Abstract
Background
Shigellosis is a major public health concern worldwide, especially in developing countries. It is an acute intestinal infection caused by bacteria of the genus Shigella, with a minimum infective dose as low as 10–100 bacterial cells. Increasing prevalence of Shigella sonnei as the etiologic agent of shigellosis in Malaysia has been reported. As there is limited information on the genetic background of S. sonnei in Malaysia, this study aimed to characterize Malaysian S. sonnei and to evaluate the prospect of using multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) for subtyping of local S. sonnei.
Methods
Forty non-repeat clinical strains of S. sonnei isolated during the years 1997–2000, and 2007–2009 were studied. The strains were isolated from stools of patients in different hospitals from different regions in Malaysia. These epidemiologically unrelated strains were characterized using biotyping, antimicrobial susceptibility testing, pulsed-field gel electrophoresis (PFGE) and MLVA.
Results
The two biotypes identified in this study were biotype a (n = 29, 73%) and biotype g (n = 11, 27%). All the 40 strains were sensitive to kanamycin, ceftriaxone and ciprofloxacin. Highest resistance rate was observed for streptomycin (67.5%), followed by tetracycline (40%) and trimethoprim-sulfamethoxazole (37.5%). All the S. sonnei biotype g strains had a core resistance type of streptomycin - trimethoprim-sulfamethoxazole - tetracycline whereas the 29 biotype a strains were subtyped into eight resistotypes. All the strains were equally distinguishable by PFGE and MLVA. Overall, PFGE analysis indicated that S. sonnei biotype a strains were genetically more diverse than biotype g strains. Cluster analysis by MLVA was better in grouping the strains according to biotypes, was reflective of the epidemiological information and was equally discriminative as PFGE.
Conclusions
The S. sonnei strains circulating in Malaysia throughout the period studied were derived from different clones given their heterogeneous nature. MLVA based on seven selected VNTR loci was rapid, reproducible and highly discriminative and therefore may complement PFGE for routine subtyping of S. sonnei.
Keywords: Shigella sonnei, Biotype, Diversity, MLVA, PFGE, Resistotype
Background
Shigellosis is an acute intestinal infection caused by bacteria of the genus Shigella. The main symptom of this infection is bloody diarrhoea and the minimum infective dose is as low as 10–100 bacterial cells due to relative resistance to stomach acid [1]. Shigellosis is a major public health concern worldwide, especially in developing countries [2]. The infection is most frequent in children, the elderly and the immunocompromised [2,3]. A more recent annual estimate of the shigellosis burden was at 90 million incidences and 108,000 deaths [4].
Shigella can be differentiated into four species or serogroups, S. dysenteriaeS. flexneriS. boydii, and S. sonnei based on biochemical properties and group-specific O antigens in the outer membrane of the cell wall. There is a shift in Shigella dominance from S. flexneri to S. sonnei in developing and developed countries [4-6]. In Malaysia, Lee and Puthucheary [7] and Banga Singh et al. [8] reported an increasing dominance of S. sonnei as the etiologic agent of shigellosis. Apart from the apparent dominance shift, several studies have described the spread of a well-defined pandemic S. sonnei clone since 1990s which is characterized by common features such as biotype g, a particular PFGE XbaI pulsotype, resistance pattern of streptomycin - trimethoprim-sulfamethoxazole - tetracycline, and the presence of a 2.2 kbp class 2 integron [9,10].
Bacterial subtyping is frequently applied for outbreak investigations and surveillance of infectious diseases. Strain-specific fingerprints generated are used to facilitate the identification of disease transmission routes and sources [11]. Biotyping is one of the earlier phenotypic subtyping methods applied to S. sonnei. The bacteria can be subdivided into five biotypes (a, d, e, f, and g) on the basis of biochemical properties. This method however is not sufficiently discriminative.
Several genotyping methods with higher discriminatory power such as pulsed-field gel electrophoresis (PFGE) have been applied to subtype S. sonnei[12-14]. Since PFGE is a gel-based method, it requires strict adherence to standardized protocol for reproducible results. Standardized PulseNet PFGE protocol has been useful for inter-laboratory comparison of results [15]. However, due to the monomorphic nature of S. sonnei, PFGE occasionally may not be able to distinguish epidemiologically unrelated S. sonnei isolates and is not appropriate for phylogenetic analysis of strains that have evolved over a longer time span [16,17].
Multilocus variable-number tandem-repeat (VNTR) analysis (MLVA) is a highly discriminative sequence-based subtyping tool and has been used to study the genetic relatedness among S. sonnei. This method is based on the inherent variability of short sequences that are organized as tandem repeats at multiple VNTR loci. The VNTR loci in S. sonnei were found to have different degrees of variability [17]. MLVA with four to eight highly variable VNTR loci exhibited a discriminatory power parallel to or higher than PFGE [18]. Furthermore, MLVA based on the combination of VNTR loci with different variability has also been successfully used for phylogenetic analysis of S. sonnei that have evolved over different timescales [17].
