Abstract
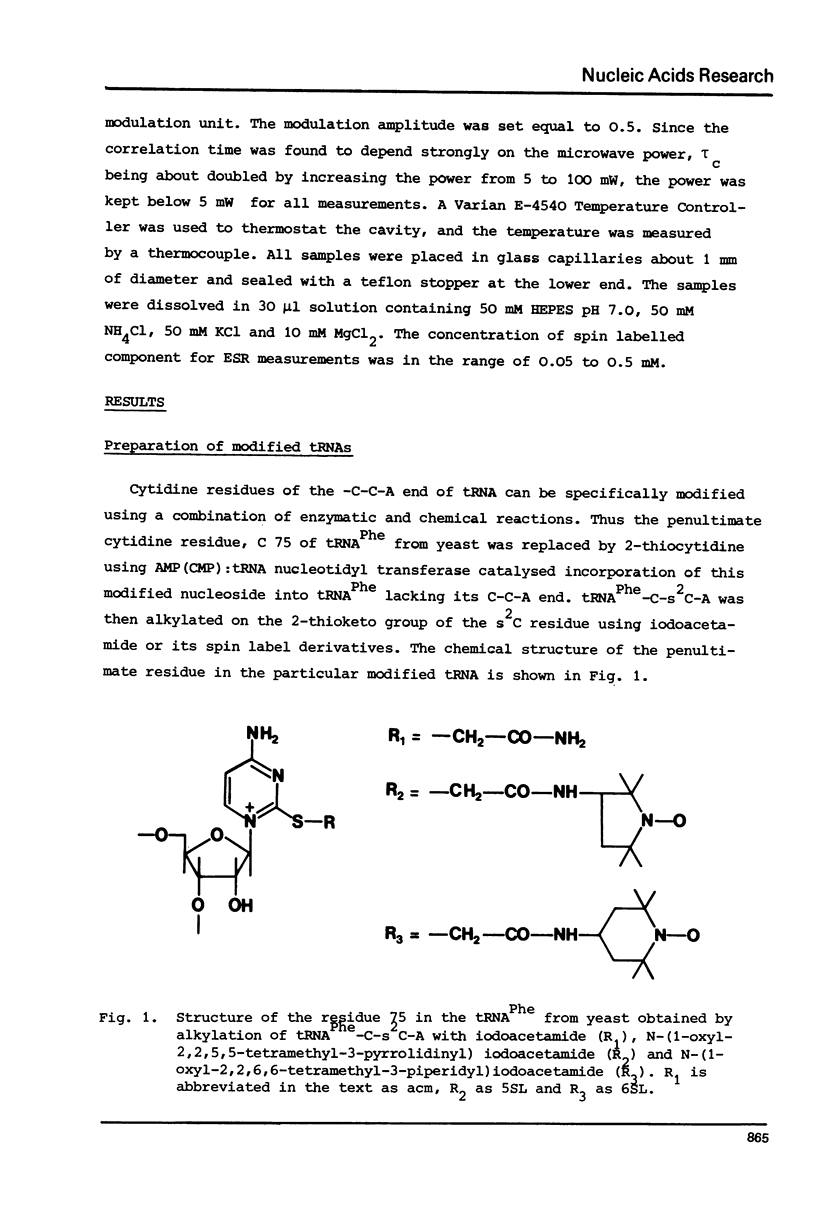
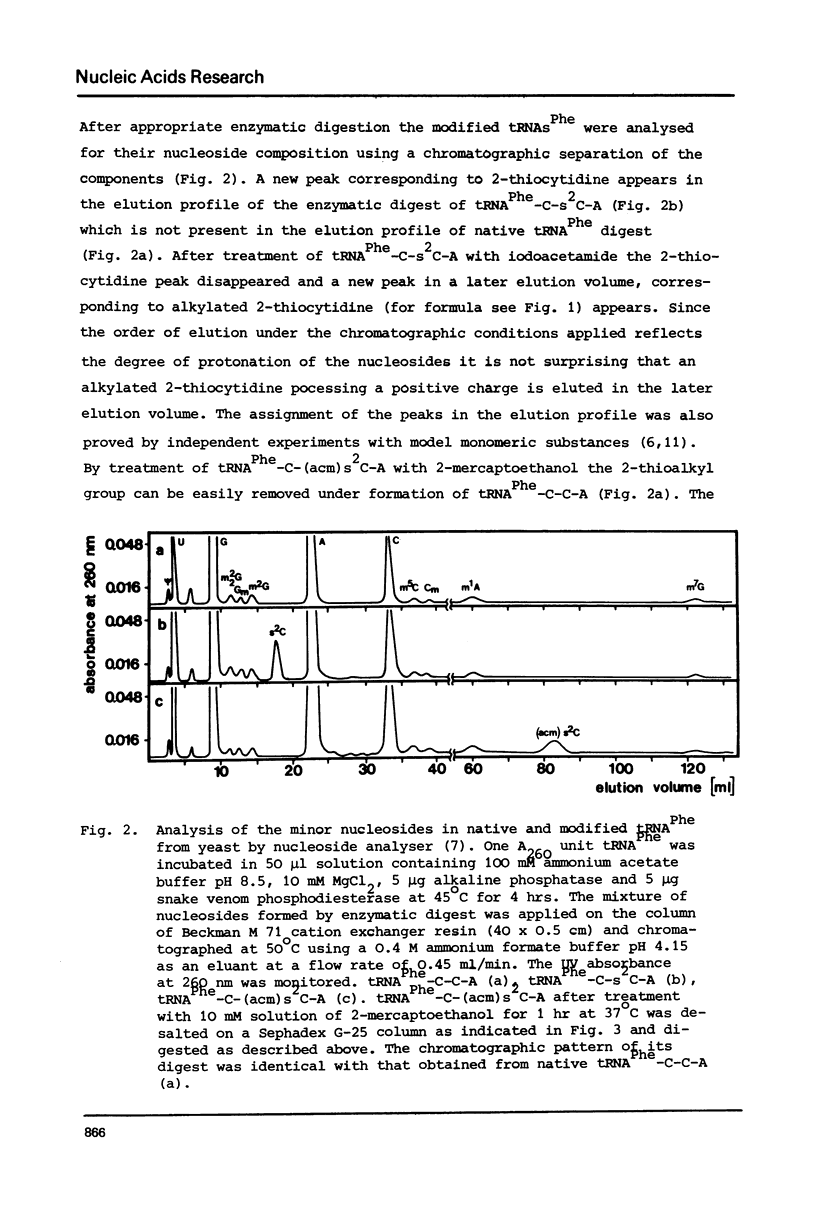
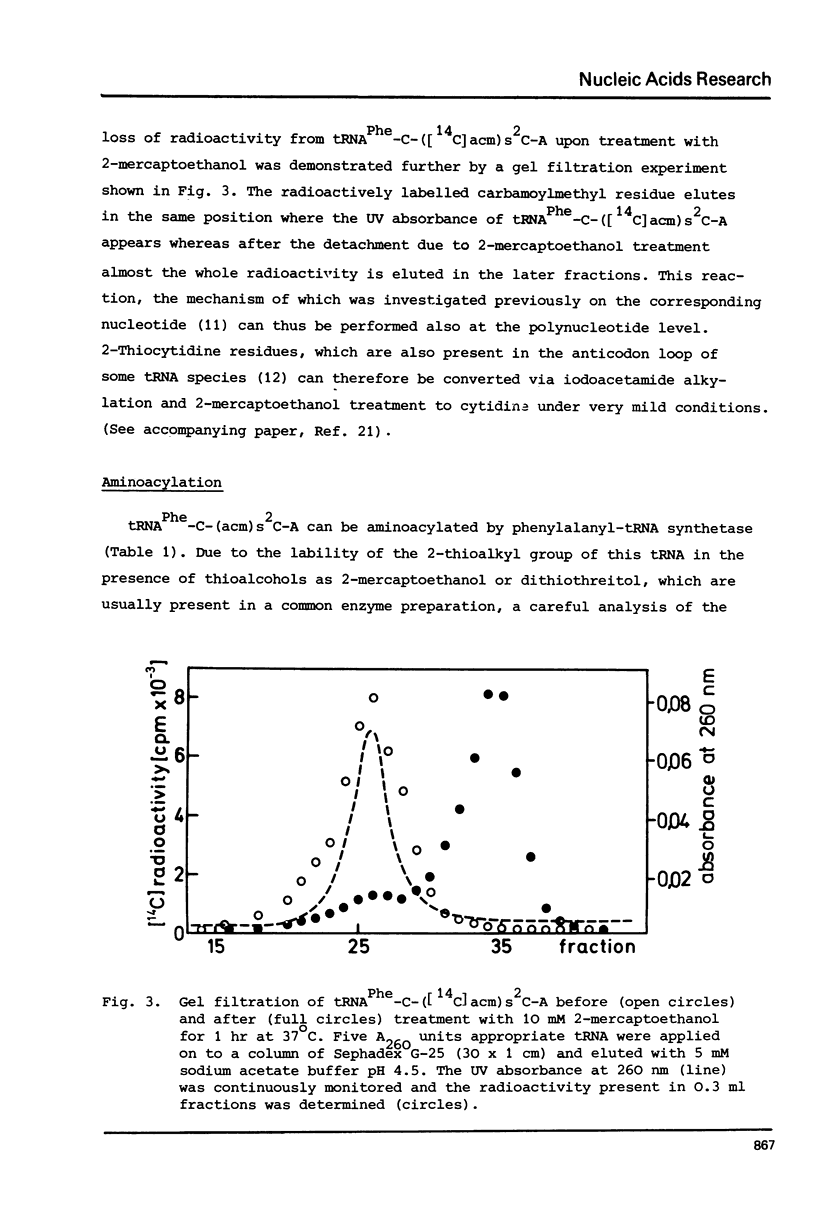
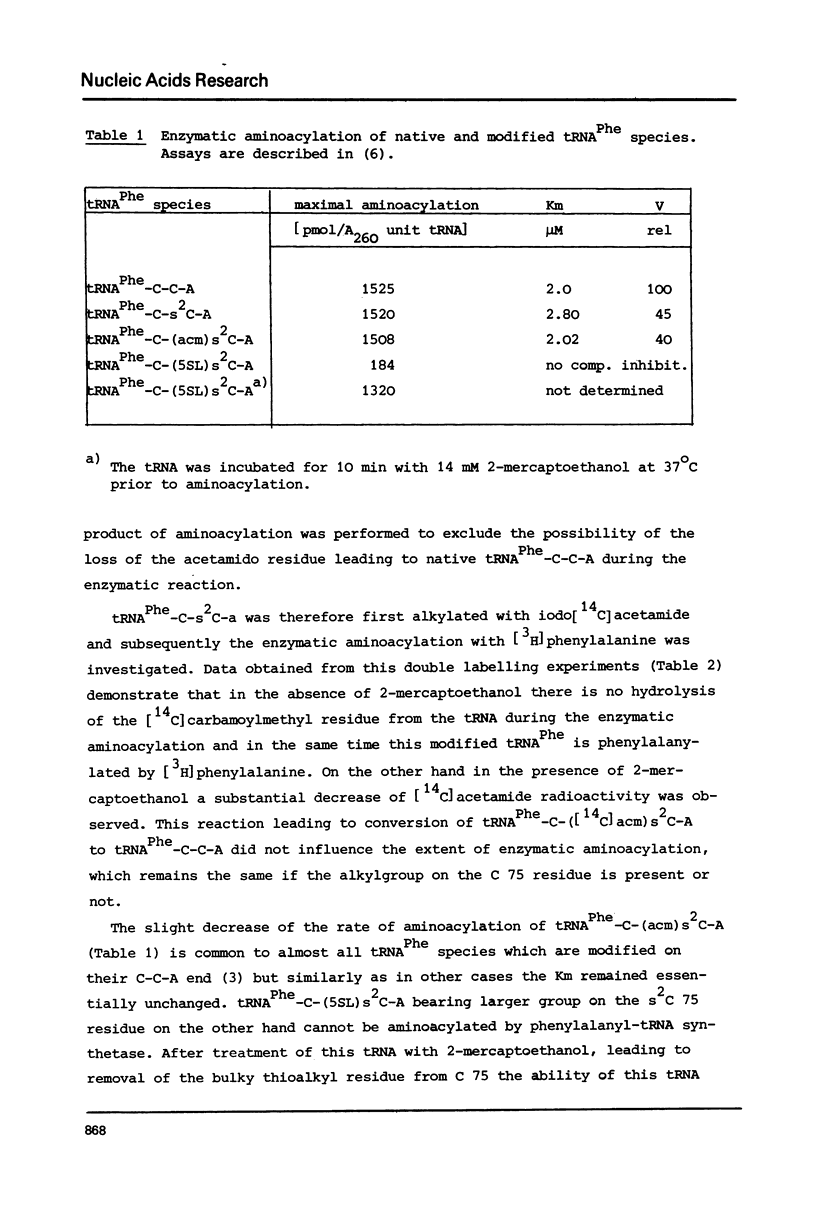
The 2-thioketo function of tRNAPhe-C-s2C-A in which the penultimate cytidine residue is replaced by 20thiocytidine can serve as a site of specific attachment of spin label. By alkylation of tRNAPhe-C-s2C-A with iodoacetamide or its spin label derivatives tRNAPhe-C-(acm)s2C-A or tRNAPheC-(SL)s2C-A are formed. The enzymatic phenylalanylation of these tRNAsPhe revealed that the 2-position of the penultimate cytidine can be modified without impairing this enzymatic reaction but there exists a sterical limitation for the subsituent on this position beyond which the tRNAPhe:phenylalanyl-tRNA synthetase recognition is not possible. Both Phe-tRNAPhe-C-(acm)s2C-A as well as Phe-tRNAPhe-C(SL)s2C-A form ternary complexes with EF-Tu.GTP. The part of the 3'-terminus of tRNAPhe where the additional substituents are attached is therefore not involved in the interaction with this elongation factor. This could be also demonstrated by ESR measurements of spin labelled tRNAsPhe. The correlation times, tauc, for tRNAPhe-C-(SL)s2C-A, Phe-tRNAPhe-C-(SL)s2C-A and Phe-tRNAPhe-C-(SL)s2C-A.EF-Tu:GTP are essentially identical indicating that the structure of the 3'-end of tRNAPhe is not influenced significantly by aminoacylation or ternary complex formation.