Abstract
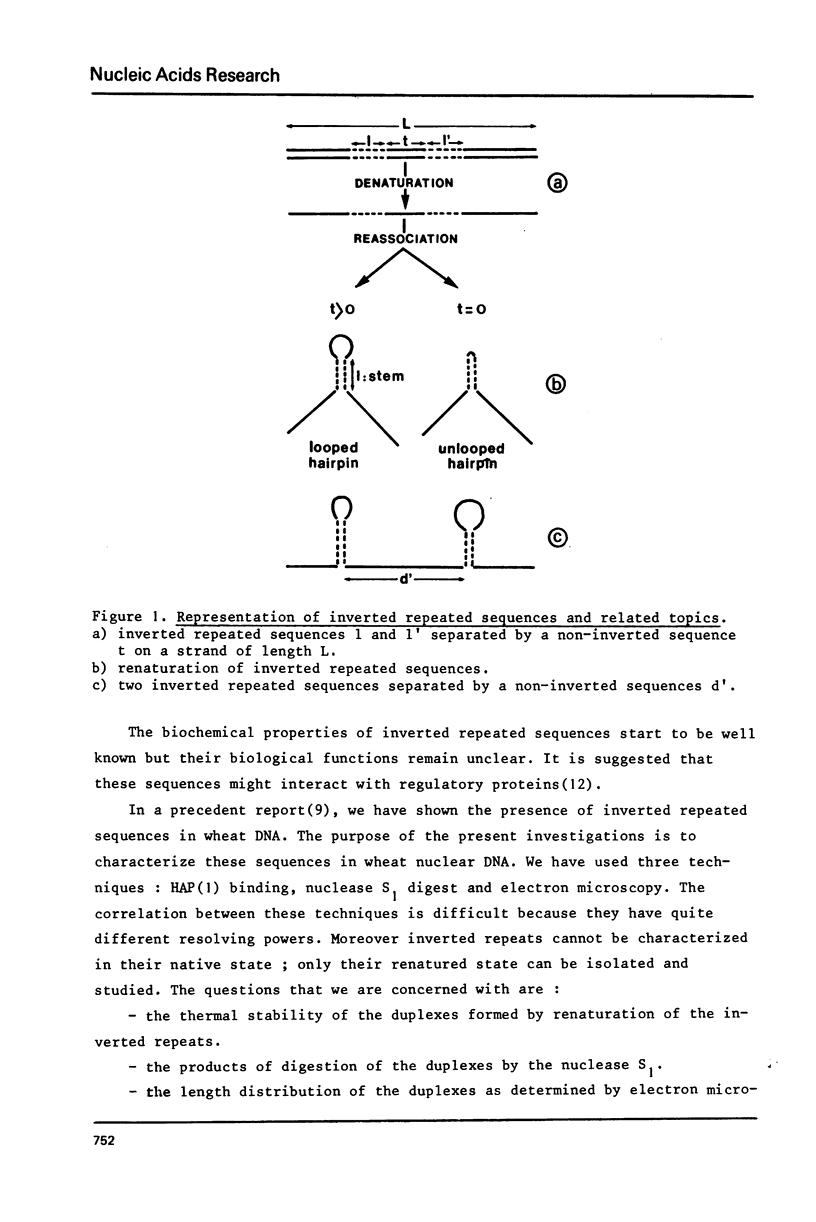
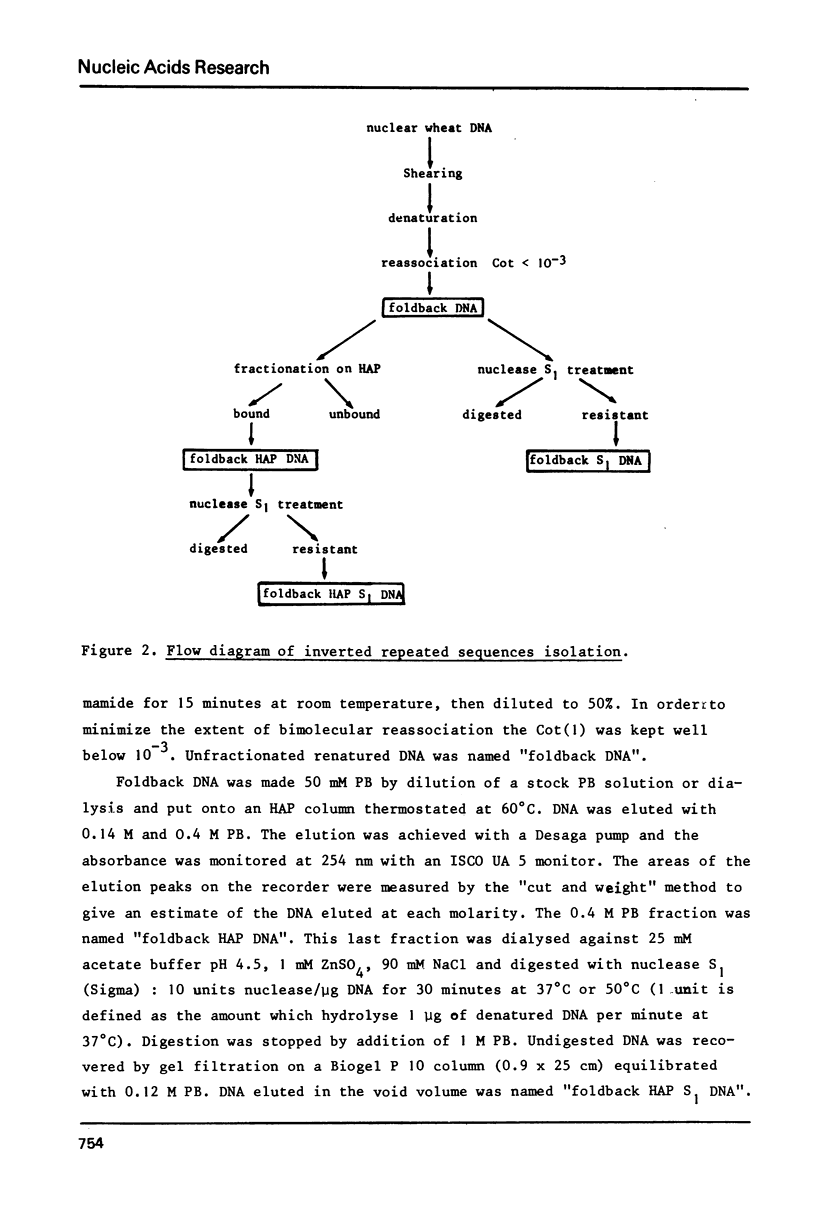
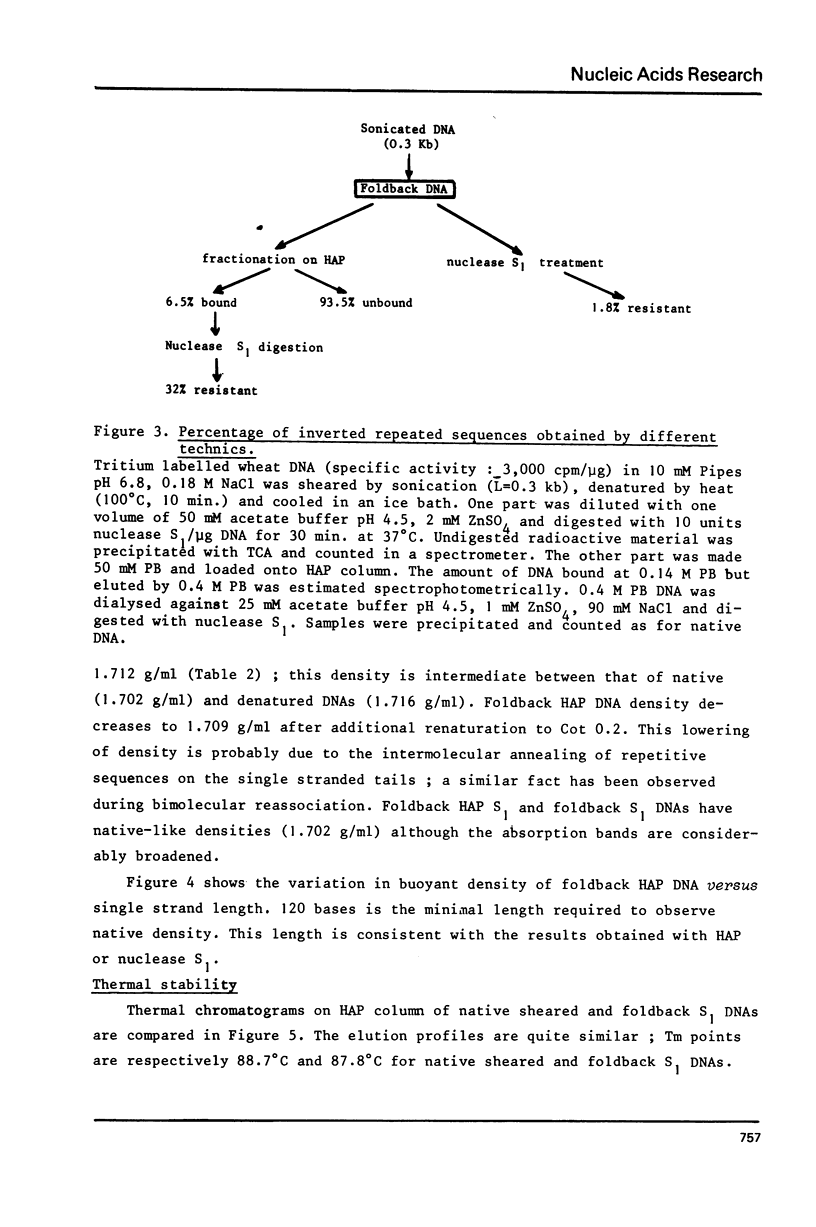
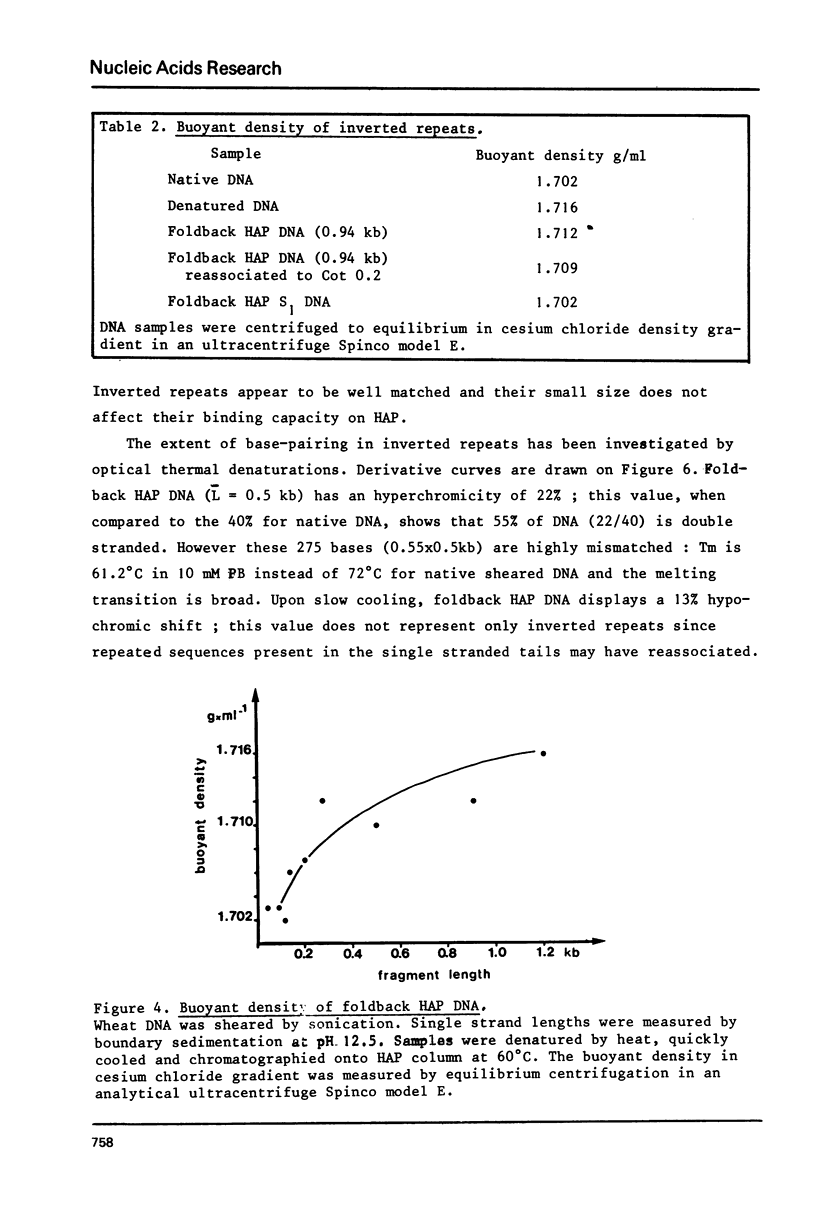
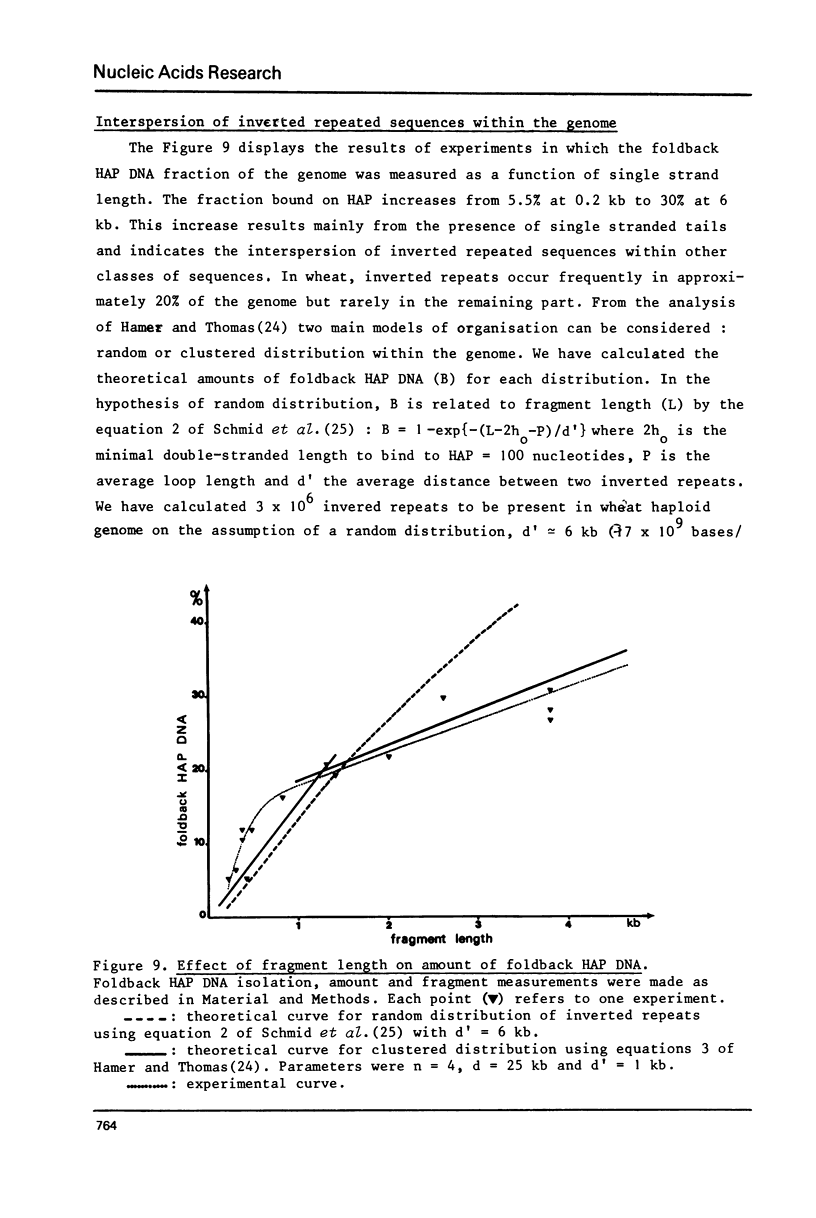
The properties of inverted repeated sequences in wheat nuclear DNA have been studied by HAP(1) chromatography, nuclease S1 digestion and electron microscopy. Inverted repeated sequences comprise 1.7% of wheat genome. The HAP studies show that the amount of "foldback HAP bound DNA" depends on DNA length. Inverted repeats appear to be clustered with an average intercluster distance of 25 kb. It is estimated that there are approximately 3 x 10(6) inverted repeats per haploid wheat genome. The sequences around inverted repeats involve all families of repetition frequencies. Inverted repeats are observed as hairpins in electron microscopy. 20% of hairpins are terminated by a single-stranded spacer ranging from 0.3 to 1.5 kb in length. Duplex regions of the inverted repeats range from 0.1 to 0.45 kb with number average values of 0.24 kb and 0.18 kb for unlooped and looped hairpin respectively. Thermal denaturations and nuclease S1 digestions have revealed a length of about 100 bases for duplex regions. The methods used to study inverted repeated sequences are compared and discussed.