Although S. sonnei is becoming an important etiologic agent of shigellosis in Malaysia, there is limited information on the genetic background of local strains. Therefore, the objective of the study was to characterize local S. sonnei strains by analyzing their biotypes, antimicrobial resistance patterns and genotypes. In addition, the prospect of using MLVA for routine subtyping of local S. sonnei in comparison with PFGE was also evaluated.
Methods
Bacterial strains
A total of 70 Shigella strains isolated from 1997–2009 were obtained from the Institute for Medical Research, Malaysia. However, only 40 strains S. sonnei from epidemiologically unrelated shigellosis cases were recovered from years 1997–2000 and 2007–2009 and analyzed. The strains were isolated from stools of patients in different hospitals in Malaysia. Each strain was exclusively from one patient. The shigellosis cases were from different regions, although mainly from northern region of Peninsular Malaysia.
Biotyping
Biotyping was performed using fermentation of rhamnose and xylose and hydrolysis of ortho-nitrophenyl-/I-D-galactopyranoside (ONPG), and biotypes were designated according to methods described by Nastasi et al.[19]. Five biotypes are defined (biotypes a, d, e, f, and g) based on the combination of the positive/negative biochemical reactions.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using Kirby-Bauer disc diffusion method according to guidelines of the Clinical and Laboratory Standards Institute [20]. E. coli ATCC 25922 was used as the quality-control strain. The antimicrobials commonly used for treatment of shigellosis were tested using antimicrobial-impregnated discs from Oxoid, Basingstoke, UK: ampicillin (A, 10 μg), chloramphenicol (C, 30 μg), streptomycin (S, 10 μg), tetracycline (T, 30 μg), ciprofloxacin (Cip, 5 μg), kanamycin (K, 30 μg), ceftriaxone (Cro, 30 μg), nalidixic acid (N, 30 μg) and trimethoprim–sulfamethoxazole (Sx, 25 μg).
Pulsed-field gel electrophoresis (PFGE)
All S. sonnei strains were analyzed with the restriction enzyme XbaI (Promega, Madison, WI, USA) using standardized PulseNet PFGE protocol, with extended run time of 25 hours in a CHEF Mapper system (Bio-Rad, U.S.A.) [15]. XbaI-digested Salmonella serotype Braenderup H9812 was used as the molecular weight standard. Computer-assisted analysis of the PFGE banding patterns was performed with BioNumerics software version 6.0 (Applied Maths, Kortrijk, Belgium). Banding patterns were compared based on the Dice coefficient, and a dendrogram based on the PFGE-XbaI profiles was constructed by unweighted pair group method with arithmetic averages (UPGMA) algorithm. A band position tolerance of 1.0% and optimization of 1.5% was applied during the comparison of banding patterns.
Multilocus variable-number tandem-repeat analysis (MLVA)
Seven VNTR loci, SS1, SS3, SS6, SS9, SS10, SS11, and SS13 previously reported by Liang et al.[18] were used. The primers and conditions are indicated in Table 1. Two multiplex polymerase chain reactions (mPCR) were carried out; mPCR1 consisted of primers for SS1, SS3, SS6, and SS9 while mPCR2 consisted of primers for SS10, SS11, and SS13. Each 10 μl PCR mixture contained 1X PCR buffer, 3 mM MgCl2, 0.2 mM of each dNTPs, 0.05 to 0.2 μM of each primer, 1 unit of GoTaq® Flexi DNA polymerase (Promega, Madison, WI, USA), and 1 μl of DNA template. Crude DNA template was prepared as follows: A pure culture of S. sonnei was plated on Luria-Bertani agar and incubated overnight at 37°C. A single colony was removed from the plate, suspended in 100 μl of sterile deionized water, boiled for 5 min and immediately cooled on ice. After centrifugation at 12,100 g for 3 min, the supernatant was transferred into a new tube for subsequent PCR analysis. PCR conditions were as follows: initial denaturation at 94°C for 5 min; 30 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 50 s and extension at 72°C for 60 s; and final extension at 72°C for 5 min.
Table 1.
