, by serving as part of the BARF1:M-CSF interdigitated salt bridge network, become less favorable/available for making salt bridges with FMS both conformationally and electrostatically. Notably, of the three α helices involved in FMS interaction, only the αB helix is shifted in the BARF1-bound M-CSF. Therefore, in the shifted conformation, it would be difficult to keep both the αB and αA/αC interactions with FMS intact simultaneously (Fig. 4B).
The second conformational change is the bending of the M-CSF dimer orientation relative to its signaling orientation. Although M-CSF is a tightly associated dimer crosslinked by a disulfide bond, it shows a propensity for bending upon binding to distinct partners (16). In its noninhibited, receptor-associated orientation, the M-CSF dimer is relatively bent away from the membrane compared with free M-CSF (16). However, in its BARF1-inhibited orientation, the M-CSF dimer is bent toward the membrane, forced by the rigid BARF1 hexamer (Fig. 4C). There is no wiggle room for the bent M-CSF to spring back and assume the ideal FMS-activating orientation, because the 2:2 composite BARF1:M-CSF interface is large and strong (picomolar affinity is relatively rare in protein–protein interactions) and is imprinted on an inelastic BARF1 hexametric scaffold (Fig. S4). The BARF1 binding generates a forceful global effect on the orientation of two M-CSF protomers, and this changed relative position of the two ligand protomers of M-CSF dimers likely would perturb the orientation of two FMS protomers, leading to attenuated signaling potency of the complex. Collectively, both M-CSF conformational changes, one disturbing the local FMS-binding segment of M-CSF and the other affecting the global orientation of M-CSF and FMS, compromise M-CSF’s ability to activate its receptor FMS.
Mechanism of BARF1 Attenuating Rather than Blocking M-CSF:FMS Signaling.
Although BARF1 compromises M-CSF:FMS signaling, it apparently does not completely abolish the binding of M-CSF to FMS (Fig. 1B) or M-CSF–induced FMS phosphorylation (Fig. 1C and Fig. S3). How is such partial inhibition achieved structurally? Although the M-CSF conformational changes induced by BARF1 binding are not ideal for potent receptor signaling, the nonoverlapping nature of the BARF1 and FMS footprints on M-CSF determines that such conformational changes still may be accommodated, albeit at the expense of signaling efficiency. Although BARF1 binding shifts part of the FMS-binding interface of M-CSF relative to the remaining interface, FMS may remodel its ligand-recognizing surface to meet the new conformations of M-CSF, likely at an entropic penalty. Additionally, even though the M-CSF dimer is bent by the rigid BARF1 hexameric ring and tilts the M-CSF–bound FMS receptor into an unfavorable conformation, the receptor may be able to rotate at a hinge adjacent to the membrane, most likely the Ig3–Ig4 hinge, to regain the signaling configuration and the formation of the required Ig4–Ig4 interface (15, 26). Such hinge movement imposes an additional energetic burden and would reduce the signaling efficiency, but it is achievable, as exemplified by the FMS-related receptor KIT (15).
In addition to the conformational changes described above, the ring-like hexameric design of BARF1 also may be a factor in partial attenuation, because it may be able to control the balance of circulating versus receptor-bound M-CSF. As shown in Fig. 4D, the hexameric assembly of the BARF1:M-CSF complex orients three M-CSF dimers differently in relation to a single cell membrane. When the soluble, circulating M-CSF binds BARF1, only one of every three M-CSF dimers can assume the orientation to dimerize FMS receptors symmetrically. Because the FMS extracellular Ig domains are linked to its transmembrane helix directly, without a long stalk region, FMS binding to an M-CSF dimer would orient the hexameric ring of the complex vertical to the membrane, preventing the other two M-CSF dimers bound to the same complex from readily accessing other FMS receptors on the same cell surface (Fig. 4D). When the membrane-bound M-CSF binds BARF1, however, the hexameric complex would have to lie flat, rendering all three bound M-CSF dimers unable to dimerize vertically standing FMS receptors on the same cell surface. Therefore, BARF1 should be able to block membrane-spanning isoforms of M-CSF efficiently but to reduce only two thirds of the circulating M-CSF molecules for signaling. The collective consequence is a reduction of the effective M-CSF population available, but a low level of signaling is still allowed.
