Abstract
The evolution of cadherins, which are essential for metazoan multicellularity and restricted to metazoans and their closest relatives, has special relevance for understanding metazoan origins. To reconstruct the ancestry and evolution of cadherin gene families, we analyzed the genomes of the choanoflagellate Salpingoeca rosetta, the unicellular outgroup of choanoflagellates and metazoans Capsaspora owczarzaki, and a draft genome assembly from the homoscleromorph sponge Oscarella carmela. Our finding of a cadherin gene in C. owczarzaki reveals that cadherins predate the divergence of the C. owczarzaki, choanoflagellate, and metazoan lineages. Data from these analyses also suggest that the last common ancestor of metazoans and choanoflagellates contained representatives of at least three cadherin families, lefftyrin, coherin, and hedgling. Additionally, we find that an O. carmela classical cadherin has predicted structural features that, in bilaterian classical cadherins, facilitate binding to the cytoplasmic protein β-catenin and, thereby, promote cadherin-mediated cell adhesion. In contrast with premetazoan cadherin families (i.e., those conserved between choanoflagellates and metazoans), the later appearance of classical cadherins coincides with metazoan origins.
The cadherin gene family is hypothesized to have had special importance for metazoan origins (1–5). Cadherins are cell-surface receptors that function in cell adhesion, cell polarity, and tissue morphogenesis (6–8). Moreover, cadherins are found in the genomes of all sequenced metazoans, including diverse bilaterians, cnidarians, and sponges, and are apparently lacking from multicellular lineages such as plants, fungi, and Dictyostelium (9). Although it once seemed likely that cadherins were unique to metazoans, 23 genes encoding the diagnostic extracellular cadherin (EC) domain (10) have since been discovered in the genome of the unicellular choanoflagellate Monosiga brevicollis, one of the closest living relatives of Metazoa (1, 11).
Proteins in the cadherin family are characterized by the presence of one or more tandem copies of the EC domain, an ∼100-aa protein domain that mediates adhesion with EC domains in other cadherins (10, 12–14). Cadherins are further assigned to different subfamilies based on the number and arrangement of additional, non-EC protein domains and sequence motifs that refine cadherin function and suggest shared ancestry (2, 3). For example, classical cadherins are distinguished by the presence of a cytoplasmic cadherin domain (CCD) at the C terminus that regulates interactions with the cytoplasmic protein β-catenin (2, 3, 12, 15). When bound to β-catenin, classical cadherins on neighboring cells interact homophilically and, thereby, promote cell-cell adhesion (16). When not bound to β-catenin, classical cadherins are rapidly degraded (17, 18). The regulation of classical cadherin function by β-catenin thereby forms the foundation of adherens junctions and is crucial for cell adhesion in all studied bilaterian tissues, including epithelia, neurons, muscles, and bones (3, 19).
The classical cadherins are one of six cadherin families (including fat, dachsous, fat-like, CELSR/flamingo, and protocadherins) that are found in most metazoans. In contrast with the cell adhesion functions of classical cadherins, CELSR/flamingo, dachsous, fat, and fat-like cadherins regulate planar cell polarity in organisms as disparate as Drosophila and mouse (20–22). Members of the protocadherin family have diverse functions that include mechanosensation in stereocilia and regulation of nervous system development (23, 24). It is not known whether the bilaterian roles of these cadherin families had already evolved in the last common ancestor of metazoans, and it is not clear how these cadherin families themselves originated.
To date, only one cadherin family—the hedgling family—is inferred to have been present in the last common ancestor of choanoflagellates and metazoans. Hedgling family members are defined by the presence of an N-terminal hedgehog signal domain (Hh-N) and are absent from Bilateria (25, 26). Differences in the cadherin repertoire of choanoflagellates and metazoans have led to the proposal that cadherins in these two lineages may have largely independent histories—that is, one or a few ancestral cadherins may have undergone independent evolutionary radiations in each lineage (2). To reconstruct the evolutionary history of cadherin families before and after the transition to metazoan multicellularity, we have analyzed the diversity of cadherins in the newly sequenced genomes of phylogenetically relevant taxa: the colony forming choanoflagellate Salpingoeca rosetta, the close choanoflagellate/metazoan outgroup Capsaspora owczarzaki, and the homoscleromorph sponge Oscarella carmela.
Results
Reconstructing the Ancestry of Cadherin Diversity.
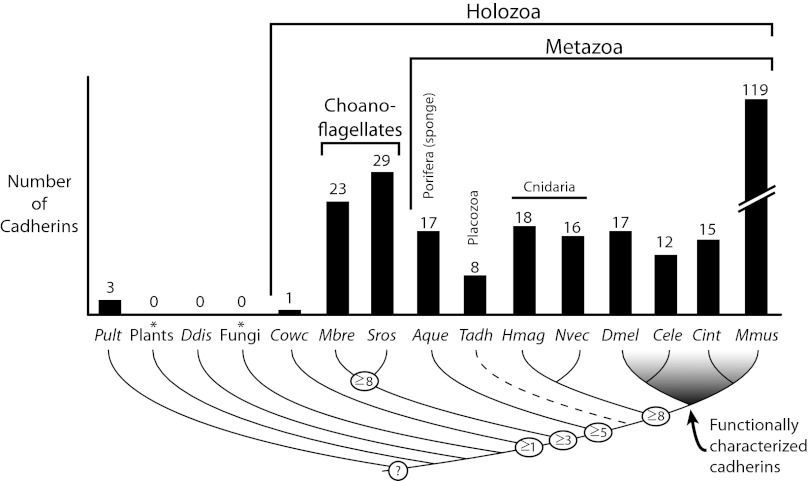
By searching the S. rosetta genome using BLAST analyses (27) and hidden Markov model (HMM)-based searches (28–30) for the EC domain (Fig. 1), we identified at least 29 predicted cadherin genes (Fig. 1 and SI Appendix, Figs. S1 and S2), all of which were verified through deep sequencing of the transcriptome (SI Appendix, Table S1). The number of cadherin genes in S. rosetta, like that in M. brevicollis (1), rivals that of most metazoans (Fig. 1), whereas the C. owczarzaki genome assembly was found to contain only a single cadherin gene.
