The genome project, with the discovery of thousands of new protein sequences every year, has created a revolution in protein physics, chemistry, and biology. This has led to a renewed and very much expanded interest in the protein folding problem, particularly among biophysical scientists. There are two parts to this problem. The first is predicting the three-dimensional structure of a protein from its amino acid sequence, often referred to as cracking the second half of the genetic code. The second, which is the subject of this Commentary, is to understand how proteins fold. This problem has recently taken on additional importance, as more and more human diseases, such as Alzheimer’s and Parkinson’s diseases, are believed to be caused by aggregation of misfolded proteins (1). There is, moreover, the intriguing possibility that evolution preserves sequences that not only form structures that adequately perform a specific biological function but also those that avoid misfolded states and fold sufficiently quickly to avoid aggregation. That is, minimizing nonnative interactions and maintaining sufficient folding speed may very well be additional selection pressures in protein evolution (2, 3).
The question of how a protein folds can be phrased more precisely as follows: What are the sequences of structural changes that occur in a polypeptide as it finds its way from the myriad of possible structures in the denatured state to the final, unique native structure? How many different folding routes exist and what are their relative probabilities? The article by Sabelko et al. in this issue of the Proceedings (4) represents a significant advance toward answering these questions. These investigators observe a time course for folding that does not have the functional form corresponding to a single exponential or a sum of a few exponentials, as has been observed previously in studies of protein folding kinetics. Instead they discovered a single, continuous nonexponential time course that can be well described by a stretched exponential function: exp[−(kt)β]. Nonexponential kinetics have played an important role in understanding conformational changes in native proteins (5–7). They are particularly interesting for protein folding because they could arise from a process that is “downhill” in free energy—i.e., one in which the overall free energy barrier separating the native from the denatured state is very small or nonexistent. For large barriers only the structures of the initial and final states are observable because structures along the folding route are too sparsely populated.† If, however, the barrier becomes very small or disappears altogether, all of the structures can in principle be detected and characterized by spectroscopy. There is then the exciting prospect of performing single molecule experiments that directly observe exploration of the energy landscape and folding routes. Finding proteins that fold with a “downhill scenario” is an essential first step in this quest. That some proteins will exhibit downhill folding, moreover, is one of the novel theoretical predictions of an energy landscape analysis of protein folding (9). This prediction requires some further explanation.
Until about a decade ago the problem of understanding how proteins fold was addressed by identifying and characterizing one or two metastable structures thought to be obligatory intermediates in a sequential process along a well-defined protein folding pathway. The prevailing view was that structural characterization of such intermediates would give the clue to the basic underlying mechanism, as in the study of organic chemical reactions (10). Unlike small-molecule chemical reactions, however, in which covalent bonds are broken and new ones formed in a structurally well-defined transition state, the many degrees of freedom of a polypeptide chain demand a rather different approach. A polypeptide of 100 amino acids has a huge number of conformations, even if only a tiny fraction of the >2100 (=1030) possible conformations are thermally occupied. Understanding the complexity of protein folding at the microscopic level and developing models that make quantitative predictions therefore requires a statistical approach—i.e., the theoretical and computational tools of modern statistical mechanics (2, 9, 11–18).
A major goal of a statistical mechanical model of protein folding is to obtain the free energy of a polypeptide as a function of just a few coordinates. This is a hard problem for computer simulations, as well as for analytical theory. The desired result is a low-dimensional free energy surface from which both the thermodynamics and kinetics of folding can be calculated. In this picture the kinetics of folding and unfolding are obtained from the dynamics of diffusion on this free energy surface (19–22).§ The most useful free energy surface would be one defined by measurable structural parameters. Natural candidates are the overall size of the molecule, for example the radius of gyration (Rg) measured in low-angle x-ray scattering experiments, the fraction of secondary structure as determined from far ultraviolet circular dichroism, and the number of peptide bonds in their native conformation, which might be measurable by vibrational spectroscopy. A useful measure of overall topology, particularly in lattice simulations, has been the fraction of native tertiary contacts between residues (Q) (2, 9–16). Q can in principle be measured in equilibrium experiments by NMR (23), but a method to monitor Q with adequate time resolution is not yet at hand.
