Abstract
Myxobacteria are amongst the top producers of natural products. The diversity and unique structural properties of their secondary metabolites is what make these social microbes highly attractive for drug discovery. Screening of products derived from these bacteria has revealed a puzzling amount of hits against infectious and non-infectious human diseases. Preying mainly on other bacteria and fungi, why would these ancient hunters manufacture compounds beneficial for us? The answer may be the targeting of shared processes and structural features conserved throughout evolution.
Keywords: Myxobacteria, Natural products, Drug discovery, Chemical space
Commentary
Natural products from plants and microbes have played a pivotal role in drug discovery for more than a century [1-3]. In recent years, myxobacteria have matched fungi, actinomycetes as well as some species of the genus Bacillus as top producers of microbial secondary metabolites [4-6]. More importantly, screening campaigns have revealed a large proportion of the myxobacteria secondary metabolism to have activities against human diseases such as cancer, bacterial and viral infections [6-8].
Myxobacteria are a group of proteobacteria which reside mainly in soil [9,10]. These social microbes move by an axonal cellular motion called gliding [11,12], and although cells grow independently, they form collective swarms to prey and generate transient structures, called fruiting bodies (Figure 1), when resources are scarce [13]. During cooperative feeding, individual cells organize in waves which travel in a rippling-like motion [12,14]. As waves of cells collide, they aggregate in mounds that grow in size forming fruiting bodies that can harbor about 105 individuals. Cells within these structures become myxospores. Sporulation is triggered by signaling at the cell-cell contact surface when nutrients are available, and the myxospores germinate to eventually develop new swarms [11]. To control these processes, myxobacteria have evolved a unique mechanism of extracellular and intracellular signals, including diverse proteins and small metabolites [15].
Figure 1 .
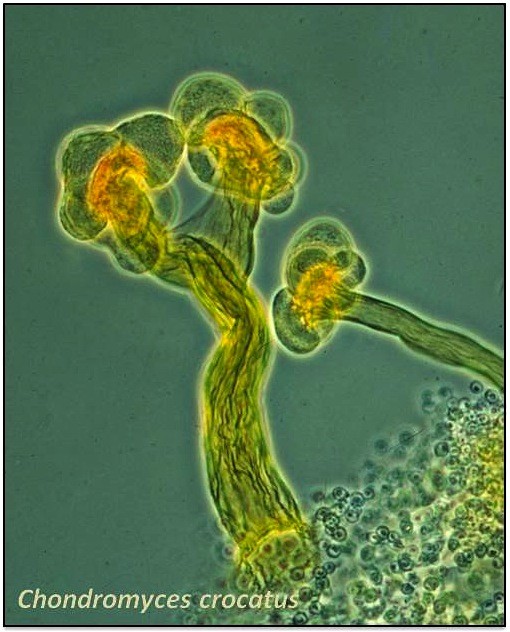
Image of fruiting bodies from the myxobacteriumChondromyces crocatus(courtesy of Hans Reichenbach).
The chemical space of the myxobacteria metabolome is rare both in diversity and biological activities [5,16,17]. Their secondary metabolites present structural elements not commonly produced by other microbes such as unusual hybrids of polyketides and non-ribosomally made peptides [5,18]. In fact, around 40% of the described myxobacterial compounds represent novel chemical structures [9]. Furthermore, most small molecules from myxobacteria are not glycosylated as opposed to products derived from actinomycetes [19] and they target molecules that are often not targeted by metabolites from other microbes. Examples include inhibitors of mitochondrial respiration and eukaryotic protein synthesis, carboxylase and polymerase inhibitors and molecules that affect microtubule assembly [17]. Although the reasons why myxobacteria display such a large array of secondary metabolites are still not well understood, it has been argued that they confer a competitive advantage in the soil environment and are used to modulate cell-cell interactions within the population [20], to protect ecological niches in their competitive environment [17], and used as weapons for predation [13].
This level of chemical complexity requires an equally complex regulatory network to function, altogether enhancing the survival and competitivity of both the individual and the population [10]. This is reflected in the genetic space employed by myxobacteria for their secondary metabolism. One of the largest bacterial genome reported to date belongs to the myxobacterium Sorangium cellulosum with around 20 secondary metabolite loci and probably more to be discovered [15]. Another well studied myxobacterium, Myxococcus xanthus, has around 18 secondary metabolite gene clusters accounting for around 9% of its genome [21] which is more than some species of actinomycetes with around 6% of genome coverage for secondary metabolite loci [22,23]. Given this large space on the level of the genome, the known diversity between different myxobacteria and the vast number of different bacterial strains available in various collections, there seems to be an immense room for exploration and exploitation.
