Abstract
Our current understanding of 3-dimensional (3D) cell migration is primarily based on results from fibrous scaffolds with randomly organized internal architecture. Manipulations that change the stiffness of these 3D scaffolds often alter other matrix parameters that can modulate cell motility independently or synergistically, making observations less predictive of how cells behave when migrating in 3D. In order to decouple microstructural influences and stiffness effects, we have designed and fabricated 3D polyethylene glycol (PEG) scaffolds that permit orthogonal tuning of both elastic moduli and microstructure. Scaffolds with log-pile architectures were used to compare the 3D migration properties of normal breast epithelial cells (HMLE) and Twist-transformed cells (HMLET). Our results indicate that the nature of cell migration is significantly impacted by the ability of cells to migrate in the third dimension. 2D ECM-coated PEG substrates revealed no statistically significant difference in cell migration between HMLE and HMLET cells among substrates of different stiffness. However, when cells were allowed to move along the third dimension, substantial differences were observed for cell displacement, velocity and path straightness parameters. Furthermore, these differences were sensitive to both substrate stiffness and the presence of the Twist oncogene. Importantly, these 3D modes of migration provide insight into the potential for oncogene-transformed cells to migrate within and colonize tissues of varying stiffness.
INTRODUCTION
A central rationale for studying cell migration is to better understand cancer biology. More specifically, standard two-dimensional (2D) environments, such as extracellular matrix (ECM) protein-coated tissue culture plastic or glass surfaces have enabled the molecular analysis of the cellular machinery that regulates cell motility [1–3]. From a pathophysiological perspective, however, cancer cells are able to invade, migrate through and colonize three-dimensional (3D) environments during later stages of cancer progression (i.e., metastasis). Thus, methods for monitoring in vitro cell motility in 3D may provide a better approximation of tumor cell behavior [4]. The mechanical and structural properties that govern the relationship between tumor cell motility and their metastatic potential of cancer cells are critically important since most cancer-related deaths result from metastatic lesions, a process that requires cells to invade new tissues and colonize a tumor away from the primary tumor site [4, 5]. While most cancer cells have unique migration properties that enable them to enter the circulatory system [6, 7], not all cancer cells metastasize to the same tissues due to their unique preferences for tissue-specific mechanics and geometry [8]. Furthermore, many of these preferences are driven by the tissue of origin in which the primary tumor arose as well as oncogene-driven transformation/migration properties acquired by the cancer cells during disease progression [4]. Thus, 2D cell migration studies, while amenable to analyzing variations in the substrate’s mechanical properties, may uncover only a portion of the puzzle. While some cell migration studies that have tested the effect of substrate stiffness in 2D and 3D[9–12], the interplay between matrix mechanics/structure, disease progression, and cell motility in 3D is not well understood. To further understand this process, cancer cell migration should be investigated on scaffolds that facilitate the analysis of the unique properties represented in the metastatic tumor microenvironment [8].
Degradable synthetic and natural biopolymers which mimic the ECM have been routinely used as model systems for investigating 3D cell motility in vitro [13–16]. Most biopolymers scaffolds are typically fibrous in nature having random pore structures or mesh sizes ranging from nanometer to micron scale. The internal architecture can be varied by modulating the solid content, gelation conditions, or by introducing entities which facilitate fiber aggregation and/or crosslinking. However, these scaffolds do not allow precise control over internal architecture[17], and changes in one parameter (e.g. fiber stiffness) results in concurrent changes in other physical parameters (e.g., pore size, fiber architecture and deformability) making cell interactions difficult to interpret [18–20]. For example in collagen scaffolds, modulating the collagen concentration of the prepolymer concurrently changes fiber morphology and alters pore-size, which in turn affects cell motility [21]. This caveat makes such fibrous scaffolds exhibit highly non-linear behavior at macro and micro-scales, due to heterogeneous pore size distribution throughout the scaffold and therefore make them less predictive of cell behavior [22]. When cells interact with and deform the local matrix via various bundling, slipping and sliding mechanisms, the resulting effective stiffness experienced by the cell may differ from the differences in the bulk stiffness. The strain-stiffening response in fibrous scaffolds also depends on the degree of crosslinking, which means that cells experience a local matrix stiffness, that may be different from the bulk measurements [23, 24]. This makes it difficult to attribute cell behavior to any one specific scaffold parameter[18–20]. Ultimately, these multi-parameter effects of changing one variable (e.g., pore size, shape, or internal geometry) inhibit the ability to deconstruct the process of cell migration under different conditions in which only one parameter is modified. Lastly, naturally-derived materials used in several scaffolds are inherently adhesive, offering little or no control over the presentation of ECM proteins, adding to the complexity of analyzing results. Since cell migration in 3D ECMs is closely tied to several microstructural parameters[16, 25], there is a need to develop scaffolds which allow precise tuning of individual parameters (internal architectures, mechanical properties etc.) to evaluate orthogonal effects of these structural cues on cell motility.