Full text
PDF




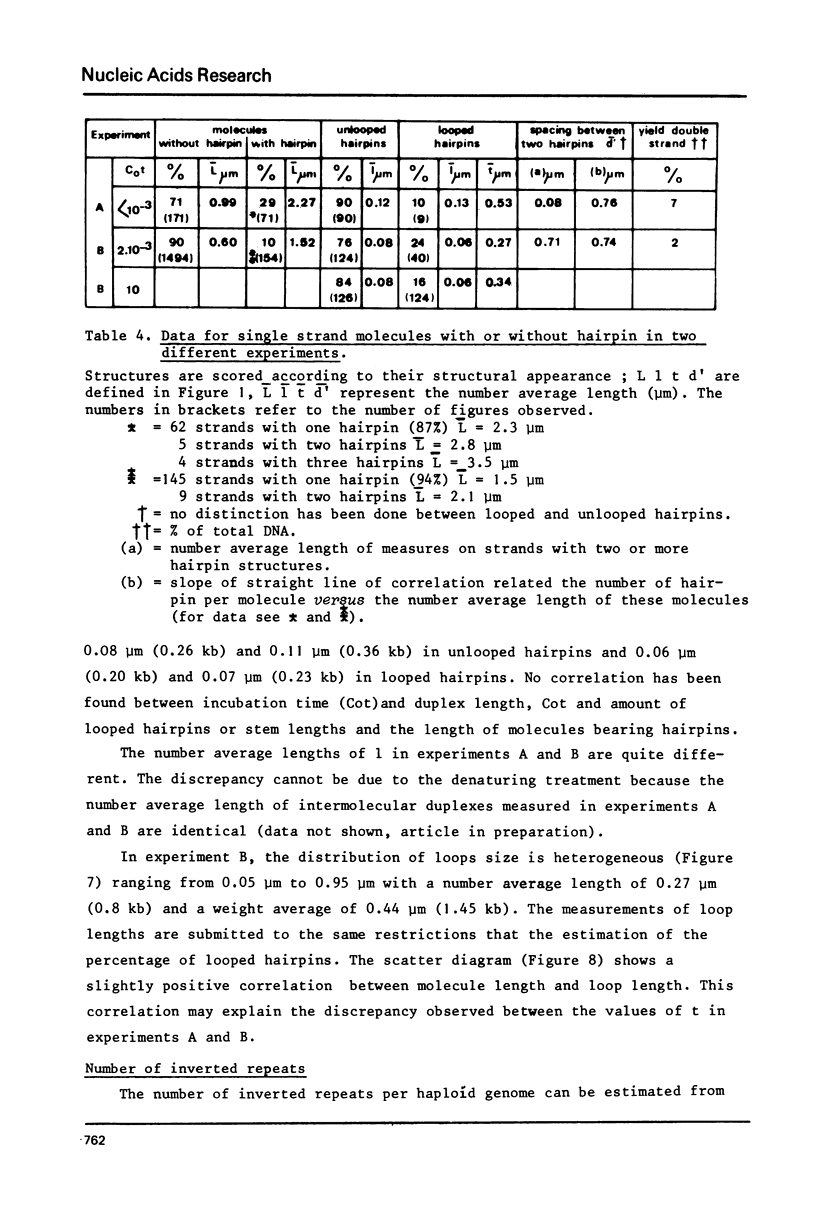





Images in this article
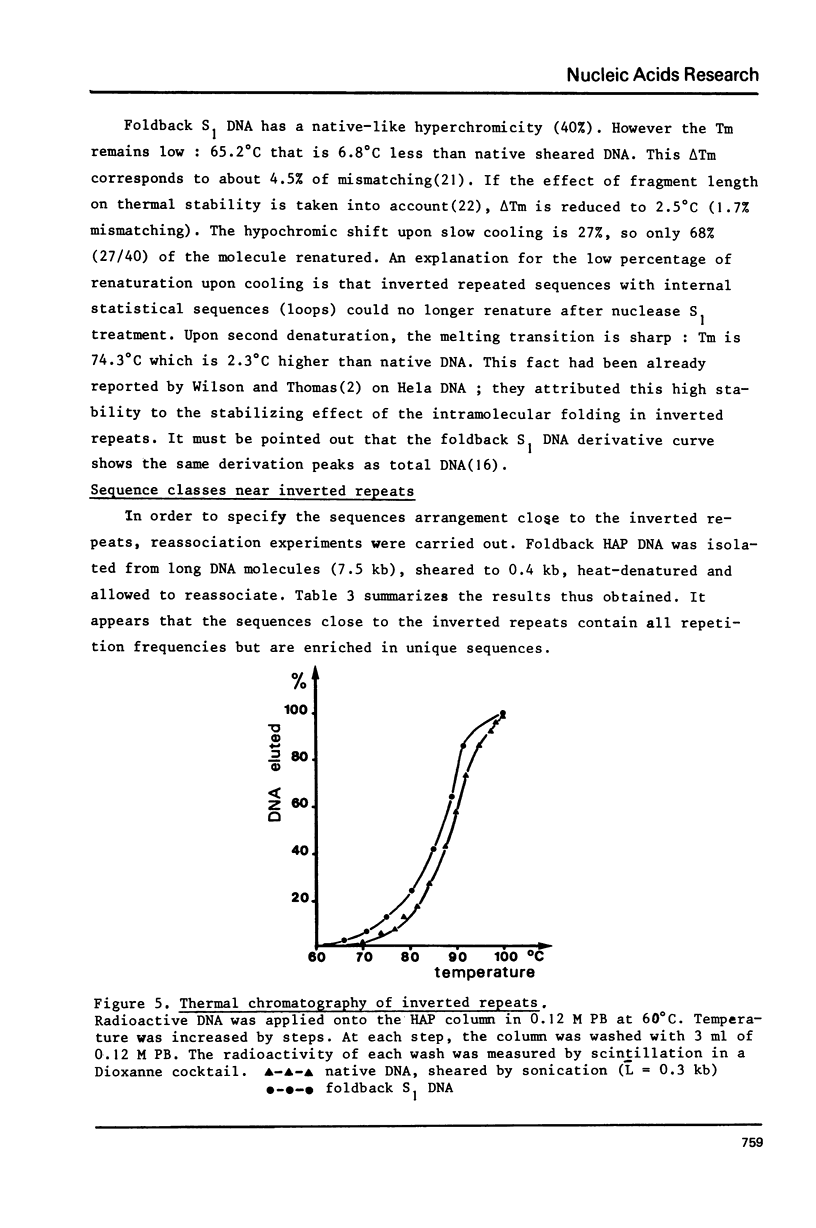
Selected References
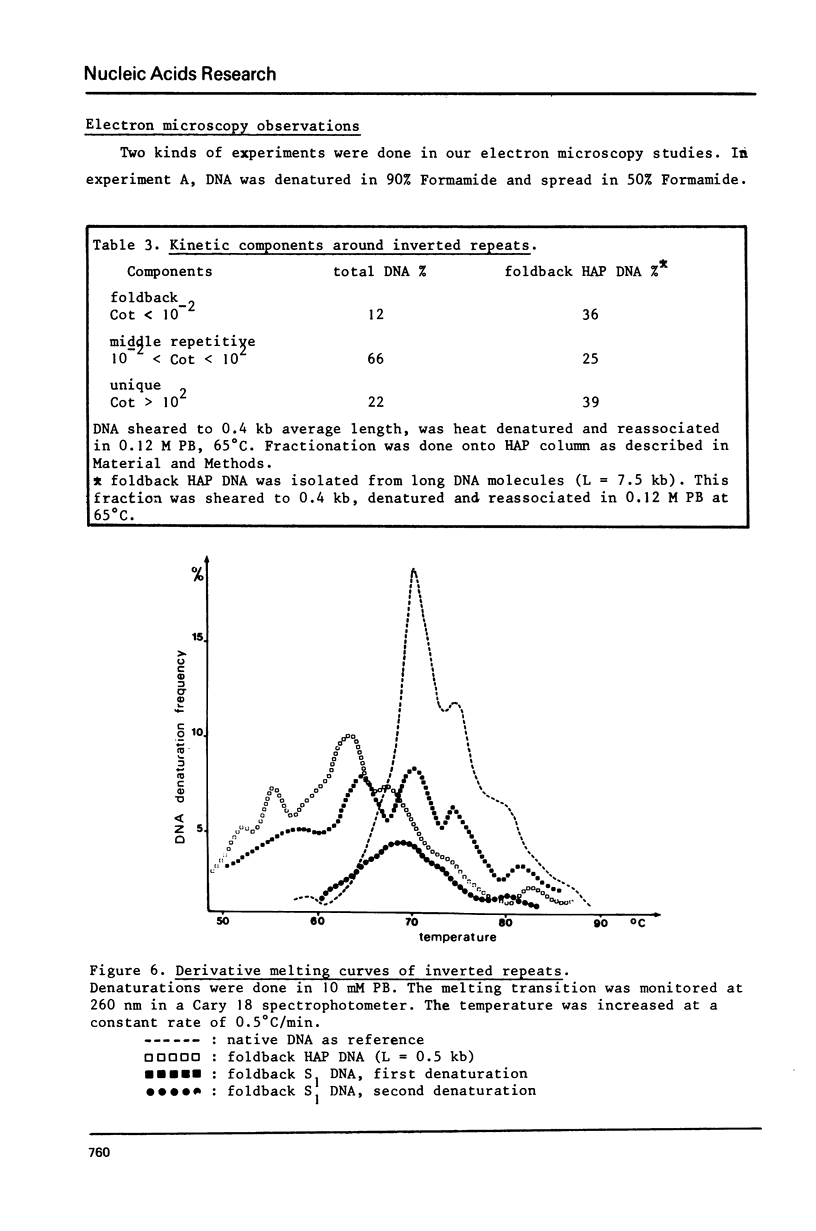
These references are in PubMed. This may not be the complete list of references from this article.
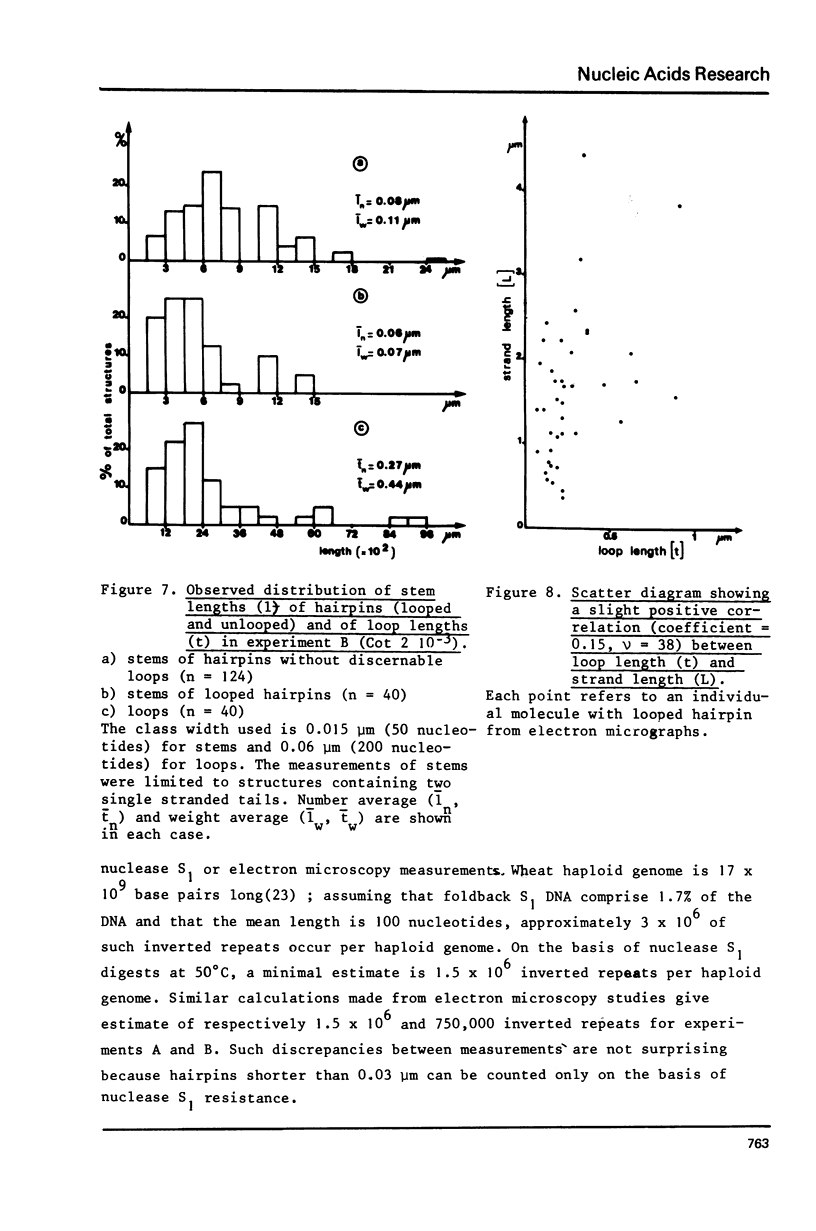
- Bell A. J., Hardman N. Characterization of foldback sequences in hamster DNA using electron microsocpy. Nucleic Acids Res. 1977 Jan;4(1):247–268. doi: 10.1093/nar/4.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Hearst J. E. An electron microscopic study of mouse foldback DNA. Cell. 1975 Aug;5(4):429–446. doi: 10.1016/0092-8674(75)90062-8. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Pardue M. L. Electron microscopy of DNA crosslinked with trimethylpsoralen: test of the secondary structure of eukaryotic inverted repeat sequences. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2644–2648. doi: 10.1073/pnas.73.8.