MLVA primers, number of alleles and Nei’s index of diversity (D)
| Locus* | Dye_Forward primer (5′ to 3′) | No. of alleles | Nei’s diversity index, D |
|---|---|---|---|
|
SS1 |
VIC_TTGCCAGTACACCTCACTCG |
13 |
0.82 |
|
SS3 |
6-FAM_CTGGGAGATGAACAGGAGGA |
16 |
0.92 |
|
SS6 |
NED_GAGTCGCTAAACGCTTGCTT |
17 |
0.90 |
|
SS9 |
PET_CGCAATCAGCAAAACAAAGA |
12 |
0.88 |
|
SS10 |
6-FAM_ACGGTGGGCTTTCTCTACCT |
6 |
0.75 |
|
SS11 |
VIC_CTGGTCCGGGAGATTATCG |
8 |
0.83 |
| SS13 | PET_AGACGCTGGCTTATGACGAT | 3 | 0.42 |
* Liang et al. (2007).
The resulting PCR products were diluted 20 times with sterile deionized water and separated on an ABI 3130XL genetic analyzer using the size standard Gene Scan LIZ 500 (Applied Biosystems). Amplicon sizes were converted to copy numbers using BioNumerics software version 6.0 (Applied Maths, Kortrijk, Belgium). Each unique allelic string was designated a unique MLVA type. A dendrogram was constructed by UPGMA clustering based on categorical coefficient. Minimum spanning tree algorithm was used to construct a minimum spanning tree (MST) to determine phylogenetic pattern. The allelic profiles of 173 strains from Taiwan, China, Indonesia, Vietnam, and Cambodia, previously reported by Chiou et al.[16] and Liang et al.[18] were extracted and compared to the present data in the MST analysis based on the seven tested loci.
The polymorphism of each VNTR locus was represented by Nei’s diversity index, calculated as 1 - ∑ (allelic frequency)2. Simpson’s index of diversity (D) and Non-Approximated Confidence Interval (C.I.N.A.) were calculated as described previously by Simpson [21] to compare the discriminatory powers of PFGE and MLVA.
Results
Biotypes of S. sonnei
Two biotypes were identified among the S. sonnei strains, i.e. biotype a (ONPG +, rhamnose +, xylose –) and biotype g (ONPG +, rhamnose –, xylose –). Out of the 40 strains tested, 29 (73%) and 11 (27%) were of biotypes a and g, respectively.
Antimicrobial susceptibility profiles
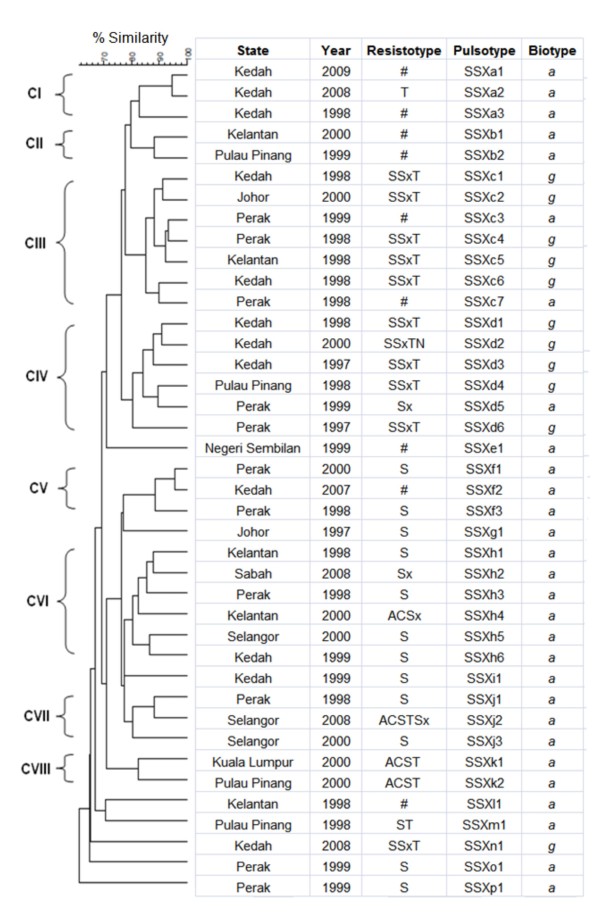
All the 40 strains were sensitive to kanamycin, ceftriaxone and ciprofloxacin. The resistance rates of other antimicrobials are as follows: streptomycin (67.5%), tetracycline, (40%), trimethoprim-sulfamethoxazole (37.5%), ampicillin (10%), chloramphenicol (10%) and nalidixic acid (2.5%). Nine strains (22.5%) were susceptible to all the antimicrobials tested. Fifteen strains (37.5%) were multidrug-resistant (MDR, defined as resistance to three or more classes of antimicrobials) with resistotype SSxT being dominant (n = 10), followed by ACST (n = 2), ACSx (n = 1), SSxTN (n = 1) and ACSTSx (n = 1). Resistotype SSxT was exclusively associated with 10 out of 11 S. sonnei biotype g strains. The remaining biotype g strain showed additional resistance towards nalidixic acid besides having a core resistance type of SSxT. On the contrary, the biotype a strains have eight different resistotypes (Figure 1).