Fig. 1.
Phylogenetic distribution and abundance of cadherins in the genomes of diverse eukaryotes. Once thought to be restricted to metazoans, cadherins are abundant in choanoflagellates and evolved before the divergence of Capsaspora owczarzaki, choanoflagellates, and metazoans (1). EC domains detected in the genome of the oomycte Pythium ultimum likely evolved through convergence or lateral gene transfer (9). The number of cadherin families inferred at ancestral nodes (determined based upon their shared domain composition and organization) is indicated (open circles). The dashed lineage of Trichoplax adhaerens reflects its uncertain phylogenetic placement. *All fungal and plant species represented in the Pfam v24.0 database (29) were analyzed. Aque, A. queenslandica; Cele, Caenorhabditis elegans; Cint, Ciona intestinalis; Cowc, C. owczarzaki; Ddis, Dictyostelium discoideum; Dmel, D. melanogaster; Hmag, Hydra magnipapillata; Mbre, M. brevicollis; Mmus, Mus musculus; Nvec, N. vectensis; Pult, P. ultimum; Sros, S. rosetta; Tadh, T. adhaerens.
To increase the taxonomic breadth of genomes available from early branching metazoan lineages, we also sequenced the genome of the sponge O. carmela by using massively parallel sequencing (Illumina). Although the genome assembly is fragmented relative to traditional Sanger assemblies (SI Appendix), multiple cadherin-domain encoding sequences were detected and two cadherin genes assembled in near entirety (GenBank accession nos. JN197609 and AEC12441). The value of this draft genome for providing unique insights into cadherin evolution is demonstrated by the fact that one of the two assembled cadherins, JN197609, has homologs in choanoflagellates, despite being absent from the genome of the only other sequenced sponge, Amphimedon queenslandica, which encodes at least 17 cadherins (Fig. 2 and ref. 31).
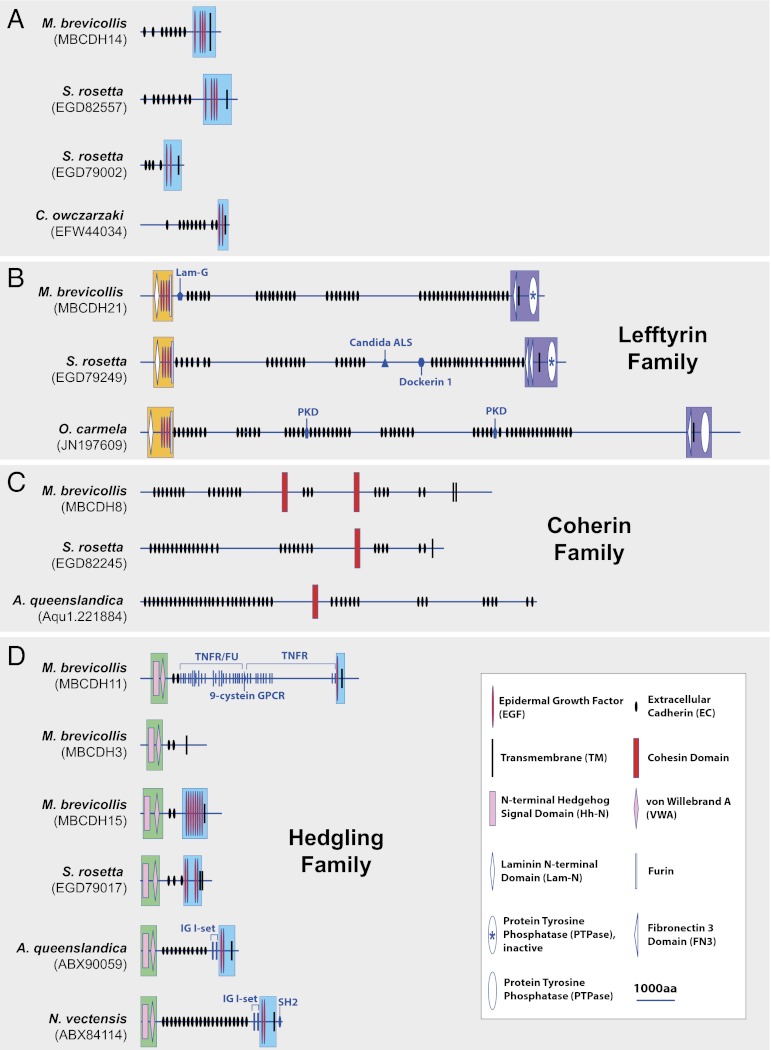
Fig. 2.