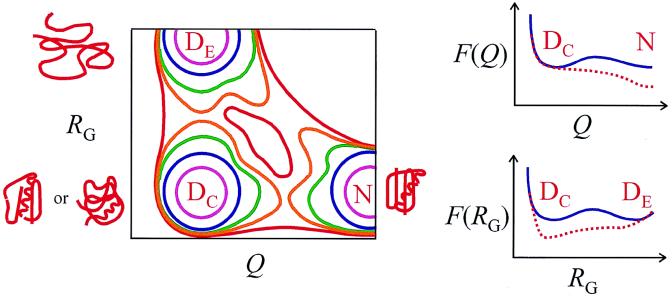
To easily visualize a free energy surface, two of these parameters can be selected. A schematic of a Q, Rg surface is shown in Fig. 1 (24). This surface shows three minima, an expanded denatured state (DE), a compact denatured state (DC), and the fully folded native state (N) (25). DE, DC, and N are considered separate thermodynamic states, because they are separated from each other by barriers greater than a few kBT (kB is the Boltzmann constant, and kBT at 300 K is 0.6 kcal/mol). If equilibration among the microstates is fast compared with the barrier crossing, then simple exponential kinetics result. If, on the other hand, the free energy barrier disappears, the kinetics will become nonexponential. A striking result of the theoretical calculation of free energy surfaces is that the barriers to folding are predicted to be quite small (2, 26, 27), even though protein folding is a very slow process by ordinary standards of chemical reaction kinetics. The barriers are small because of near cancellation along the reaction coordinate of large entropy terms, arising from the many possible configurations, by comparable stabilizing (free) energy terms from interresidue contacts. For example, when a microscopic model that assumes negligible nonnative interactions is used, the free energy barrier to folding the 80 amino acid protein monomeric λ repressor is calculated to be only 5–7 kBT at the temperature where the populations of native and denatured states are equal (27). If a chemical denaturant is used to achieve equal populations, the folding rate is ∼800 s−1 at 310 K (28). Assuming that this rate can be calculated approximately from koexp(−ΔG‡/RT) and that the barrier height remains 5–7 kBT, the pre-exponential factor ko is only ∼105 s−1 to 106 s−1 (29), which may be compared with kBT/h = 6 × 1012 s−1 of gas phase chemical reaction kinetics.
Figure 1.
Schematic of one and two-dimensional free energy surfaces. (Left) Free energy as a function of the fraction of native contacts (Q) and the radius of gyration (Rg). This surface has three significant minima, one for an expanded denatured state (DE), one for a compact denatured state (DC), and one for the native state (N). To simplify the brief discussion of this surface the somewhat unrealistic assumption is made that Q is the same for DE and DC, and Rg is the same for DC and N. For this free energy surface Q may be a good reaction coordinate for forming the native state N from DC or DE, and Rg may be a good reaction coordinate for collapse of the expanded denatured state DE to DC or N, but not vice versa. If so, then the kinetics of folding and unfolding, as well as the kinetics of collapse and expansion of the denatured protein, might be described as diffusion on one-dimensional free energy surfaces (Right).† One of the major outstanding issues is whether DE has a topology that is near native (assumed for a “molten globule”) or is more nearly a “random globule” with a distribution of topologies, in which case DC should be considered a trap on the free energy surface containing misfolded structures. (Upper Right) Free energy as a function of Q for folding over a barrier (continuous blue curve) and for downhill folding (dotted red curve) in a type 0 scenario in the classification of Bryngelson et al. (9). (Lower Right) Free energy as a function of Rg. Submillisecond time-resolved small-angle x-ray scattering experiments are being performed that hold the promise of resolving these kinetics (L. Pollack, M. W. Tate, N. C. Darnton, J. B. Knight, S. M. Gruner, W.A.E., and R. H. Austin, unpublished work).