Figure 1.
Dendrogram ofS. sonneiPFGE patterns generated by UPGMA clustering method using Dice coefficient. Key: all sensitive (#), ampicillin (A), chloramphenicol (C), streptomycin (S), tetracycline (T), and trimethoprim–sulfamethoxazole (Sx).
Genotypes based on PFGE
The genetic relatedness of the strains was determined by PFGE. XbaI-digested S. sonnei DNA generated 40 reproducible unique PFGE patterns, each with 15 – 24 DNA bands, with a Dice coefficient, F ranging from 0.70 to 0.98. Based on 80% similarity, eight clusters (CI – CVIII) were observed (Figure 1). Most clusters contained strains from different geographical regions. Similarly, most clusters contained strains isolated in different years except cluster CVIII which contained two biotype a strains isolated in year 2000 with identical resistotype ACST. Cluster CI contained three biotype a strains from Kedah. Cluster CII contained two biotype a strains which were sensitive to all the antimicrobials tested. Ten of the 11 biotype g strains were dispersed in cluster CIII and cluster CIV. Cluster III contained five biotype g and two biotype a strains. One of the biotype a strain even shared 93.3% similarity with a biotype g strain. Another five biotype g strains were grouped in cluster CIV with one biotype a strain. Cluster V contained three biotype a strains. Two strains in this cluster were closely related to each other (95.5% similarity) despite being isolated seven years apart from two different locations (Perak and Kedah). Cluster VI and cluster VII contained six and three biotype a strains, respectively. Seven biotype a and one biotype g strains were not assigned into any of these clusters.
Genotyping by MLVA
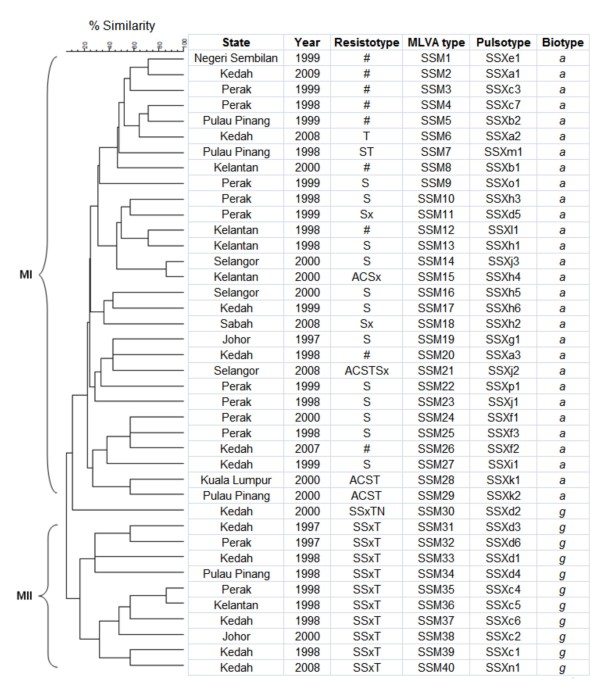
MLVA based on seven VNTR loci was performed to further characterize the S. sonnei strains. The number of alleles and diversity of each locus are listed in Table 1. Among the seven loci tested, SS1, SS3, SS6, SS9, and SS11 were more variable compared to SS10 and SS13. Locus SS13 showed the lowest diversity (D = 0.42) while SS3 was the most diverse (D = 0.92). From our result, the biotype a strains shared a single common allele at locus SS13 whereas two common alleles were detected at this locus for S. sonnei biotype g. The combination of the seven VNTR loci with high and low degree of variability was sufficient for molecular subtyping of the strains. The 40 S. sonnei strains were discriminated into 40 different MLVA types. Two pairs of single-locus variants, SSM14 and SSM15, and SSM35 and SSM36 were detected. The strains in each of the pair were isolated in the same year but from different regions. Two main clusters, MI and MII were observed from the dendrogram generated (Figure 2). Cluster MI consists of biotype a strains while cluster MII consists only biotype g strains. There was no clear demarcation of the strains isolated from different years and localities. From the MST, it is apparent that the S. sonnei in this study were heterogeneous as the tree branched out extensively. Most MLVA types have a distance of two to four loci from each other and the biotype a and g strains were differentiated into two main branches (Figure 3). The low variability locus SS13 established the ‘trunk’ of the MST, separating the biotype a and g strains while the six remaining highly variable loci further discriminated the strains in each branch, establishing the ‘twig and leaves’ of the tree.
Figure 2.
