Most bacteria are capable of two general modes of growth, a planktonic or community-based lifestyle. In planktonic growth, organisms exist as independent, free-living cells, swimming or suspended in a liquid medium depending on their motility capacity. When environmental situations dictate, bacteria can grow together as a group and form biofilm structures, in which the cells live clustered in a matrix-encased community (1). Biofilm bacteria are usually depicted as being sessile and entrapped within an extracellular matrix, remaining nonmotile and in a unique state of growth (2). In PNAS, the work by Houry et al. (3) challenges this view of biofilm bacteria with the discovery that a subpopulation consisting of 0.1–1% of cells within a biofilm, referred to as stealth swimmers, has the ability to remain motile. Previous work has shown a role for motility in the initial attachment to surfaces, the construction of 3D structures, and the final step of biofilm disassembly (4). The work by Houry et al. (3) uses time lapse imaging of fluorescently labeled Bacillus thuringiensis to uncover the presence of the motile subpopulation, and they also discover that the motile bacilli move through the biofilm biomass. These swimmers could tunnel into the structured biofilm matrix, generating short-lived pores that facilitate the diffusion of nutrients and macromolecules. In support of these observations, motile-deficient strains are unable to replicate the phenotype. Through exogenous addition experiments, motile B. thuringiensis are found to penetrate an established biofilm and are visualized at the base of the structure, providing evidence that the presence of a swimming population is not a biofilm-specific mutational event. The work by Houry et al. (3) also concludes that the contribution of these swimmers originates from the planktonic population. These important observations indicate that current models of microbial motility in the context of biofilm development need to be revisited.
The matrix of a biofilm is quite complex and consists of exopolysaccharide, extracellular DNA, proteins, and other cell envelope material (5). Despite this intricate framework, the stealth swimmers are able to navigate through this seemingly impenetrable material. However, the age of the biofilm was found to dictate the ability of swimmers to tunnel into biomass. Biofilms are known to evolve over time and change matrix composition (6), and the altered resistance to swimmer penetration reinforces the fact that biofilms are not static entities. The work by Houry et al. (3) observes that changing viscoelastic properties in older biofilms because of increased levels of exopolysaccharide results in attenuated permeability to swimmer cells. It is also observed that the tunneling ability is not intrinsic to all motile bacteria, suggesting that the strength of an organism’s motility or some other unique structural feature of these biofilms could limit stealth swimmer activity. Other than exopolysaccharide effects, it will be interesting to examine how extracellular DNA, protein components, and amyloid fibers in the biofilm matrix influence the ability of stealth swimmers to migrate.
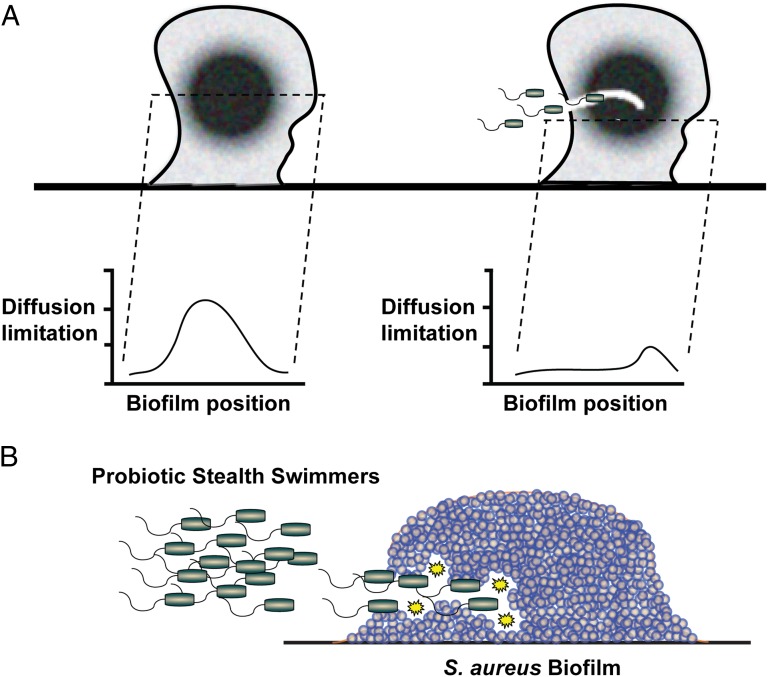
The biofilm matrix has been shown to act as a molecular sieve that limits the diffusion of signaling molecules, nutrients, oxygen, waste products, and antimicrobials (5). Combined with the effects of hydrodynamic forces, biofilms develop into heterogeneous structures that are an ensemble of distinct microenvironments with unique physical and genetic properties. The work by Houry et al. (3) shows that pore formation by stealth swimmers facilitates the diffusion of macromolecules throughout an established biofilm community by tracking the migration of fluorescently labeled tracers (Fig. 1A). This observation may have profound effects on how we view biofilm heterogeneity and the pioneering approaches to deliver small molecules to the most diffusion-limited regions of biofilm structures.
Fig. 1.