The prediction of small free energy barriers, and therefore the possibility of experimentally observing downhill folding, has been a significant factor in motivating biophysical scientists to develop new techniques with better time resolution to study protein folding (30, 31). Sabelko et al. (4) took advantage of the nanosecond time resolution of the laser temperature-jump method (31) to study the kinetics of folding from the cold denatured state. They studied two proteins, a double mutant of a small (76 amino acid) single-domain protein, ubiquitin, and a large (415 amino acid) two-domain protein, phosphoglycerate kinase. The latter protein reaches the native state slowly (>10 s) and has a more complex free energy surface than the one shown in Fig. 1. The free energy surface for ubiquitin, on the other hand, appears to be much simpler, with only three minima (apart from those produced by proline isomerization, which causes minor slow kinetic phases), as in Fig. 1 (32). For this protein Sabelko et al. observe two exponential relaxations at 2°C with time constants of ∼20 μs and ∼5 ms (τ1/2 ≈ 3 ms), the second relaxation corresponding to formation of the native structure. At a slightly higher temperature (8°C), the first relaxation remains exponential and becomes slightly faster, while the rate of the second increases dramatically (τ1/2 ≈ 100 μs) and the time course becomes nonexponential. They assign the 20-μs phase to “nonspecific” contraction of the polypeptide chain. Sabelko et al. are faced with an interesting problem in interpreting the second relaxation. How can one simultaneously explain the existence of a significant barrier between the denatured and native states in equilibrium NMR experiments at 8°C, yet observe nonexponential kinetics in the formation of the native state [from DE, since DC is expected to be at much higher free energy than either DE or N (32) (Fig. 1)]? Sabelko et al. resolve this puzzle by proposing a change in the shape of the free energy surface: denatured molecules of the free energy minimum (DE) at the initial temperature find themselves on the native side of a free energy maximum (the barrier top) at the elevated temperature. Folding is then downhill and the kinetics are nonexponential.¶ The position of the free energy barrier shifts to lower Q (Fig. 1) at the elevated temperature because of strengthening of the hydrophobic effect, so that the stabilizing forces overcome the decrease in configurational entropy earlier along the reaction coordinate.
Sabelko et al. (4) also make a more subtle point that energetic “ruggedness” or “roughness” may determine the extent of the deviation from an exponential time course. Roughness arises from nonnative interactions or native interactions in wrong topologies. By averaging over the many degrees of freedom in reducing the description of the system to a one- or two-dimensional free energy surface, most of the roughness is absorbed into the effective diffusion coefficient used in calculating the kinetics (2, 9, 21). This diffusion coefficient may vary along the reaction coordinate (21) and would result in stretched exponential kinetics if it continuously decreases (6). One might also expect that sequences with many favorable nonnative contacts, such as very hydrophobic sequences, would exhibit dynamics on the free energy surface with a smaller effective diffusion coefficient (34). An interesting consequence of these considerations is the possibility of observing large curvature in plots of the log(folding rate) versus the free energy driving force for downhill folders at constant temperature. Even though increasing the strength of the interresidue interaction would increase the free energy bias toward the native state, the net effect at very high bias might be to decrease the overall rate because of a much lower diffusion constant. That is, the folding rate at constant temperature could exhibit a maximum. In the classification by Bryngelson et al. (9) this would correspond to a continuous progression from a type I to a type 0a to a type 0b folding scenario. One candidate for such behavior is a double mutant of the 80-residue monomeric λ repressor mentioned earlier, which has a folding rate extrapolated to zero denaturant of ∼105 s−1 (35), close to the estimated maximum rate of protein folding of ∼106 s−1 (36).