Predicted domain architecture of modern representatives of premetazoan cadherins. At least three cadherin families evolved before the origin of metazoans. (A) The single cadherin discovered in the genome of C. owczarzaki has a cassette of EGF repeats positioned proximal to a single transmembrane domain (blue box) that is also found in choanoflagellate and sponge cadherins. The phylogenetic relationships among cadherins with this feature are not yet clear. The lefftyrin (B) and coherin (C) families are present only in choanoflagellates and sponges. Lefftyrins are distinguished by an N-terminal “LEF” cassette (orange box) with a Lam-N domain, four EGF repeats, and a Furin repeat and a C-terminal “FTY” cassette (purple box) with one or two Fibronectin 3 domains, a transmembrane domain, and a tyrosine phosphatase domain. Coherins contain a diagnostic bacterial/archaeal-like cohesin (50) domain. (D) The hedgling family (1, 26) is present in choanoflagellates, sponges and cnidarians and is absent from bilaterians. All hedglings contain an N-terminal Hedgehog signal domain linked to a von Willebrand A domain (green box) and most contain a series of EGF repeats proximal to the transmembrane domain (blue box). Candida ALS, Candida Agglutinin-like sequence; IG I-set, Ig I-set; KU, BPTI/Kunitz family of serine protease inhibitors; Lam-G, Laminin G domain; 9-cystein GPCR, 9-cystein G protein coupled receptor; PKD, polycystic kidney disease; SH2, src homogy domain 2; TNFR, tumor necrosis factor receptor.
To reconstruct the evolutionary relationships among cadherins from nonmetazoans and early branching metazoans, we grouped cadherins from C. owczarzaki, choanoflagellates, and sponges according to shared structural features (i.e., domain composition and arrangement). Mapping of the phylogenetic distribution of cadherin families reveals that they have origins that predate the evolution of Metazoa. Although the earliest branching lineage to contain a predicted cadherin (Owcz_Cdh1) is C. owczarzaki, the evolutionary connection between this and cadherin families from choanoflagellate and metazoans is uncertain (Fig. 2A). Owcz_Cdh1 has at least 10 predicted EC domains, two membrane-proximal epidermal growth factor (EGF) domains, and a transmembrane (TM) domain. This domain organization resembles that of cadherins in the choanoflagellates M. brevicollis (accession no. MBCDH14) and S. rosetta (accession nos. EGD82557 and EGD79002) but is not sufficiently complex to definitively indicate that these proteins are orthologous.
In contrast, two cadherin families are clearly shared by choanoflagellates and sponges to the exclusion of all other lineages analyzed in this study. The first, lefftyrins, are defined by the presence of an amino-terminal “LEF” cassette [containing a Laminin N-terminal (Lam-N) domain, four EGF domains, and a Furin domain] and a carboxyl-terminus “FTY” cassette [containing one or two Fibronectin 3 (FN3) domains, a TM domain and a cytoplasmic protein tyrosine phosphatase (PTPase) domain; Fig. 2B]. The M. brevicollis lefftyrin family member, MBCDH21, also has an N-terminal Laminin G (Lam-G) domain that has prompted previous comparisons with metazoan classical cadherins and fat cadherins (1, 4). Cadherins in the second family, the coherins (Fig. 2C), are united by the presence of at least one cohesin domain (not to be confused with the eukaryotic cohesin protein that regulates sister chromatid separation). The presence of cohesin domains (SI Appendix, Fig. S3) in coherins is diagnostic because they are otherwise found only in bacteria and archaea (32).
Members of the remaining premetazoan family of cadherins, the hedglings (Fig. 2D), are found in choanoflagellates, sponges, and the cnidarian Nematostella vectensis (1, 25, 26), but are absent from C. owczarzaki and bilaterians. Hedglings contain an amino-terminal Hedgehog signal domain (Hh-N; ref. 33) that was thought to be exclusive to the secreted signaling portion of the metazoan-specific Hedgehog protein. The amino-terminal Hh-N domain in all hedglings is adjacent to a von Willebrand factor A (VWA) domain and, with the exception of one M. brevicollis hedgling (accession no. MBCDH3), all hedglings have a carboxyl-terminal cassette with between one and eight extracellular EGF domains positioned proximal to the TM region. Although the first identified choanoflagellate hedgling, MBCDH11 from M. brevicollis, contains additional domains (including TNFR, Furin, and 9-cystein GPCR), all other choanoflagellate hedglings detected in this study and all known metazoan hedglings lack these domains. Thus, hedgling in the last common ancestor of metazoans more likely resembled hedglings from metazoans (e.g., Aque_hedgling and Nvec_hedgling) and S. rosetta (accession no. EGD79017) than MBCDH11. The inference that the last common ancestor of choanoflagellates and metazoans contained lefftyrins, cohesins, and hedgling cadherins reveals the evolutionary foundations for the subsequent origin of metazoan-specific cadherins.
Metazoan Classical Cadherin/β-Catenin Adhesion Complex.