Dendrogram generated fromS. sonneiMLVA suptyping by the UPGMA clustering method using categorical coefficient. Key: all sensitive (#), ampicillin (A), chloramphenicol (C), streptomycin (S), tetracycline (T), and trimethoprim–sulfamethoxazole (Sx).
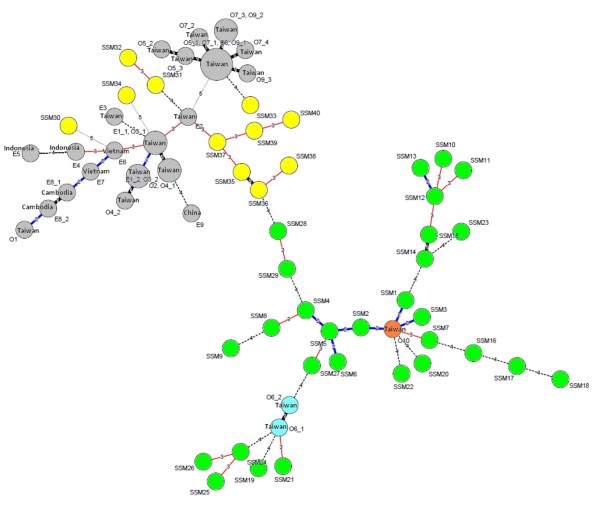
Figure 3.
Minimum spanning tree for the 40 MalaysianShigella sonneistrains and strains from five other countries. For Malaysian strains, each circle represents one strain with a unique MLVA profile. Each circle is noted with the MLVA type of the strain. MLVA types in yellow colour are Malaysian biotype g strains whereas those in green are Malaysian biotype a strains. For the profiles of strains from five other countries, the circle size is not proportional to the number of strains belonging to the MLVA profile but it is proportional to the number of outbreaks or unrelated episodes sharing the same profile. Circles in grey colour denote global clonal group A, orange denotes group B, and blue denotes group C. Additional information on these foreign strains is provided [see Additional file 1]. The distance between two MLVA types is numbered. A distance of one locus between two closest MLVA types is marked by a thick black line, a distance of two loci is marked by a blue line, and a distance of three loci is indicated by a thin red line. A distance of four loci is indicated by a broken line and a distance of five loci is indicated by a light broken line.
S. sonnei is one of the major etiologic agents of travel-associated diarrhoea [22]. Therefore our results were compared with MLVA profiles of 173 strains from five other Asian countries to determine if these strains were closely related. Analyses of MST (Figure 3) show that most of the Malaysian biotype g strains were related to the Taiwanese strains than those strains isolated from neighbouring countries (Indonesia, Vietnam, and Cambodia). Only one Malaysian biotype g strain was distantly related to the Vietnamese strains, sharing common alleles at two VNTR loci. The biotype a strains were also related to strains from Taiwan. In particular, three Malaysian biotype a strains shared common alleles at five VNTR loci with the Taiwanese strains.
Comparison of discriminatory power of MLVA and PFGE in genotyping of S. sonnei
PFGE and MLVA displayed excellent resolution in discriminating the Malaysian S. sonnei. Both methods were equally discriminative with a Simpson’s index of diversity, D of 1.0 (C.I.N.A. 0.989-1.011).
Discussion
The predominant biotype identified in this study was biotype a (n = 29, 73%) and the remaining strains were of biotype g (n = 11, 27%). These two biotypes were the most commonly reported biotypes in Australia from 1990–2009 [23]. In many other countries, an increasing prevalence of S. sonnei biotype g was reported. Izumiya et al.[14] reported the prevalence of S. sonnei biotype g among travel associated cases in Japan while almost all biotype a isolates were from patients with travel history to Southeast Asia. In Korea, S. sonnei isolated during 1977–1986 were of biotype a, whereas isolates in 1991–2000 were of biotype g[24]. Similarly, S. sonnei isolated from cases acquired in Ireland, Italy, United States, and a few African countries also suggested the increasing prevalence of S. sonnei biotype g since the 1990s [9,25,26]. More Malaysian strains however need to be analyzed to obtain a more accurate representation of the distribution and prevalence of S. sonnei biotypes in Malaysia.
Overall, PFGE analysis in this study indicated that S. sonnei biotype a strains were genetically more diverse than biotype g strains. The non-rhamnose fermenting biotype g strains were given further attention as there is a spread of a pandemic clone of biotype g strains across different continents [9,10]. Ten out of 11 Malaysian biotype g strains in this study exhibited similar characteristics of the pandemic clone that is biotype g, resistance pattern of SSxT, and similar but distinguishable PFGE patterns. A number of these biotype g strains were highly similar in PFGE banding patterns. The banding patterns, by visual comparison, were similar to some biotype g strains from Ireland and Italy [13]. These Malaysian strains could be phylogenetically linked to the pandemic biotype g clone. The biotype g strains were isolated from the period 1997 to 2000 and year 2008, indicating that these strains persisted in Malaysia for at least a decade.