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Rosenfeld A., Hearst J. E. Characterization of the most rapidly renaturing sequences in mouse main-band DNA. J Mol Biol. 1973 Dec 15;81(3):299–325. doi: 10.1016/0022-2836(73)90143-5. [DOI] [PubMed] [Google Scholar]
- Cohen S. N. Transposable genetic elements and plasmid evolution. Nature. 1976 Oct 28;263(5580):731–738. doi: 10.1038/263731a0. [DOI] [PubMed] [Google Scholar]
- Darlix J. L., Horaist M. Existence and possible roles of transcriptional barriers in T7 DNA early region as shown by electron microscopy. Nature. 1975 Jul 24;256(5515):288–292. doi: 10.1038/256288a0. [DOI] [PubMed] [Google Scholar]
- Deininger P. L., Schmid C. W. An electron microscope study of the DNA sequence organization of the human genome. J Mol Biol. 1976 Sep 25;106(3):773–790. doi: 10.1016/0022-2836(76)90264-3. [DOI] [PubMed] [Google Scholar]
- Deonier R. C., Hadley R. C. Distribution of inverted IS-length sequences in the E. coli K12 genome. Nature. 1976 Nov 11;264(5582):191–193. doi: 10.1038/264191a0. [DOI] [PubMed] [Google Scholar]