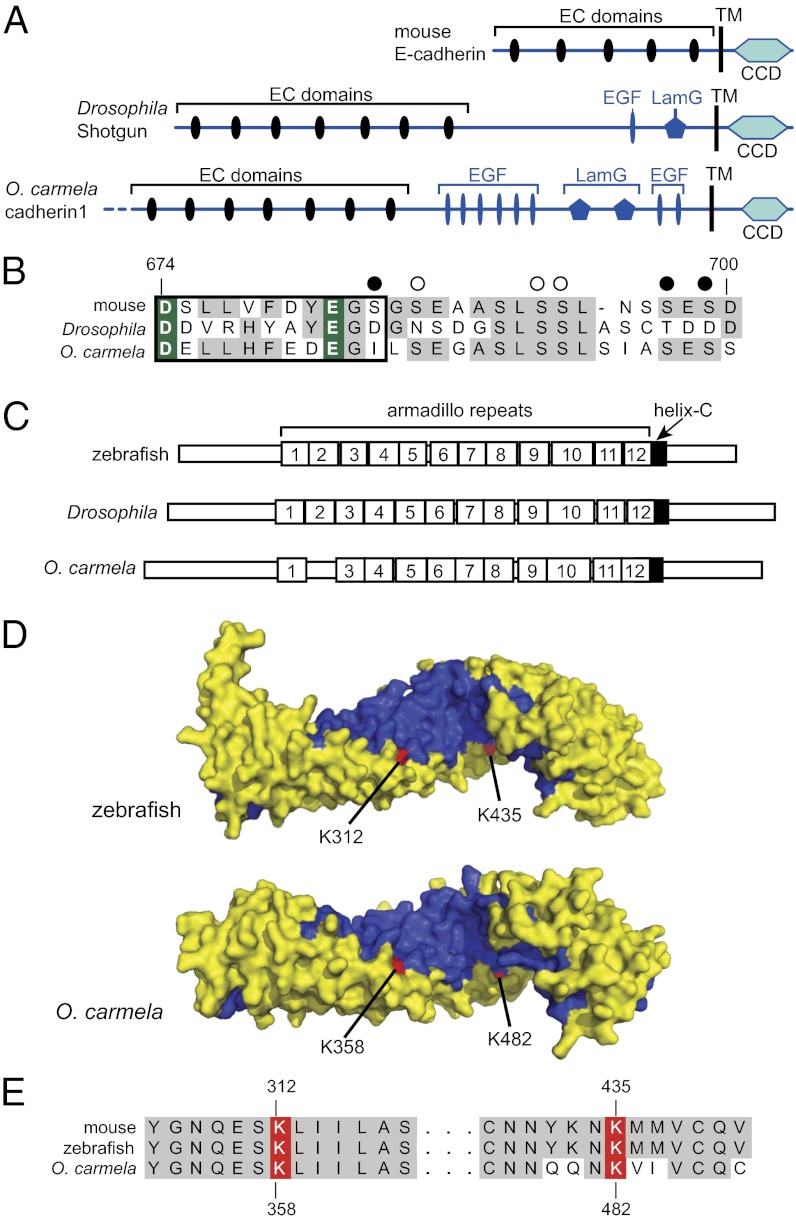
Among the cadherins that evolved along the metazoan stem lineage, classical cadherins have the clearest potential link to metazoan origins, both because of their ubiquity in modern metazoan lineages and because of their central roles in bilaterian cell adhesion (4). To investigate whether the adhesive functions of classical cadherins might extend to the earliest branching lineages of metazoans, we examined the possibility that the regulatory interaction between classical cadherins and β-catenin is conserved in sponges. The single detected classical cadherin homolog in O. carmela, OcCdh1 (GenBank accession no. AEC12441), encodes at least seven EC domains and a CCD domain, as well as multiple EGF and Lam-G domains that are typical of classical cadherins in invertebrates (e.g., Drosophila melanogaster N-cadherin and Shotgun; Fig. 3A and refs. 3 and 34). By aligning the amino acid sequence of the CCD of OcCdh1 with those of other classical cadherins, we found that two residues (D675 and E682) necessary for binding and modulating interactions with β-catenin (35) in bilaterians are conserved (Fig. 3B).
Fig. 3.
A conserved β-catenin/classical cadherin protein complex in a sponge. (A) The genome of the sponge O. carmela encodes a classical cadherin, Oc_cdh1, identified by the presence of the diagnostic cadherin cytoplasmic domain (CCD). Oc_cdh1 also has EGF and Lam-G domains in a membrane-proximal position that is typical of invertebrate classical cadherins (4). The dashed line at the N terminus of Oc_cdh1 indicates that the gene model is incomplete because of the draft nature of the genome assembly. (B) An alignment of a portion of the Oc_cdh1 CCD with bilaterian CCDs demonstrates the conservation of two residues (Aspartate and Glutamate, highlighted in green) required for binding to β-catenin (SI Appendix, Fig. S4 depicts the full alignment and includes the only known CCD from the demosponge A. queenslandica, in which critical β-catenin binding residues are also conserved). Conserved residues are shaded gray and Casein Kinase II and Glycogen Synthase Kinase 3b phosphorylation sites essential for the regulation of adhesion dynamics are indicated by filled or open circles, respectively (35, 38, 39). (C) The O. carmela genome also encodes a single β-catenin ortholog (Oc_bcat) with 11 predicted armadillo (arm) repeats and a helix-C domain; each arm repeat is numbered according to its similarity (determined by best-reciprocal Blast) with the 12 arm repeats from other metazoan β-catenin homologs (SI Appendix, Fig. S4). (D) Through comparison of a surface representation of the 3D structure of zebrafish β-catenin (37) with a structural model of Oc_bcat, we predict the conservation of a positively charged groove lined by the third helix (blue) of each arm repeat. Within this groove there are two lysine residues whose orientation resembles that of conserved lysines from zebrafish β-catenin. (E) These lysines align with Lysine-312 and Lysine-435 of mouse β-catenin, each of which are required for binding to mouse E-cadherin (35, 38, 39) at Aspartate-647 and Glutamate-682 (highlighted in B). Ocar_cdh1 was initially discovered from a yeast two-hybrid screen using full-length Ocar_bcat as bait (SI Appendix, Table S2; see SI Appendix for further discussion). CCD, cadherin cytoplasmic domain; EC, extracellular cadherin; EGF, epidermal growth factor domain; Lam-G, Laminin G domain; TM, transmembrane domain.