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arai K. I., Kawakita M., Kaziro Y. Studies on polypeptide elongation factors from Escherichia coli. II. Purification of factors Tu-guanosine diphosphate, Ts, and Tu-Ts, and crystallization of Tu-guanosine diphosphate and Tu-Ts. J Biol Chem. 1972 Nov 10;247(21):7029–7037. [PubMed] [Google Scholar]
- Baksht E. K., Gal A., de Groot N., Hochberg A. A., Sprinzl M., Cramer F. Properties of phenylalanine transfer ribonucleic acid with modified 3'-terminal end in protein biosynthesis using a rabbit reticulocyte cell-free system: effect of the replacement of cytidine residues from the CpCpA end of tRNA by 5-iodocytidine or 2-thiocytidine. Nucleic Acids Res. 1977 Jul;4(7):2205–2212. doi: 10.1093/nar/4.7.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraburtty K. Effect of sodium bisulfite modification on the arginine acceptance of E. coli tRNA Arg. Nucleic Acids Res. 1975 Oct;2(10):1793–1804. doi: 10.1093/nar/2.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark B. F. Correlation of biological activities with structural features of transfer RNA. Prog Nucleic Acid Res Mol Biol. 1977;20:1–19. doi: 10.1016/s0079-6603(08)60468-7. [DOI] [PubMed] [Google Scholar]