- Dickson R. C., Abelson J., Barnes W. M., Reznikoff W. S. Genetic regulation: the Lac control region. Science. 1975 Jan 10;187(4171):27–35. doi: 10.1126/science.1088926. [DOI] [PubMed] [Google Scholar]
- Dott P. J., Chuang C. R., Saunders G. F. Inverted repetitive sequences in the human genome. Biochemistry. 1976 Sep 7;15(18):4120–4125. doi: 10.1021/bi00663a032. [DOI] [PubMed] [Google Scholar]
- Hamer D. H., Thomas C. A., Jr Palindrome theory. J Mol Biol. 1974 Mar 25;84(1):139–144. doi: 10.1016/0022-2836(74)90217-4. [DOI] [PubMed] [Google Scholar]
- Hardman N., Jack P. L. Characterization of foldback sequences in Physarum polycephalum nuclear DNA using the electron microscope. Eur J Biochem. 1977 Apr 1;74(2):275–283. doi: 10.1111/j.1432-1033.1977.tb11391.x. [DOI] [PubMed] [Google Scholar]
- Huguet T., Jouanin L. The heterogeneity of wheat nuclear DNA. Biochim Biophys Acta. 1972 Apr 12;262(4):431–440. doi: 10.1016/0005-2787(72)90486-8. [DOI] [PubMed] [Google Scholar]
- JOHNSTON F. B., STERN H. Mass isolation of viable wheat embryos. Nature. 1957 Jan 19;179(4551):160–161. doi: 10.1038/179160b0. [DOI] [PubMed] [Google Scholar]