We next investigated whether O. carmela β-catenin (Oc_bcat; GenBank accession no. HQ234356) has diagnostic protein domains and residues indicative of the ability to interact with classical cadherins. Oc_bcat contains at least 11 of the 12 conserved armadillo (arm) repeats (36, 37) that are typical of eumetazoan β-catenin proteins (Fig. 3C) and shows 66.4% amino acid sequence identity with human β-catenin over the conserved arm-repeat region. Furthermore, Oc_bcat has two lysine residues (homologous to positions K312 and K435 in mouse) required for the interaction of mouse β-catenin with E-cadherin (Fig. 3 D and E and refs. 35, 38, and 39).
By threading the full-length sequence of Oc_bcat onto the crystal structure of zebrafish β-catenin (Fig. 3D), we predict that the third helix of each arm repeat in Oc_bcat orients along the surface of a positively charged groove that has been shown to contact E-cadherin directly in mouse (35, 38, 39). Moreover, the conserved lysines of β-catenin that are required to mediate interactions with E-cadherin are oriented similarly in the 3D models of the full-length zebrafish (37) and Oc_bcat. Furthermore, an unbiased yeast two-hybrid screen of O. carmela proteins using Oc_bcat as the “bait” recovered OcCdh1 as a binding partner (SI Appendix). Further study is required to determine whether OcCdh1 and Oc_bcat have the capacity to bind to each other directly in vivo and, thereby, contribute to cell adhesion in O. carmela.
Discussion
Cadherins represent a compelling case study for how large metazoan gene families evolve. Like members of most metazoan signaling and adhesion protein families, cadherins are typically large, multidomain proteins. Such protein families evolve through duplication and divergence and through the shuffling of protein domains among different protein families (40, 41). By using a phylogenetically informed comparative genomic approach, we were able to reconstruct a concrete portrait of the minimal cadherin diversity in the metazoan stem lineage. Furthermore, by reconstructing the ancestral domain composition of early-evolving cadherin families, we have been able to predict their evolutionary relationships with other, later-evolving modern protein families.
Premetazoan Cadherin Diversity.
An initially surprising result from the genome of M. brevicollis was that the genomes of choanoflagellates and most metazoans have comparable numbers of cadherin genes (1), despite vast differences in their biology. This result is further supported by our analysis of the S. rosetta genome, which has at least 29 predicted cadherin genes. In contrast, our analyses of cadherin relationships among metazoans, choanoflagellates, and C. owczarzaki suggest that as few as three modern cadherin families were present in the last common ancestor of choanoflagellates and metazoans, and that potentially only one cadherin was present in the last common ancestor of C. owczarzaki, choanoflagellates, and metazoans (Fig. 4A). However, these inferences may represent an underestimate because of limited available data. For example, C. owczarzaki is the only known member of its lineage, it diverged from choanoflagellates and metazoans more than 650 Mya, and it is a symbiont (42) that is likely to have evolved from a free-living ancestor; hence, aspects of its biology and genome content may be reduced.
Fig. 4.
An emerging model of cadherin evolution. (A) At least five modern families of cadherins—hedglings, coherins, lefftyrins, CELSR/flamingo and classical cadherins—evolved before the diversification of modern metazoans. Of these families, only the CELSR/flamingo and classical cadherin families are clearly conserved in all metazoan lineages (2, 4, 31). In contrast, among metazoans, hedgling is restricted to sponges and cnidarians. All of the cadherin families that evolved before the divergence of choanoflagellates and metazoans (“premetazoan” cadherin families) have been lost or have evolved beyond recognition in bilaterians. The relationships among the single cadherin detected in the genome of C. owczarzaki (Cowc_Cdh1) and other modern cadherin families are uncertain (indicated by dotted circle, also see Fig. 2A). (B) In addition to having EC domains, members of many cadherin families contain domains that provide clues to their evolutionary origins and to their relationships with other modern protein families (see Discussion).
The contrast between the large number of cadherins in modern lineages and the low diversity of cadherins inferred in the metazoan-stem lineage raises the intriguing possibility that modern cadherin diversity arose from a handful of ancestral cadherin families that still exist today (however, it is notable that all of the premetazoan cadherin families detected are absent from Bilateria). Alternatively, although future studies of a broader diversity of choanoflagellates and early branching metazoans may reveal additional members of the premetazoan cadherin repertoire, it is also possible that cadherins present in the ancestors of metazoans and choanoflagellates were subsequently lost (or evolved beyond recognition) in both lineages.
Radiation of Cadherins in Choanoflagellate and Metazoan Lineages.
The study of cadherin families conserved in choanoflagellates and metazoans promises to provide an unprecedented perspective on cadherin function before the evolution of metazoan multicellularity. Three cadherin families—lefftyrins, coherins, and hedglings—were present in the last common ancestor of metazoans and choanoflagellates and seem to have evolutionary connections to diverse metazoan signaling and adhesion gene families (Fig. 4B). For example, lefftyrins, so far known only from choanoflagellates and the sponge O. carmela, contain a Lam-N domain that is otherwise found in the proteins laminin, netrin, and usherin. These proteins are united by the fact that they function in the extracellular matrix (43–46). Furthermore, the carboxyl-terminal FTY cassette of lefftyrins is diagnostic of metazoan receptor PTPases, which help regulate cellular responses to interactions with neighboring cells and the extracellular matrix (47–49). C. owczarzaki is the most divergent outgroup of metazoa that has cadherins, and we have discovered that its genome also encodes a metazoan-like receptor PTPase that lacks EC domains (GenBank accession no. EFW39745). Thus, it seems that lefftyrins may have evolved through a domain-shuffling event that brought PTPase and EC domains together in the choanoflagellate/metazoan stem lineage.