The amount of different small molecules from myxobacteria targeting other soil bacteria and fungi, around 29% and 54% respectively, and their higher production rates during exponential growth seems to reinforce the idea of a broad use of secondary metabolites for hunting [13,17]. Any predatory microorganism would benefit greatly from such a diverse armament but why would a large amount of these metabolites be active against human diseases and pathogens? An attractive explanation is that many of these products target shared processes or structural features conserved throughout evolution [24-26]. For example, the LSm1-7 protein complex in mammalian cells was shown to be required for efficient hepatitis C virus (HCV) translation and replication [25]. The Brome mosaic virus (BMV), a plant virus that can replicate in yeast, utilizes the respective yeast homologues for the same processes [27-29]. Likewise, the bacteriophage Q, a plus-strand RNA virus as HCV and BMV, requires Hfq, the homologue of LSm1 in bacteria for its expansion [30]. Thus there is a functional conservation of cellular and viral regulatory elements across kingdoms and virus groups that may be exploited for antiviral drug development. Indeed, a recent screen against processing body proteins that include the LSm1-7 complex revealed several hits from a myxobacterial metabolite library that overlapped with antiviral activities (Martinez et al., unpublished). To learn more about the bioactivity profile of these potent compounds, systematic testing in a broad panel of bioassays as offered by e.g. academic consortia such as EU-OPENSCREEN would be strategically worthwhile. However, to develop a metabolite hit into an applicable pharmaceutical compound is not an easy task, especially given the complexity of their natural product chemistry, side effects and poor bioavailability. Therefore, to make better use of natures pharmaceutical factories, new technologies such as engineering of microorganisms to synthesize complex molecular structures, in silico tools to predict the target profile and anticipate potential side effects of those metabolites, and targeted delivery strategies for example via nanoparticles are under the spotlight and will play an increasing role in the future [31-35].
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Juana Diez, Email: juana.diez@upf.edu.
Javier P Martinez, Email: javier.martinez@upf.edu.
Jordi Mestres, Email: jmestres@imim.es.
Florenz Sasse, Email: florenz.sasse@helmholtz-hzi.de.
Ronald Frank, Email: ronald.frank@helmholtz-hzi.de.
Andreas Meyerhans, Email: andreas.meyerhans@upf.edu.
Acknowledgements
The authors of this article are supported by grants from the Spanish Ministry of Science and Innovation BFU2010-20803 to JD, SAF2010-21336 to AM, BIO2011-26669 to JM and the Spanish Instituto de Salud Carlos III. The EU-OPENSCREEN preparatory phase project receives funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement n 261861.
References
- Davies J, Ryan KS. Introducing the parvome: bioactive compounds in the microbial world. ACS Chem Biol. 2012;7(2):252–259. doi: 10.1021/cb200337h. [DOI] [PubMed] [Google Scholar]
- Mishra BB, Tiwari VK. Natural products: an evolving role in future drug discovery. Eur J Med Chem. 2011;46(10):4769–4807. doi: 10.1016/j.ejmech.2011.07.057. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguelles-Arias A, Ongena M, Halimi B, Lara Y, Brans A, Joris B, Fickers P. Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb Cell Fact. 2009;8:63. doi: 10.1186/1475-2859-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode HB, Muller R. Analysis of myxobacterial secondary metabolism goes molecular. J Ind Microbiol Biotechnol. 2006;33(7):577–588. doi: 10.1007/s10295-006-0082-7. [DOI] [PubMed] [Google Scholar]
- Weissman KJ, Muller R. Myxobacterial secondary metabolites: bioactivities and modes-of-action. Nat Prod Rep. 2010;27(9):1276–1295. doi: 10.1039/c001260m. [DOI] [PubMed] [Google Scholar]
- Gentzsch J, Hinkelmann B, Kaderali L, Irschik H, Jansen R, Sasse F, Frank R, Pietschmann T. Hepatitis C virus complete life cycle screen for identification of small molecules with pro- or antiviral activity. Antiviral Res. 2011;89(2):136–148. doi: 10.1016/j.antiviral.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Nickeleit I, Zender S, Sasse F, Geffers R, Brandes G, Sorensen I, Steinmetz H, Kubicka S, Carlomagno T, Menche D. et al. Argyrin a reveals a critical role for the tumor suppressor protein p27(kip1) in mediating antitumor activities in response to proteasome inhibition. Cancer Cell. 2008;14(1):23–35. doi: 10.1016/j.ccr.2008.05.016. [DOI] [PubMed] [Google Scholar]
- Reichenbach H. Myxobacteria, producers of novel bioactive substances. J Ind Microbiol Biotechnol. 2001;27(3):149–156. doi: 10.1038/sj.jim.7000025. [DOI] [PubMed] [Google Scholar]
- Velicer GJ, Vos M. Sociobiology of the myxobacteria. Annu Rev Microbiol. 2009;63:599–623. doi: 10.1146/annurev.micro.091208.073158. [DOI] [PubMed] [Google Scholar]