Stealth swimmers tunnel into bacterial biofilms. The generation of transient pores by the swimmers improves diffusion limitations (A). Through the use of engineered swimmers that express antimicrobial agents, probiotic biofilm treatments that target problematic chronic infections, such as those infections caused by S. aureus (B), could be developed.
The work by Houry et al. (3) also observes that stealth swimming functions in polymicrobial situations. B. thuringiensis swimmers were able to tunnel into biofilms produced by both Gram-positive (Staphylococcus aureus and Enterococcus faecalis) and -negative (Yersinia enterocolitica, Pseudomonas aeruginosa, and Listeria monocytogenes) pathogens. In addition, other motile Bacillus species are observed to tunnel into S. aureus biofilms, suggesting that stealth swimming in heterologous biofilms could be a widespread phenomenon in the environment.
Biofilms are notoriously difficult to eradicate with chemotherapeutic approaches (7), and physical methods, such as surgical debridement or removal of colonized devices, must be performed to treat the chronic infection (8). The extracellular matrix serves as a protective barrier for microbes deep within the community (9), limiting the diffusion of antimicrobials and rendering them ineffective (10). The work by Houry et al. (3) shows that transient pores generated by stealth swimmers can render S. aureus biofilms over 100 times more sensitive to the common biocide benzalkonium chloride. Houry et al. (3) also observe that a B. thuringiensis strain engineered to produced lysostaphin, an endopeptidase that cleaves the pentaglycine cross-links specific to the S. aureus cell wall (11), is effective at eradicating established S. aureus biofilms of multiple different strain types (Fig. 1B). Considering the importance of S. aureus and other pathogens in chronic biofilm infections (8), the ability to eliminate a recalcitrant biofilm
Pore formation by stealth swimmers facilitates the diffusion of macromolecules throughout an established biofilm community.
using a probiotic approach suggests that this application of stealth swimmers warrants additional investigation.
On some level, it is surprising that a minor population of stealth swimmers cells within the total biofilm can have such profound impact on community behavior. The fact that diverse populations of cells exist in the biofilm to carry out different functions can be related to classic ecological theory. Diversity can improve both the productivity and stress resistance of biological communities, and this concept forms the basis of the insurance hypothesis, which states that the presence of diverse subpopulations increases the range of environmental conditions in which community members will survive or thrive (12). Insurance effects could be translated by swimmer cells to the rest of the biofilm structure by providing improved nutrient availability and generating channels to release toxic waste products accumulating deep within the biofilm layers.
The work by Houry et al. (3) that discovers the behavior of stealth swimmers raises questions about the biofilm development program and outlines potential avenues for combating resistant biofilm infections. It remains to be determined why only a small part of the population exists as swimmer cells and how different matrix components, biofilm age, and environmental conditions influence the ability of stealth swimmer to tunnel through complex biofilm structures. The fact that bacterial mutants with altered matrix material, such as a B. subtilis abrB mutant, are refractive to swimmer activity (3) suggests that our understanding of the contributions of the biofilm matrix components to this mechanism is in need of additional investigation. Signaling molecules may also play an underappreciated role in dictating swimmer functioning, and the interrelationship between quorum sensing systems and stealth swimmers is another potentially exciting future research direction to investigate. The challenge ahead is to expand on these intriguing findings, determine their generality across the biofilm landscape, and develop potential applications that will take advantage of this newfound knowledge.
Footnotes
The authors declare no conflict of interest.
See companion article on page 13088.
References
- 1.Davey ME, O’toole GA. Microbial biofilms: From ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemon KP, Earl AM, Vlamakis HC, Aguilar C, Kolter R. Biofilm development with an emphasis on Bacillus subtilis. Curr Top Microbiol Immunol. 2008;322:1–16. doi: 10.1007/978-3-540-75418-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houry A, et al. Bacterial swimmers that infiltrate and take over the biofilm matrix. Proc Natl Acad Sci USA. 2012;109:13088–13093. doi: 10.1073/pnas.1200791109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shrout JD, Tolker-Nielsen T, Givskov M, Parsek MR. The contribution of cell-cell signaling and motility to bacterial biofilm formation. MRS Bull. 2011;36:367–373. doi: 10.1557/mrs.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 6.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 7.Parsek MR, Singh PK. Bacterial biofilms: An emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 8.Zimmerli W, Trampuz A, Ochsner PE. Prosthetic-joint infections. N Engl J Med. 2004;351:1645–1654. doi: 10.1056/NEJMra040181. [DOI] [PubMed] [Google Scholar]
- 9.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 10.Stewart PS. Diffusion in biofilms. J Bacteriol. 2003;185:1485–1491. doi: 10.1128/JB.185.5.1485-1491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schindler CA, Schuhardt VT. Lysostaphin: A new bacteriolytic agent for the Staphylococcus. Proc Natl Acad Sci USA. 1964;51:414–421. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boles BR, Thoendel M, Singh PK. Self-generated diversity produces “insurance effects” in biofilm communities. Proc Natl Acad Sci USA. 2004;101:16630–16635. doi: 10.1073/pnas.0407460101. [DOI] [PMC free article] [PubMed] [Google Scholar]