Whereas lefftyrins may represent a case of protein family evolution through the process of domain shuffling, the newly discovered coherin family may have evolved through horizontal gene transfer. Coherins, which are restricted to choanoflagellates and sponges, are defined by the presence of EC domains and the cohesin domain. The cohesin domain is otherwise known only from archaea and bacteria. In the bacterial genus Clostridium, the cohesin domain functions in the assembly of the cellulosome, a complex of enzymes used to degrade plant cell walls (50). The possible evolutionary connection between coherins and the prokaryotic cohesin domain-containing proteins highlights the complexities of the evolutionary processes that shaped cadherin evolution during the early ancestry of Metazoa. Unless the cohesin domain of coherins evolved by convergent evolution with its prokaryotic counterpart, then it must have been acquired by horizontal gene transfer (32); this explanation seems quite plausible when considering that the earliest metazoan ancestors likely were bacterivorous (51). Either way, the presence of a cohesin domain in coherins is compelling evidence of the homology of these proteins between sponges and choanoflagellates.
Premetazoan Cadherin Functions.
Our understanding of the scope of cadherin function derives from their study in morphologically complex bilaterians, but C. owczarzaki is unicellular (42) and choanoflagellates exist as either single cells or simple undifferentiated colonies (52–54). Cadherins in these organisms may have functions that are unrelated to cadherin functions known from bilaterians. For example, even in colony-forming S. rosetta, adjacent cells are linked by cytoplasmic bridges and lack structures that resemble the cadherin-based adherens junctions of metazoans (53). However, it is possible to identify some analogous functions that might be served by cadherins in nonmetazoans. For example, cadherins in unicellular lineages could have adhesive functions other than the regulation of stable cell-cell adhesion, such as during bacterial prey capture, attachment to ECM, attachment to environmental substrates, or gamete recognition (although sex is undocumented in choanoflagellates).
One biological context in which cadherin function may be conserved between choanoflagellates and metazoans is in the collar cells of sponges. Like choanoflagellates, sponge collar cells have a motile flagellum used to generate water flow for the capture of bacterial prey on a surrounding microvillar collar where they are phagocytosed. It is reasonable to hypothesize that cadherin families restricted to sponges and choanoflagellates (i.e., lefftyrins and coherins), in particular, may have functions specific to the biology of collar cells. Such functions may include roles in the regulation of microvillar collar integrity or bacterial prey capture. Indeed, one cadherin (MBCDH1) has been shown to localize to the microvillar collar of M. brevicollis (1). Furthermore, there is precedent for a physiologically important interaction between bacteria and cadherins in metazoans: Some pathogenic bacteria interact with classical cadherins in gut epithelia, thereby stimulating the host cells to phagocytose the invading pathogen (55–57).
Linking Cadherin Evolution to the Origin of Metazoa.
A challenge for relating cadherin gene family evolution to metazoan morphological evolution is that, until now, none of the functionally characterized cadherin families of bilaterians have been studied in nonbilaterians. Of all of the modern cadherin families, the classical cadherin family is perhaps the strongest candidate for having played a role in the evolution of metazoan multicellularity (2, 4). The CCD of classical cadherins binds to β-catenin to regulate cell-cell adhesion in all studied bilaterian tissues. Here, we show that the genome of the sponge O. carmela encodes a typical nonchordate classical cadherin with a CCD domain-containing cytoplasmic tail that is predicted to be capable of binding to O. carmela β-catenin. Thus, it is plausible that an evolutionarily conserved classical cadherin/β-catenin adhesion complex was a feature of the cell biology of the last common ancestor of all modern metazoans.
The ubiquity of certain cadherin families in lineages that diverged more than 600 Mya indicates that these protein families have conserved (and essential) roles in organisms with vastly different biology. As we learn about their functions, we stand to gain insight into ancestral features of metazoans and their single-celled relatives—similarities that are fundamental to their basic cell biology.
Materials and Methods
The genomes of C. owczarzaki and S. rosetta were sequenced and assembled by the Broad Institute (Massachusetts Institute of Technology/Harvard; http://www.broadinstitute.org/annotation/genome/multicellularity_project/MultiHome.html), and the S. rosetta gene models were refined by using Illumina RNA-seq data. The O. carmela genome was sequenced by using paired-end Illumina reads at the Vincent J. Coates Genomic Sequencing Laboratory at the University of California, Berkeley and an early draft was assembled in-house. To identify new cadherins in these genomes, we performed protein homology-based searches (i.e., Blast; ref. 27) and domain-based searches (e.g., Pfam; ref. 29 and Smart; ref. 30). Any protein containing an EC domain was defined as a cadherin, and most of these also had a transmembrane domain. Cadherin families were identified based on the shared composition and arrangement of their protein domains. Structural predictions for Ocar_bcat were inferred by using LOOPP (58) to thread the full-length sequence onto the crystal structure of full-length zebrafish β-catenin.
For detailed experimental procedures, see SI Appendix.