- Jelinek W., Darnell J. E. Double-stranded regions in heterogeneous nuclear RNA from Hela cells. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2537–2541. doi: 10.1073/pnas.69.9.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A. C., Borstad L., Fraser M. J., Denhardt D. T. Isolation of repeated and self-complementary sequences from E. coli DNA. Nucleic Acids Res. 1974 Nov;1(11):1539–1548. doi: 10.1093/nar/1.11.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Ptashne M., Barrell B. G., Donelson J. Sequence of a repressor-binding site in the DNA of bacteriophage lamda. Nature. 1974 Aug 2;250(465):394–397. doi: 10.1038/250394a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Isolation of inverted repeat sequences, including IS1, IS2, and IS3, in Escherichia coli plasmids. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2316–2320. doi: 10.1073/pnas.73.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays E. Double-stranded RNA in chromatin transcripts formed by exogenous RNA polymerase. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1121–1125. doi: 10.1073/pnas.73.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryskov A. P., Saunders G. F., Farashyan V. R., Georgiev G. P. Double-helical regions in nuclear precursor of mRNA (pre-mRNA). Biochim Biophys Acta. 1973 Jun 8;312(1):152–164. doi: 10.1016/0005-2787(73)90060-9. [DOI] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Deininger P. L. Sequence organization of the human genome. Cell. 1975 Nov;6(3):345–358. doi: 10.1016/0092-8674(75)90184-1. [DOI] [PubMed] [Google Scholar]
- Schmid C. W., Manning J. E., Davidson N. Inverted repeat sequences in the Drosophila genome. Cell. 1975 Jun;5(2):159–172. doi: 10.1016/0092-8674(75)90024-0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Flavell R. B. Nucleotide sequence organisation in the rye genome. Biochim Biophys Acta. 1977 Jan 3;474(1):82–97. doi: 10.1016/0005-2787(77)90216-7. [DOI] [PubMed] [Google Scholar]
- Smith M. J., Britten R. J., Davidson E. H. Studies on nucleic acid reassociation kinetics: reactivity of single-stranded tails in DNA-DNA renaturation. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4805–4809. doi: 10.1073/pnas.72.12.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell H. M. Molecular mechanism for genetic recombination. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2483–2487. doi: 10.1073/pnas.69.9.