In this study, more than half of the strains were resistant towards streptomycin (67.5%) and all strains were susceptible to kanamycin. This is in agreement with an earlier study in Malaysia by Hoe et al.[12] on Malaysian strains collected during the period 1997–2000. Resistance to trimethoprim-sulfamethoxazole and tetracycline are commonly reported in S. sonnei[5,14,24,27]. However the resistance rates to trimethoprim-sulfamethoxazole and tetracycline in this study were relatively low at 37.5% and 40%, respectively as compared to those observed in Taiwan, Thailand, Japan and Korea [5,14,24,27]. Most of the strains in this study remained susceptible to ceftriaxone, ciprofloxacin, ampicillin, chloramphenicol and nalidixic acid. These results were consistent with a study on S. sonnei from Northeast Malaysia [8]. The low resistance rate to nalidixic acid in Malaysia is in concordance with a report by Izumiya et al.[14] where resistance to nalidixic acid was less frequent in S. sonnei originated from South-east Asia. MDR S. sonnei strains persisted throughout the study periods. Selective antibiotic pressure may lead to the persistence of MDR S. sonnei strains in Malaysia [12] as ampicillin and trimethoprim-sulfamethoxazole are used for the treatment of Shigella infection locally [28]. To the best of our knowledge, all the 40 infected patients recovered from their illness.
PFGE and MLVA showed comparable discrimination in subtyping of S. sonnei. Both techniques further differentiated the S. sonnei biotype a and g strains. However MLVA was better at grouping the strains on the basis of biotypes, and the overall percentage of similarity among strains was low when subtyping was done using MLVA. Furthermore, MLVA subtyping was more rapid and less laborious compared to PFGE. Interpretation of result was less subjective and results were more readily comparable between laboratories. All these suggest that MLVA may be a suitable complement to PFGE or even an alternative for routine subtyping of S. sonnei.
Most of the PFGE and MLVA clusters contained strains from multiple geographical locations, some clusters even contained strains isolated from distant parts of the country although they were epidemiologically unrelated. This concurred with the report of Pichel et al.[3] where epidemiologically unrelated Argentinean S. sonnei from very distant geographical areas were clustered together by PFGE. Epidemiologically unrelated MLVA single-locus variants, and strains isolated seven years apart at different locations yet with highly similar PFGE pattern were also observed. These observations where strains with no apparent epidemiological linkage were clustered together may be due to travel of individuals within Malaysia, long-persisted dissemination of different clones of S. sonnei throughout the country, person-to-person transmission of a particular strain over an extended period with minor genetic changes, or a combination of these events.
Based on the MST analysis, S. sonnei in the present study were heterogeneous, and a number of strains in this study were related to S. sonnei from Taiwan and to a lesser extent, to those from neighbouring countries. Since S. sonnei is frequently found responsible for travel-associated diarrhoea, it is not uncommon that transmission of S. sonnei across countries and even continents occurred easily. A study on the global distribution of S. sonnei clones divided the strains from 50 countries, including Malaysia into three major clonal groups [29]. The 173 strains from five countries that were used for comparison in this study belonged to clonal groups A (n = 148) and C (n = 17) which were globally spread and clonal group B (n = 8) which was found in Europe, Africa and Asia [29] (Figure 3). One hundred and fifty one of these strains originated from 10 shigellosis outbreaks (O1 – O10) in Taiwan between 1996 and 2004. Another 22 strains were collected from nine epidemiologically unrelated episodes (E1 – E9) in Taiwan, China, Indonesia, Vietnam and Cambodia between 1998 to 2005 [16,18]. Based onVNTR loci SS1 and SS6, the Malaysian strains in the present study also belonged to these three major clonal groups. These observations suggest the S. sonnei strains circulating in Malaysia belonged to different clones that were spread worldwide. Although our observation may not represent the precise clonal structure of S. sonnei in Malaysia due to the limited sample size, the present study has nevertheless demonstrated the wide genetic diversity of S. sonnei circulating in this country.
Conclusions
Shigellosis is endemic in Malaysia. Our results suggested that S. sonnei circulating in Malaysia were heterogeneous and generally derived from different clones. MLVA based on seven selected VNTR loci was rapid, reproducible and highly discriminative and therefore may complement PFGE for routine subtyping of S. sonnei.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XPK carried out the experiments, analysis and interpretation of data and drafted the manuscript. CSC provided the information for VNTR primers and participated in design and interpretation of data and helped in drafting of the manuscript. HW provided partial funding for the project and helped in editing of the manuscript. NA co-supervised the project, and participated in the design of project, and helped in drafting of the manuscript. Norazah A provided some of the strains and helped in editing the manuscript. KLT conceived study, participated in its design, coordinated and supervised the project and co-wrote the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Information on the 173 S. sonnei strains from five other countries. Information provided are extracted from previous study by Chiou et al. (2006), Liang et al. (2007) and Filliol-Toutain et al. (2011).