Supplementary Material
Acknowledgments
We thank M. Abedin, S. Brenner, A. Brooks, M. Eisen, W. J. Nelson, M. Paris, D. Scannell, B. Steele, S. Q. Schneider, L. Tonkin, and Q. Zhou for technical support, advice, and helpful discussions. This work was supported in part by funding from an American Cancer Society Postdoctoral Fellowship (to S.A.N.), American Cancer Society Research Scholar Grant 116795-RSG-09-044-01-DDC (to N.K.), the National Aeronautics and Space Administration Astrobiology program (to N.K., S.A.N., and D.J.R.), the Hellman Family Fund (to N.K.), and a National Defense Science and Engineering Graduate fellowship from the Department of Defense (to D.J.R.). N.K. is a Fellow in the Integrated Microbial Biodiversity program of the Canadian Institute for Advanced Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. PRJNA20341 (Capsaspora genome); PRJNA37927S (Salpingoeca genome); EGD72656, EGD73963, EGD74518, EGD74667, EGD74707, EGD74783, EGD75074, EGD75359, EGD75381, EGD75404, EGD75405, EGD75586, EGD75710, EGD76846, EGD77346, EGD78086, EGD78170, GD78171, EGD78831, EGD78839, EGD78969, EGD78970, EGD79002, EGD79017, EGD79249, EGD80879, EGD80917, EGD81200, EGD82245, and EGD82557 (S. rosetta cadherins); EFW44034 (Capsaspora owczarzaki cadherins), JN197609 (Oscarella carmela lefftyrin), AEC12441 (Oscarella carmela cadherin 1), and HQ234356 (Oscarella carmela β-catenin)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1120685109/-/DCSupplemental.
References
- 1.Abedin M, King N. The premetazoan ancestry of cadherins. Science. 2008;319:946–948. doi: 10.1126/science.1151084. [DOI] [PubMed] [Google Scholar]
- 2.Hulpiau P, van Roy F. New insights into the evolution of metazoan cadherins. Mol Biol Evol. 2011;28:647–657. doi: 10.1093/molbev/msq233. [DOI] [PubMed] [Google Scholar]
- 3.Hynes RO, Zhao Q. The evolution of cell adhesion. J Cell Biol. 2000;150:F89–F96. doi: 10.1083/jcb.150.2.f89. [DOI] [PubMed] [Google Scholar]
- 4.Oda H, Takeichi M. Evolution: Structural and functional diversity of cadherin at the adherens junction. J Cell Biol. 2011;193:1137–1146. doi: 10.1083/jcb.201008173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rokas A. The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu Rev Genet. 2008;42:235–251. doi: 10.1146/annurev.genet.42.110807.091513. [DOI] [PubMed] [Google Scholar]
- 6.Angst BD, Marcozzi C, Magee AI. The cadherin superfamily: Diversity in form and function. J Cell Sci. 2001;114:629–641. doi: 10.1242/jcs.114.4.629. [DOI] [PubMed] [Google Scholar]
- 7.Saburi S, McNeill H. Organising cells into tissues: New roles for cell adhesion molecules in planar cell polarity. Curr Opin Cell Biol. 2005;17:482–488. doi: 10.1016/j.ceb.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Simons M, Mlodzik M. Planar cell polarity signaling: From fly development to human disease. Annu Rev Genet. 2008;42:517–540. doi: 10.1146/annurev.genet.42.110807.091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lévesque CA, et al. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol. 2010;11:R73. doi: 10.1186/gb-2010-11-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Overduin M, et al. Solution structure of the epithelial cadherin domain responsible for selective cell adhesion. Science. 1995;267:386–389. doi: 10.1126/science.7824937. [DOI] [PubMed] [Google Scholar]
- 11.King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- 12.Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- 13.Posy S, Shapiro L, Honig B. Sequence and structural determinants of strand swapping in cadherin domains: Do all cadherins bind through the same adhesive interface? J Mol Biol. 2008;378:954–968. doi: 10.1016/j.jmb.2008.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro L, et al. Structural basis of cell-cell adhesion by cadherins. Nature. 1995;374:327–337. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- 15.Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shapiro L, Weis WI. Structure and biochemistry of cadherins and catenins. Cold Spring Harb Perspect Biol. 2009;1:a003053. doi: 10.1101/cshperspect.a003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen YT, Stewart DB, Nelson WJ. Coupling assembly of the E-cadherin/beta-catenin complex to efficient endoplasmic reticulum exit and basal-lateral membrane targeting of E-cadherin in polarized MDCK cells. J Cell Biol. 1999;144:687–699. doi: 10.1083/jcb.144.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber AH, Stewart DB, Laurents DV, Nelson WJ, Weis WI. The cadherin cytoplasmic domain is unstructured in the absence of beta-catenin. A possible mechanism for regulating cadherin turnover. J Biol Chem. 2001;276:12301–12309. doi: 10.1074/jbc.M010377200. [DOI] [PubMed] [Google Scholar]
- 19.Okazaki M, et al. Molecular cloning and characterization of OB-cadherin, a new member of cadherin family expressed in osteoblasts. J Biol Chem. 1994;269:12092–12098. [PubMed] [Google Scholar]
- 20.Casal J, Lawrence PA, Struhl G. Two separate molecular systems, Dachsous/Fat and Starry night/Frizzled, act independently to confer planar cell polarity. Development. 2006;133:4561–4572. doi: 10.1242/dev.02641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodrich LV, Strutt D. Principles of planar polarity in animal development. Development. 2011;138:1877–1892. doi: 10.1242/dev.054080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viktorinová I, König T, Schlichting K, Dahmann C. The cadherin Fat2 is required for planar cell polarity in the Drosophila ovary. Development. 2009;136:4123–4132. doi: 10.1242/dev.039099. [DOI] [PubMed] [Google Scholar]