- Kaiser D. Coupling cell movement to multicellular development in myxobacteria. Nat Rev. 2003;1(1):45–54. doi: 10.1038/nrmicro733. [DOI] [PubMed] [Google Scholar]
- Nan B, Chen J, Neu JC, Berry RM, Oster G, Zusman DR. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci USA. 2011;108(6):2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Wei X, Ebright R, Wall D. Antibiotic production by myxobacteria plays a role in predation. J Bacteriol. 2011;193(18):4626–4633. doi: 10.1128/JB.05052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleman JE, Kirby JR. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol Rev. 2009;33(5):942–957. doi: 10.1111/j.1574-6976.2009.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, Bartels D, Bekel T, Beyer S, Bode E. et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol. 2007;25(11):1281–1289. doi: 10.1038/nbt1354. [DOI] [PubMed] [Google Scholar]
- Bon RS, Waldmann H. Bioactivity-guided navigation of chemical space. Acc Chem Res. 2010;43(8):1103–1114. doi: 10.1021/ar100014h. [DOI] [PubMed] [Google Scholar]
- Weissman KJ, Muller R. A brief tour of myxobacterial secondary metabolism. Bioorg Med Chem. 2009;17(6):2121–2136. doi: 10.1016/j.bmc.2008.11.025. [DOI] [PubMed] [Google Scholar]
- Silakowski B, Kunze B, Muller R. Multiple hybrid polyketide synthase/non-ribosomal peptide synthetase gene clusters in the myxobacterium Stigmatella aurantiaca. Gene. 2001;275(2):233–240. doi: 10.1016/S0378-1119(01)00680-1. [DOI] [PubMed] [Google Scholar]
- Rix U, Fischer C, Remsing LL, Rohr J. Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat Prod Rep. 2002;19(5):542–580. doi: 10.1039/b103920m. [DOI] [PubMed] [Google Scholar]
- Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9(5):445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Bode HB, Muller R. The impact of bacterial genomics on natural product research. Angew Chem Int Ed. 2005;44(42):6828–6846. doi: 10.1002/anie.200501080. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D. et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417(6885):141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol. 2003;21(5):526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- Hong J. Role of natural product diversity in chemical biology. Curr Opin Chem Biol. 2011;15(3):350–354. doi: 10.1016/j.cbpa.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller N, Mina LB, Galao RP, Chari A, Gimenez-Barcons M, Noueiry A, Fischer U, Meyerhans A, Diez J. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci USA. 2009;106(32):13517–13522. doi: 10.1073/pnas.0906413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K, Kromer JO, Wittmann C, Alves-Rodrigues I, Meyerhans A, Diez J, Heinzle E. Metabolite profiling studies in Saccharomyces cerevisiae: an assisting tool to prioritize host targets for antiviral drug screening. Microb Cell Fact. 2009;8:12. doi: 10.1186/1475-2859-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez J, Ishikawa M, Kaido M, Ahlquist P. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc Natl Acad Sci USA. 2000;97(8):3913–3918. doi: 10.1073/pnas.080072997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas A, Alves-Rodrigues I, Noueiry A, Ahlquist P, Diez J. Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J Virol. 2006;80(1):246–251. doi: 10.1128/JVI.80.1.246-251.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry AO, Diez J, Falk SP, Chen J, Ahlquist P. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol Cell Biol. 2003;23(12):4094–4106. doi: 10.1128/MCB.23.12.4094-4106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fernandez MT Franze, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968;219(5154):588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- Andexer JN, Kendrew SG, Nur-e-Alam M, Lazos O, Foster TA, Zimmermann AS, Warneck TD, Suthar D, Coates NJ, Koehn FE. et al. Biosynthesis of the immunosuppressants FK506, FK520, and rapamycin involves a previously undescribed family of enzymes acting on chorismate. Proc Natl Acad Sci USA. 2011;108(12):4776–4781. doi: 10.1073/pnas.1015773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestres J, Seifert SA, Oprea TI. Linking pharmacology to clinical reports: cyclobenzaprine and its possible association with serotonin syndrome. Clin Pharmacol Ther. 2011;90(5):662–665. doi: 10.1038/clpt.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Garg S. Pure drug and polymer based nanotechnologies for the improved solubility, stability, bioavailability and targeting of anti-HIV drugs. Adv Drug Deliv Rev. 2010;62(45):491–502. doi: 10.1016/j.addr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- Villaverde A. Nanotechnology, bionanotechnology and microbial cell factories. Microb Cell Fact. 2010;9:53. doi: 10.1186/1475-2859-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MQ, Gaisser S, Nur EAM, Sheehan LS, Vousden WA, Gaitatzis N, Peck G, Coates NJ, Moss SJ, Radzom M. et al. Optimizing natural products by biosynthetic engineering: discovery of nonquinone Hsp90 inhibitors. J Med Chem. 2008;51(18):5494–5497. doi: 10.1021/jm8006068. [DOI] [PubMed] [Google Scholar]