Hence, BARF1 may use a three-pronged approach in downregulating M-CSF:FMS signaling: reducing the effective M-CSF concentration, disturbing the FMS-binding surface, and bending M-CSF dimer to the disadvantage of FMS dimerization (Fig. 4D). Each of these prongs may apply a limited penalty but still allow signaling at reduced efficiency. The exact contribution of each prong to the reduction of signaling remains to be determined. The multilevel, sterically nonexclusive strategy of BARF1 down-regulation is unprecedented and is a unique and interesting regulatory mechanism. Simply to block M-CSF:FMS signaling, a more direct approach and consequently a more efficient strategy would be for an EBV-encoded antagonist fully disguised as a receptor or ligand mimic to occupy the active surfaces required for signaling. This strategy has been used by various types of viruses in immune modulation, especially for sequestrating cytokines (27, 28). In fact, when BARF1 first was identified as a binding partner for M-CSF, it was speculated to be an FMS homolog (13). However, our study suggests that the BARF1 regulatory mechanism deviates from this initial proposed model. The reasons why EBV has designed such a complex mechanism using hexameric inhibitory scaffold for FMS signal attenuation still remain to be discerned; however, the apparent complex regulatory mechanism may be advantageous in fine tuning and maintaining low levels of M-CSF:FMS signaling to modulate pathogenesis of EBV infections. The role of the BARF1 pathogenesis in natural infections soon may be be discerned by experimental infection of nonhuman primates. A mutation in the BARF1 gene, which will allow the direct testing of BARF1 function in infection, was described recently in the EBV-related rhesus lymphocryptovirus (8). Finally, the BARF1:M-CSF structure provides additional evidence that BARF1 modulates M-CSF–mediated immune responses and may provide a basis for beginning to understand any putative role of BARF1 in the alteration of cellular growth (17, 29–32).
Experimental Procedures
Recombinant Protein Preparation and Crystallization.
The BARF1 and M-CSF proteins were expressed from HEK293S N-acetylglucosaminyltransferase I–negative cells (33) using the BacMam method previously described (20). The N-linked glycans of these proteins were trimmed with endoglycosidase F1 (Sigma) before crystallization. Details are described in SI Experimental Procedures.
Data Collection and Structure Determination.
The crystallographic data were measured at the Argonne National Laboratory Beamline 21-ID-G. The data were processed, and the structure was determined as described in SI Experimental Procedures. Crystallographic statistics are summarized in Table S1.
Isothermal Titration Calorimetry.
Calorimetric measurements were implemented on a VP-ITC calorimeter (MicroCal) as described in SI Experimental Procedures. The data were processed and fitted with the MicroCal Origin 5.0 software.
Phosphorylation Assay.
COS7 or HEK293H cells were infected with full-length human or mouse FMS BacMam virus for 6 h, then were serum-starved overnight and were stimulated with 75 nM each (COS7 cells) or 25 nM each (HEK293H cells) of M-CSF or BARF1:M-CSF for 2 min. The stimulated cells were washed with cold PBS and lysed with Triton-X lysis buffer. The lysates were spun down, and the supernatant was collected. Equal amounts of the samples were run on a SDS/PAGE gel and transferred PVDF membranes and immunoblotted with anti-M-CSF receptor and anti–phospho-M-CSF receptor (Tyr723) (Cell Signaling Technology). The blots were developed using the WesternBreeze Immunodetection Kit (Invitrogen). Alternatively, membranes were incubated with an HRP-conjugated goat anti-rabbit secondary antibody (Cell Signaling Technology) and visualized by ECL (Amersham). Blots were scanned and band intensities were quantified using ImageJ 1.45p software (National Institutes of Health). Student t tests were performed to determine statistically significance differences in band intensities.
Supplementary Material
Acknowledgments
We thank P. J. Focia and Z. Wawrzak for support in data collection. Data were measured at the LS-CAT beamline 21-ID-D at the Advanced Photon Source, Argonne, IL. X.H. is supported by National Institutes of Health (NIH) Grant R01 GM078055. R.L. is the Dan and Bertha Spear Research Professor and is supported by NIH Grant R01 CA073507. The Structural Biology Facility is supported by the Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structural factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4FA8).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205309109/-/DCSupplemental.