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szala S., Michalska J., Paterak H., Bieniek B., Chorazy M. Inverted sequences in rat DNA. FEBS Lett. 1977 May 1;77(1):94–98. doi: 10.1016/0014-5793(77)80200-7. [DOI] [PubMed] [Google Scholar]
- Tuffet A., Huguet T. Séquences en double chaîne dans l'ARN transcrit in vitro par la RNA polymérase de E. coli sur l'ADN nucléaire de blé. C R Acad Sci Hebd Seances Acad Sci D. 1975 Sep 29;281(13):933–936. [PubMed] [Google Scholar]
- Ullman J. S., McCarthy B. J. The relationship between mismatched base pairs and the thermal stability of DNA duplexes. I. Effects of depurination and chain scission. Biochim Biophys Acta. 1973 Feb 4;294(1):405–415. doi: 10.1016/0005-2787(73)90095-6. [DOI] [PubMed] [Google Scholar]
- Wagner R. E., Jr, Radman M. A mechanism for initiation of genetic recombination. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3619–3622. doi: 10.1073/pnas.72.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V., Dure L. S., 3rd Developmental biochemistry of cotton seed embryogenesis and germination. VII. Characterization of the cotton genome. J Mol Biol. 1976 Mar 15;101(4):503–536. doi: 10.1016/0022-2836(76)90242-4. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of ribosomal RNA and DNA. I. Processing of Xenopus laevis ribosomal RNA and structure of single-stranded ribosomal DNA. J Mol Biol. 1974 Oct 25;89(2):379–395. doi: 10.1016/0022-2836(74)90526-9. [DOI] [PubMed] [Google Scholar]
- Wilson D. A., Thomas C. A., Jr Hydroxyapatite chromatography of short double-helical DNA. Biochim Biophys Acta. 1973 Dec 21;331(3):333–340. doi: 10.1016/0005-2787(73)90019-1. [DOI] [PubMed] [Google Scholar]
- Wilson D. A., Thomas C. A., Jr Palindromes in chromosomes. J Mol Biol. 1974 Mar 25;84(1):115–138. doi: 10.1016/0022-2836(74)90216-2. [DOI] [PubMed] [Google Scholar]
- Wuilmart C., Urbain J., Givol D. On the location of palindromes in immunoglobulin genes. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2526–2530. doi: 10.1073/pnas.74.6.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G., Blomberg C. Kinetic aspects of the DNA helix - cruciform transition. J Theor Biol. 1976 Dec;63(2):347–353. doi: 10.1016/0022-5193(76)90038-2. [DOI] [PubMed] [Google Scholar]