Finally, there is the exciting prospect of studying downhill folding in experiments on single molecules. For proteins with a single significant barrier in their ensemble behavior, one naively expects that, with the current time resolution in single-molecule fluorescence experiments of ∼0.010–1.0 ms, individual protein molecules will appear to stochastically jump between the native structure and the structures of the denatured state, since the time for the polypeptide backbone to change a pair of dihedral angles is only ∼1 ns (37). Unless information is obtained on structure distributions in the denatured state (38), this experiment could therefore be disappointing, because it might turn out to be just a difficult way of determining the folding and unfolding rates for a two-state kinetic system. For a downhill folding protein, however, it may be possible with only a modest improvement in time resolution to directly observe the intermediate structures and monitor the complete sequence of structural events that lead to folding and unfolding of the native structure (by using, e.g., Förster excitation energy transfer (38, 39) or contact fluorescence quenching). Slowing could be achieved by increasing the viscosity of the solution. Such experiments could make a major contribution to the ultimate goal of describing the distribution of folding routes from the denatured to the native state. The results of single-molecule and ensemble kinetic experiments will provide a reality check on computer films that we shall be watching in the not too distant future of individual protein molecules folding from random coil structures all the way to the native structure in all-atom molecular dynamics simulations (40).
Acknowledgments
I thank Stephen Hagen, Victor Muñoz, Heinrich Roder, Attila Szabo, and Robert Zwanzig for helpful discussions.
Footnotes
A commentary on this article begins on page 6031.
By assuming a linear relation between changes in activation and equilibrium free energies produced by mutations, important inferences can be made about the structures of the ensemble of molecules that pass over the barrier top along the minimum free energy path between the native and denatured states—i.e., the transition state ensemble (8).
Whether a low-dimensional free energy surface can be found that describes the kinetics of folding remains a subject of some controversy (20). A somewhat surprising and encouraging result is the success of a one-dimensional free energy surface in describing the dynamics of simplified models of proteins, but still very complicated systems (21, 22).
With data over a wider range of times it should be possible to prove that the process is truly nonexponential [see, e.g., ref. 33, in which the stretching parameter β for the conformational relaxation in myoglobin is ∼0.1 (6)]. At longer times, moreover, the exponential tail might be observable because of folding of those molecules that fall back into the denatured well DE after the temperature jump, rather than move downhill to the native state. Also, why the ∼20-μs phase has such a large relative amplitude and is exponential at all temperatures is an important remaining question.
References
- 1.Lansbury P T. Proc Natl Acad Sci USA. 1999;96:3342–3344. doi: 10.1073/pnas.96.7.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Onuchic J, Luthey-Schulten A, Wolynes P G. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 3.Mirny L A, Abkevich V I, Shakhnovich E I. Proc Natl Acad Sci USA. 1998;95:4976–4981. doi: 10.1073/pnas.95.9.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabelko J, Ervin J, Gruebele M. Proc Natl Acad Sci USA. 1999;96:6031–6036. doi: 10.1073/pnas.96.11.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frauenfelder H, Sligar S G, Wolynes P G. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 6.Hagen S J, Eaton W A. J Chem Phys. 1996;104:3395–3398. [Google Scholar]
- 7.Henry E R, Jones C M, Hofrichter J, Eaton W A. Biochemistry. 1997;36:6511–6528. doi: 10.1021/bi9619177. [DOI] [PubMed] [Google Scholar]