- Kruse T. A., Clark B. F. The effect of specific structural modification on the biological activity of E. coli arginine tRNA. Nucleic Acids Res. 1978 Mar;5(3):879–892. doi: 10.1093/nar/5.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kućan Z., Freude K. A., Kućan I., Chambers R. W. Aminoacylation of bisulphite-modified yeast tyrosine transfer RNA. Nat New Biol. 1971 Aug 11;232(2):177–179. doi: 10.1038/newbio232177a0. [DOI] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- Schneider D., Solfert R., von der Haar F. Large scale purification of tRNA ser , tRNA tyr and tRNA phe from Baker's yeast. Hoppe Seylers Z Physiol Chem. 1972 Aug;353(8):1330–1336. doi: 10.1515/bchm2.1972.353.2.1330. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Goddard J. P. Loss of methionine acceptor activity resulting from a base change in the anticodon of Escherichia coli formylmethionine transfer ribonucleic acid. J Biol Chem. 1973 Feb 25;248(4):1341–1345. [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Structural requirements for aminoacylation of Escherichia coli formylmethionine transfer RNA. Biochemistry. 1977 Sep 20;16(19):4256–4265. doi: 10.1021/bi00638a020. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H., Sundari R. M. Structural requirements for recognition of Escherichia coli initiator and non-initiator transfer ribonucleic acids by bacterial T factor. J Biol Chem. 1974 Nov 25;249(22):7102–7110. [PubMed] [Google Scholar]