References
- 1.Kieff ED, Rickinson AB. 2007. Epstein-Barr virus and its replication. Fields Virology, ed. Knipe D, et al. (Lippincott, Williams and Wilkins, Philadelphia), 5th Ed, Vol. 2, pp 2603–2654.
- 2.Rickinson AB, Kieff ED. 2007. Epstein-Barr virus. Fields Virology, ed. Knipe D, et al. (Lippincott, Williams and Wilkins, Philadelphia), 5th Ed, Vol. 2, pp 2655–2700.
- 3.Wei MX, de Turenne-Tessier M, Decaussin G, Benet G, Ooka T. Establishment of a monkey kidney epithelial cell line with the BARF1 open reading frame from Epstein-Barr virus. Oncogene. 1997;14:3073–3081. doi: 10.1038/sj.onc.1201128. [DOI] [PubMed] [Google Scholar]
- 4.Sheng W, Decaussin G, Sumner S, Ooka T. N-terminal domain of BARF1 gene encoded by Epstein-Barr virus is essential for malignant transformation of rodent fibroblasts and activation of BCL-2. Oncogene. 2001;20:1176–1185. doi: 10.1038/sj.onc.1204217. [DOI] [PubMed] [Google Scholar]
- 5.Wei MX, Ooka T. A transforming function of the BARF1 gene encoded by Epstein-Barr virus. EMBO J. 1989;8:2897–2903. doi: 10.1002/j.1460-2075.1989.tb08438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danve C, Decaussin G, Busson P, Ooka T. Growth transformation of primary epithelial cells with a NPC-derived Epstein-Barr virus strain. Virology. 2001;288:223–235. doi: 10.1006/viro.2001.1072. [DOI] [PubMed] [Google Scholar]
- 7.Cohen JI, Lekstrom K. Epstein-Barr virus BARF1 protein is dispensable for B-cell transformation and inhibits alpha interferon secretion from mononuclear cells. J Virol. 1999;73:7627–7632. doi: 10.1128/jvi.73.9.7627-7632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohashi M, Orlova N, Quink C, Wang F. Cloning of the Epstein-Barr virus-related rhesus lymphocryptovirus as a bacterial artificial chromosome: A loss-of-function mutation of the rhBARF1 immune evasion gene. J Virol. 2011;85:1330–1339. doi: 10.1128/JVI.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seto E, et al. Epstein-Barr virus (EBV)-encoded BARF1 gene is expressed in nasopharyngeal carcinoma and EBV-associated gastric carcinoma tissues in the absence of lytic gene expression. J Med Virol. 2005;76:82–88. doi: 10.1002/jmv.20327. [DOI] [PubMed] [Google Scholar]
- 10.Decaussin G, Sbih-Lammali F, de Turenne-Tessier M, Bouguermouh A, Ooka T. Expression of BARF1 gene encoded by Epstein-Barr virus in nasopharyngeal carcinoma biopsies. Cancer Res. 2000;60:5584–5588. [PubMed] [Google Scholar]
- 11.Takada K. Role of EBER and BARF1 in nasopharyngeal carcinoma (NPC) tumorigenesis. Semin Cancer Biol. 2012;22:162–165. doi: 10.1016/j.semcancer.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 12.Hoebe EK, et al. Purified hexameric Epstein-Barr virus-encoded BARF1 protein for measuring anti-BARF1 antibody responses in nasopharyngeal carcinoma patients. Clin Vaccine Immunol. 2011;18:298–304. doi: 10.1128/CVI.00193-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strockbine LD, et al. The Epstein-Barr virus BARF1 gene encodes a novel, soluble colony-stimulating factor-1 receptor. J Virol. 1998;72:4015–4021. doi: 10.1128/jvi.72.5.4015-4021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanley ER, et al. Biology and action of colony—stimulating factor-1. Mol Reprod Dev. 1997;46:4–10. doi: 10.1002/(SICI)1098-2795(199701)46:1<4::AID-MRD2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 15.Yuzawa S, et al. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Liu H, Focia PJ, Shim AH, He X. Structure of macrophage colony stimulating factor bound to FMS: Diverse signaling assemblies of class III receptor tyrosine kinases. Proc Natl Acad Sci USA. 2008;105:18267–18272. doi: 10.1073/pnas.0807762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, et al. Anti-apoptotic role of BARF1 in gastric cancer cells. Cancer Lett. 2006;238:90–103. doi: 10.1016/j.canlet.