Contributor Information
Xiu Pei Koh, Email: xp_527@yahoo.com.
Chien Shun Chiou, Email: nipmcsc@cdc.gov.tw.
Noni Ajam, Email: noni@um.edu.my.
Haruo Watanabe, Email: haruwata@nih.go.jp.
Norazah Ahmad, Email: norazah@imr.gov.my.
Kwai Lin Thong, Email: thongkl@um.edu.my.
Acknowledgements
This study was supported by grants from Universiti Malaya (PS316/2010B; RG017-09BIO) and the National Institute of Infectious Disease, Japan (57-02-03-1015). We thank Institute for Medical Research (IMR) for providing some of the clinical strains for the study.
References
- Levinson W. Medical Microbiology and Immunology: Examination and Board Review. Eighth edition. The McGraw-Hill Companies, New York; 2004. Basic bacteriology & brief summaries of medically important organisms; pp. 30–476. [Google Scholar]
- World Health Organization. Future needs and directions for Shigella vaccines. Wkly Epidemiol Rec. 2006;81:51–58. [PubMed] [Google Scholar]
- Pichel M, Gonzalez Fraga S, Terragno R, Mulki J, Gentile A, Kremer C, Mola AM, Noseda R, Binsztein N. Short report: analysis of clonal relationship among Shigella sonnei isolates circulating in Argentina. Epidemiol Infect. 2007;135:681–687. doi: 10.1017/S0950268806007230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Initiative for Vaccine Research (IVR): Diarrhoeal Diseases (Updated February 2009). Shigellosis. http://www.who.int/vaccine_research/diseases/diarrhoeal/en/index6.html.
- Chompook P, Samosornsuk S, von Seidlein L, Jitsanguansuk S, Sirima N, Sudjai S, Mangjit P, Kim DR, Wheeler JG, Todd J. Estimating the burden of shigellosis in Thailand: 36-month population-based surveillance study. Bull World Health Organ. 2005;83(10):739–746. [PMC free article] [PubMed] [Google Scholar]
- Vinh H, Nhu NT, Nga TV, Duy PT, Campbell JI, Hoang NV, Boni MF, My PVT, Parry C, Nga TTT, Van Minh P, Thuy CT, Diep TS, Phuong LT, Chinh MT, Loan HT, Tham NTH, Lanh MN, Mong BL, Anh VTC, Bay PVB, Chau NVV, Farrar J, Baker S. A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect Dis. 2009;9:204. doi: 10.1186/1471-2334-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WS, Puthucheary SD. Species distribution and antibiotic resistance of Shigella isolates in an urban community in Malaysia. Med J Malaysia. 2003;58:262–267. [PubMed] [Google Scholar]
- Banga Singh K-K, Ojha SC, Deris ZZ, Abdul Rahman R. A 9-year study of shigellosis in Northeast Malaysia: antimicrobial susceptibility and shifting species dominance. J Public Health. 2011;19:231–236. doi: 10.1007/s10389-010-0384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammina C, Aleo A, Romani C, Nastasi A. Shigella sonnei biotype g carrying class 2 integrons in southern Italy: a retrospective typing study by pulsed field gel electrophoresis. BMC Infect Dis. 2006;6:117. doi: 10.1186/1471-2334-6-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjbar R, Aleo A, Giammanco GM, Dionisi AM, Sadeghifard N, Mammina C. Genetic relatedness among isolates of Shigella sonnei carrying class 2 integrons in Tehran, Iran, 2002–2003. BMC Infect Dis. 2007;22:62. doi: 10.1186/1471-2334-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Greens J, Feil E, Gerner-Smidt P, Brisse S, Struelens M. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect. 2007;13(suppl 3):1–46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- Hoe CH, Yasin RM, Koh YT, Thong KL. Antimicrobial susceptibility and pulsed-field gel electrophoresis of Shigella sonnei strains in Malaysia (1997–2000) J Appl Microbiol. 2005;99:133–140. doi: 10.1111/j.1365-2672.2005.02581.x. [DOI] [PubMed] [Google Scholar]
- DeLappe N, Doran G, Connor JO, Mammina C, Cormican M. Use of pulsed-field gel electrophoresis for comparison of similar but distinguishable isolates of Shigella sonnei collected in Ireland and Italy. J Clin Microbiol. 2006;44(10):3808–3810. doi: 10.1128/JCM.01286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumiya H, Tada Y, Ito K, Morita-Ishihara T, Ohnishi M, Terajima J, Watanabe H. Characterization of Shigella sonnei isolates from travel associated cases in Japan. J Med Microbiol. 2009;58:1486–1491. doi: 10.1099/jmm.0.011809-0. [DOI] [PubMed] [Google Scholar]
- Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3(1):59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- Chiou CS, Wei HL, Wang YW, Liao JC, Li CC. Usefulness of inter-IS1 spacer polymorphisms for subtyping of Shigella sonnei isolates. J Clin Microbiol. 2006;44:3928–3933. doi: 10.1128/JCM.01069-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou CS, Watanabe H, Wang YW, Wang WL, Terajima J, Thong KL, Phung DC, Tung SK. Utility of multilocus variable-number tandem-repeat analysis as a molecular tool for phylogenetic analysis of Shigella sonnei. J Clin Microbiol. 2009;47:1149–1154. doi: 10.1128/JCM.01607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang SY, Watanabe H, Terajima J, Li CC, Liao JC, Tung SK, Chiou CS. Multilocus variable-number tandem-repeat analysis for molecular typing of Shigella sonnei. J Clin Microbiol. 2007;45:3574–3580. doi: 10.1128/JCM.00675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nastasi A, Pignato S, Mammina C, Giammanco G. rRNA gene restriction patterns and biotypes of Shigella sonnei. Epidemiol Infect. 1993;110:23–30. doi: 10.1017/S0950268800050640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twentieth Informational Supplement (Update). CLSI document M100-S20. Clinical and Laboratory Standards Institute, Wayne, Pennsylvannia; 2010. [Google Scholar]
- Simpson EH. Measurement of species diversity. Nature. 1949;163:688. doi: 10.1038/163688a0. [DOI] [Google Scholar]
- Ekdahl K, Andersson Y. The epidemiology of travel-associated shigellosis-regional risks, seasonality and serogroups. J Infect. 2005;51:222–229. doi: 10.1016/j.jinf.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Valcanis M. In: Victorian Infectious Diseases Bulletin. Volume 13 Issue 4. Clothier H, Rowe S, Easton M, McBryde E, Fielding J, Lester R, editor. Health Protection Branch: Wellbeing, Integrated Care and Ageing, Victorian Government Department of Health, Lonsdale Street, Melbourne; 2010. Laboratory testing of Shigella; pp. 114–118. [Google Scholar]
- Seol SY, Kim YT, Jeong YS, Oh JY, Kang HY, Moon DC, Kim J, Lee YC, Cho KT, Lee JC. Molecular characterization of antimicrobial resistances in Shigella sonnei isolates in Korea. J Med Microbiol. 2006;55:871–877. doi: 10.1099/jmm.0.46441-0. [DOI] [PubMed] [Google Scholar]
- McIver CJ, White PA, Jones LA, Karagiannis T, Harkness J, Marriott D, Rawlinson WD. Epidemic strains of Shigella sonnei biotype g carrying integrons. J Clin Microbiol. 2002;40:1538–1540. doi: 10.1128/JCM.40.4.1538-1540.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammina C, Pontello M, Dal Vecchio A, Nastasi A. Shigella sonnei Working Group. Identification of Shigella sonnei biotype g isolates carrying class 2 integrons in Italy (2001 to 2003) J Clin Microbiol. 2005;43:2467–2470. doi: 10.1128/JCM.43.5.2467-2470.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TS, Chang CY, Chang LL, Chen WM, Wang TK, Chang SF. One predominant type of genetically closely related Shigella sonnei prevalent in four sequential outbreaks in school children. Diagn Microbiol Infect Dis. 2003;45:173–181. doi: 10.1016/S0732-8893(02)00524-2. [DOI] [PubMed] [Google Scholar]
- Ministry of Health. National Antibiotic Guideline 2008 (Updated 25 March 2009) http://www.pharmacy.gov.my/aeimages//File/National_Antibiotic_Guideline_2008_edit(2).pdf.
- Filliol-Toutain I, Chiou C-S, Mammina C, Gerner-Smidt P, Thong K-L, Phung DC, Pichel M, Ranjbar R, Gassama Sow A, Cooper K, Ribot E, Binsztein N, Liang S-Y. Global distribution of Shigella sonnei clones. Emerg Infect Dis. 2011 Oct [cited 2011 Oct 1]. Available from http://wwwnc.cdc.gov/eid/article/17/10/pdfs/10-1486.pdf. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information on the 173 S. sonnei strains from five other countries. Information provided are extracted from previous study by Chiou et al. (2006), Liang et al. (2007) and Filliol-Toutain et al. (2011).