- Sprinzl M. On the structure of phenylalanine tRNA from yeast. Spin-label studies. Eur J Biochem. 1974 Dec 2;49(3):595–605. doi: 10.1111/j.1432-1033.1974.tb03863.x. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Scheit K. H., Cramer F. Preparation in vitro of a 2-thiocytidine-containing yeast tRNA Phe -A 73 -C 74 -S 2 C 75 -A 76 and its interaction wiith p-hydroxymercuribenzoate. Eur J Biochem. 1973 Apr;34(2):306–310. doi: 10.1111/j.1432-1033.1973.tb02759.x. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., von der Haar F., Schlimme E., Sternbach H., Cramer F. Incorporation of 5-iodocytidine into yeast tRNAphe with tRNA nucleotidyl transferase in vitro. Eur J Biochem. 1972 Feb 15;25(2):262–266. doi: 10.1111/j.1432-1033.1972.tb01692.x. [DOI] [PubMed] [Google Scholar]
- Von Der Haar F., Gaertner E. Phenylalanyl-tRNA synthetase from baker's yeast: role of 3'-terminal adenosine of tRNA-Phe in enzyme-substrate interaction studied with 3'-modified tRNA-Phe species. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1378–1382. doi: 10.1073/pnas.72.4.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar F. Affinity elution as a purification method for aminoacyl-tRNA synthetases. Eur J Biochem. 1973 Apr 2;34(1):84–90. doi: 10.1111/j.1432-1033.1973.tb02731.x. [DOI] [PubMed] [Google Scholar]