- 8.Fersht A R. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: Freeman; 1998. [Google Scholar]
- 9.Bryngelson J D, Onuchic J N, Socci N D, Wolynes P G. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 10.Laurents D V, Baldwin R L. Biophys J. 1998;75:428–434. doi: 10.1016/S0006-3495(98)77530-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shakhnovich E I. Curr Opin Struct Biol. 1997;7:29–40. doi: 10.1016/s0959-440x(97)80005-x. [DOI] [PubMed] [Google Scholar]
- 12.Pande V S, Grosberg A Y, Tanaka T, Rokhsar D S. Curr Opin Struct Biol. 1998;8:68–79. doi: 10.1016/s0959-440x(98)80012-2. [DOI] [PubMed] [Google Scholar]
- 13.Dobson C M, Sali A, Karplus M. Angew Chem Int Ed Engl. 1998;37:868–893. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 14.Chan H S, Dill K A. Proteins. 1998;30:2–33. doi: 10.1002/(sici)1097-0134(19980101)30:1<2::aid-prot2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 15.Brooks C L., III Curr Opin Struct Biol. 1998;8:222–226. doi: 10.1016/s0959-440x(98)80043-2. [DOI] [PubMed] [Google Scholar]
- 16.Thirumalai D, Klimov D. Curr Opin Struct Biol. 1999;9:197–207. doi: 10.1016/S0959-440X(99)80028-1. [DOI] [PubMed] [Google Scholar]
- 17.Garel T, Orland H, Pitard E. In: Spin Glasses and Random Fields. Young A P, editor. Singapore: World Scientific; 1998. pp. 387–443. [Google Scholar]
- 18.Hao M H, Scheraga H A. Acc Chem Res. 1998;31:433–440. [Google Scholar]
- 19.Bryngelson J D, Wolynes P G. J Phys Chem. 1989;93:6902–6915. [Google Scholar]
- 20.Frauenfelder H, Deisenhofer J, Wolynes P G, editors. Simplicity and Complexity in Proteins and Nucleic Acids. Berlin: Dahlem Univ. Press; 1999. , in press. [Google Scholar]
- 21.Socci N D, Onuchic J N, Wolynes P G. J Chem Phys. 1996;104:5860–5868. [Google Scholar]
- 22.Klimov D K, Thirumalai D. Phys Rev Lett. 1997;79:317–320. [Google Scholar]
- 23.Van Nuland N A J, Forge V, Balbach J, Dobson C M. Acc Chem Res. 1998;31:773–780. [Google Scholar]
- 24.Socci N D, Onuchic J N, Wolynes P G. Proteins. 1998;32:136–158. [PubMed] [Google Scholar]
- 25.Dill K A, Shortle D. Annu Rev Biochem. 1991;60:795–825. doi: 10.1146/annurev.bi.60.070191.004051. [DOI] [PubMed] [Google Scholar]
- 26.Thirumalai D. J Phys I. 1995;5:1457–1467. [Google Scholar]
- 27.Portman J J, Takada S, Wolynes P G. Phys Rev Lett. 1998;81:5237–5240. [Google Scholar]
- 28.Burton R E, Huang G S, Daugherty M A, Calderone T L, Oas T G. Nat Struct Biol. 1997;4:305–310. doi: 10.1038/nsb0497-305. [DOI] [PubMed] [Google Scholar]
- 29.Chan C-K, Hu Y, Takahashi S, Rousseau D L, Eaton W A, Hofrichter J. Proc Natl Acad Sci USA. 1997;94:1779–1784. doi: 10.1073/pnas.94.5.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones C M, Henry E R, Hu Y, Chan C-K, Luck S D, Bhuyan A, Roder H, Hofrichter J, Eaton W A. Proc Natl Acad Sci USA. 1993;90:11860–11864. doi: 10.1073/pnas.90.24.11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winkler, J. R. & Gray, H. B., eds. (1998) Acc. Chem. Res. “Protein Folding Special Issue”31, 697–780.
- 32.Khorasanizadeh S, Peters I D, Roder H. Nat Struct Biol. 1996;3:193–205. doi: 10.1038/nsb0296-193. [DOI] [PubMed] [Google Scholar]
- 33.Jackson T A, Lim M, Anfinrud P A. Chem Phys. 1994;180:131–140. [Google Scholar]
- 34.Zwanzig R. Proc Natl Acad Sci USA. 1988;85:2029–2030. doi: 10.1073/pnas.85.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burton R E, Huang G S, Daugherty M A, Calderone T L, Oas T G. Nat Struct Biol. 1997;4:305–310. doi: 10.1038/nsb0497-305. [DOI] [PubMed] [Google Scholar]
- 36.Hagen S J, Hofrichter J, Szabo A, Eaton W A. Proc Natl Acad Sci USA. 1996;93:1615–1617. doi: 10.1073/pnas.93.21.11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muñoz V, Thompson P A, Hofrichter J, Eaton W A. Nature (London) 1997;390:196–199. doi: 10.1038/36626. [DOI] [PubMed] [Google Scholar]
- 38.Jia, Y., Talaga, D. S., Lau, W. L., Lu, H. S. M., DeGrado, W. F. & Hochstrasser, R. Chem. Phys., in press.
- 39.Weiss S. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 40.Duan Y, Kollman P A. Science. 1998;282:740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]