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 18.Tarbouriech N, Ruggiero F, de Turenne-Tessier M, Ooka T, Burmeister WP. Structure of the Epstein-Barr virus oncogene BARF1. J Mol Biol. 2006;359:667–678. doi: 10.1016/j.jmb.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 19.Pandit J, et al. Three-dimensional structure of dimeric human recombinant macrophage colony-stimulating factor. Science. 1992;258:1358–1362. doi: 10.1126/science.1455231. [DOI] [PubMed] [Google Scholar]
- 20.Dukkipati A, Park HH, Waghray D, Fischer S, Garcia KC. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr Purif. 2008;62:160–170. doi: 10.1016/j.pep.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Chen X, Focia PJ, He X. Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases. EMBO J. 2007;26:891–901. doi: 10.1038/sj.emboj.7601545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verstraete K, et al. Structural insights into the extracellular assembly of the hematopoietic Flt3 signaling complex. Blood. 2011;118:60–68. doi: 10.1182/blood-2011-01-329532. [DOI] [PubMed] [Google Scholar]
- 23.Shim AH, et al. Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proc Natl Acad Sci USA. 2010;107:11307–11312. doi: 10.1073/pnas.1000806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leppänen VM, et al. Structural determinants of growth factor binding and specificity by VEGF receptor 2. Proc Natl Acad Sci USA. 2010;107:2425–2430. doi: 10.1073/pnas.0914318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiesmann C, et al. Crystal structure at 1.7 A resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell. 1997;91:695–704. doi: 10.1016/s0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y, Yuzawa S, Schlessinger J. Contacts between membrane proximal regions of the PDGF receptor ectodomain are required for receptor activation but not for receptor dimerization. Proc Natl Acad Sci USA. 2008;105:7681–7686. doi: 10.1073/pnas.0802896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue X, et al. Structural basis of chemokine sequestration by CrmD, a poxvirus-encoded tumor necrosis factor receptor. PLoS Pathog. 2011;7:e1002162. doi: 10.1371/journal.ppat.1002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander JM, et al. Structural basis of chemokine sequestration by a herpesvirus decoy receptor. Cell. 2002;111:343–356. doi: 10.1016/s0092-8674(02)01007-3. [DOI] [PubMed] [Google Scholar]
- 29.Chang MS, et al. Cell-cycle regulators, bcl-2 and NF-kappaB in Epstein-Barr virus-positive gastric carcinomas. Int J Oncol. 2005;27:1265–1272. [PubMed] [Google Scholar]
- 30.Sheng W, Decaussin G, Ligout A, Takada K, Ooka T. Malignant transformation of Epstein-Barr virus-negative Akata cells by introduction of the BARF1 gene carried by Epstein-Barr virus. J Virol. 2003;77:3859–3865. doi: 10.1128/JVI.77.6.3859-3865.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stevens SJ, et al. Diagnostic value of measuring Epstein-Barr virus (EBV) DNA load and carcinoma-specific viral mRNA in relation to anti-EBV immunoglobulin A (IgA) and IgG antibody levels in blood of nasopharyngeal carcinoma patients from Indonesia. J Clin Microbiol. 2005;43:3066–3073. doi: 10.1128/JCM.43.7.3066-3073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens SJ, et al. Noninvasive diagnosis of nasopharyngeal carcinoma: Nasopharyngeal brushings reveal high Epstein-Barr virus DNA load and carcinoma-specific viral BARF1 mRNA. Int J Cancer. 2006;119:608–614. doi: 10.1002/ijc.21914. [DOI] [PubMed] [Google Scholar]
- 33.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: High-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.