- 23.Morishita H, Yagi T. Protocadherin family: Diversity, structure, and function. Curr Opin Cell Biol. 2007;19:584–592. doi: 10.1016/j.ceb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Kazmierczak P, et al. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449:87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- 25.King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adamska M, et al. The evolutionary origin of hedgehog proteins. Curr Biol. 2007;17:R836–R837. doi: 10.1016/j.cub.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 29.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fahey B, Degnan BM. Origin of animal epithelia: Insights from the sponge genome. Evol Dev. 2010;12:601–617. doi: 10.1111/j.1525-142X.2010.00445.x. [DOI] [PubMed] [Google Scholar]
- 32.Peer A, Smith SP, Bayer EA, Lamed R, Borovok I. Noncellulosomal cohesin- and dockerin-like modules in the three domains of life. FEMS Microbiol Lett. 2009;291:1–16. doi: 10.1111/j.1574-6968.2008.01420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall TM, Porter JA, Beachy PA, Leahy DJ. A potential catalytic site revealed by the 1.7-A crystal structure of the amino-terminal signalling domain of Sonic hedgehog. Nature. 1995;378:212–216. doi: 10.1038/378212a0. [DOI] [PubMed] [Google Scholar]
- 34.Iwai Y, et al. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- 35.Huber AH, Weis WI. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 36.Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 37.Xing Y, et al. Crystal structure of a full-length beta-catenin. Structure. 2008;16:478–487. doi: 10.1016/j.str.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gooding JM, Yap KL, Ikura M. The cadherin-catenin complex as a focal point of cell adhesion and signalling: New insights from three-dimensional structures. Bioessays. 2004;26:497–511. doi: 10.1002/bies.20033. [DOI] [PubMed] [Google Scholar]
- 39.Graham TA, Weaver C, Mao F, Kimelman D, Xu W. Crystal structure of a beta-catenin/Tcf complex. Cell. 2000;103:885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- 40.Doolittle RF. The origins and evolution of eukaryotic proteins. Philos Trans R Soc Lond B Biol Sci. 1995;349:235–240. doi: 10.1098/rstb.1995.0107. [DOI] [PubMed] [Google Scholar]
- 41.Lundin LG. Gene duplications in early metazoan evolution. Semin Cell Dev Biol. 1999;10:523–530. doi: 10.1006/scdb.1999.0333. [DOI] [PubMed] [Google Scholar]
- 42.Hertel LA, Bayne CJ, Loker ES. The symbiont Capsaspora owczarzaki, nov. gen. nov. sp., isolated from three strains of the pulmonate snail Biomphalaria glabrata is related to members of the Mesomycetozoea. Int J Parasitol. 2002;32:1183–1191. doi: 10.1016/s0020-7519(02)00066-8. [DOI] [PubMed] [Google Scholar]
- 43.Colognato H, Yurchenco PD. Form and function: The laminin family of heterotrimers. Dev Dyn. 2000;218:213–234. doi: 10.1002/(SICI)1097-0177(200006)218:2<213::AID-DVDY1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 44.Eudy JD, et al. Mutation of a gene encoding a protein with extracellular matrix motifs in Usher syndrome type IIa. Science. 1998;280:1753–1757. doi: 10.1126/science.280.5370.1753. [DOI] [PubMed] [Google Scholar]
- 45.Serafini T, et al. The netrins define a family of axon outgrowth-promoting proteins homologous to C. elegans UNC-6. Cell. 1994;78:409–424. doi: 10.1016/0092-8674(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 46.Vuolteenaho R, Chow LT, Tryggvason K. Structure of the human laminin B1 chain gene. J Biol Chem. 1990;265:15611–15616. [PubMed] [Google Scholar]
- 47.Petrone A, Sap J. Emerging issues in receptor protein tyrosine phosphatase function: Lifting fog or simply shifting? J Cell Sci. 2000;113:2345–2354. doi: 10.1242/jcs.113.13.2345. [DOI] [PubMed] [Google Scholar]
- 48.Blanchetot C, Tertoolen LG, Overvoorde J, den Hertog J. Intra- and intermolecular interactions between intracellular domains of receptor protein-tyrosine phosphatases. J Biol Chem. 2002;277:47263–47269. doi: 10.1074/jbc.M205810200. [DOI] [PubMed] [Google Scholar]
- 49.Tonks NK. Protein tyrosine phosphatases: From genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 50.Carvalho AL, et al. Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex. Proc Natl Acad Sci USA. 2003;100:13809–13814. doi: 10.1073/pnas.1936124100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nichols SA, Dayel MJ, King N. Genomic, phylogenetic and cell biological insights into metazoan origins. In: Telford MJ, Littlewood D, editors. Animal evolution: Genomes, fossils and trees. Oxford: Oxford Univ Press; 2009. pp. 24–32. [Google Scholar]
- 52.Leadbeater BSC. Life-history and ultrastructure of a new marine species of Proterospongia (Choanoflagellida) J Mar Biol Assoc U K. 1983;63:135–160. [Google Scholar]
- 53.Dayel MJ, et al. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev Biol. 2011;357:73–82. doi: 10.1016/j.ydbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karpov S, Coupe S. A revision of choanoflagellate genera Kentrosiga Schiller, 1953 and Desmarella Kent, 1880. Acta Protozool. 1998;37:23–27. [Google Scholar]
- 55.Mengaud J, Ohayon H, Gounon P, Cossart P, Cossart P, Mege R-M. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell. 1996;84:923–932. doi: 10.1016/s0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- 56.Boyle EC, Finlay BB. Bacterial pathogenesis: Exploiting cellular adherence. Curr Opin Cell Biol. 2003;15:633–639. doi: 10.1016/s0955-0674(03)00099-1. [DOI] [PubMed] [Google Scholar]
- 57.Blau K, et al. Flamingo cadherin: A putative host receptor for Streptococcus pneumoniae. J Infect Dis. 2007;195:1828–1837. doi: 10.1086/518038. [DOI] [PubMed] [Google Scholar]
- 58.Tobi D, Elber R. Distance-dependent, pair potential for protein folding: Results from linear optimization. Proteins. 2000;41:40–46. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.