Current fabrication methods for 3D scaffolds, such as freeze-drying [9], electrospinning [26],and gas foaming [27] do not allow precise control of the internal structural features and topology. Free form fabrication techniques like layer-by-layer stereolithography, two-photon polymerization, inverted colloidal crystals (ICC) etc. are capable of providing precise microstructural properties required to understand cell motility in 3D [28–30]. In this work, we used versatile layer-by-layer microfabrication system coined “digital micro-mirror device based projection printing” (DMD-PP) in combination with photocrosslinkable PEG to produce a precise log-pile micro-architecture. PEG-based biomaterials have been used extensively for tissue engineering of bone, vasculature and other tissues because of their high water content, biocompatibility and tunable mechanical properties [31, 32]. Previously, we have used the DMD-PP method to create 3D PEG-based biological scaffolds with complex internal architectures at microscale resolutions [33–38]. PEG-based optically transparent 3D scaffolds, are also compatible with standard microscopy techniques and there are minimal variations among scaffolds using the DMD-PP method. The DMD-PP fabrication method used in this work, unlike conventional manufacturing techniques, is “design driven” and not “process driven” and therefore facilitates systematic tuning of individual parameters, including 3D micro-architecture and elastic modulus. Although, we utilize 3D scaffolds of distinct elastic modulus to evaluate 3D migration properties of normal and cancerous breast epithelial cells, the methodology is broadly applicable for modeling 3D cell motility and various in vivo microenvironments.
MATERIALS AND METHODS
PEG prepolymer preparation
Poly (ethylene glycol) diacrylate (PEGDA, mol. wt.= 700kDa), acrylic acid and 2,2,6,6-tetramethylpiperidine 1-oxyl (TEMPO, free-radical quencher) were purchased from Sigma-Aldrich and used as received. Photoinitiator Irgacure 2959 and TINUVIN 234 UV-dye were obtained from Ciba Chemistry. TINUVIN 234, a UV-absorbing agent, was used to reduce the curing depth of the monomers and adjust the thickness of the microstructures in the DMD-PP fabrication process. TEMPO was used to enhance the contrast of the UV-curing process and optimizes feature resolution at the projection plane. Two prepolymer solutions were mixed as described below: (1) PEGDA 19%(v/v) + Acrylic acid [1% (v/v)] + Irgacure 2959 [1% (w/v)] + TINUVIN 234 [0.1% (w/v)] + TEMPO [0.01% (w/v)] + DI water [80% (v/v)] (2) PEGDA 95%(v/v) + Acrylic acid [5% (v/v)] + Irgacure 2959 [1% (w/v)] + TINUVIN 234 [0.1% (w/v)] + TEMPO [0.01% (w/v)]. The prepolymer solution was mixed thoroughly, sonicated for 30 min, and degassed for 15 min.
Scaffold fabrication method
The DMD-PP process was used to fabricate 3D scaffolds through UV photolithography in a layer-by-layer fashion (Fig. 1A)[33, 35]. Briefly, a digital light processing (DLP) chip (1920 × 1080 Discovery 4000, Texas Instrument, TX, USA) was used to create dynamic photomasks from computer-aided design (CAD) (AutoCAD, CA, USA), which represents the cross-sectional images of the 3D microstructure (Fig.1A). The DLP chip projects the patterns (e.g. line array in the log-pile structure) onto the prepolymer solution using a uniform UV light source (EXFO, Quebec, QC, Canada). Each layer of the pattern was irradiated for 20 seconds with an intensity of 50 mW/cm2 through a UV-grade optical lens (Edmunds Optics, Barrington, NJ, USA) onto the focal point of the projection lens. After the fabrication of each layer, the Z axis of the fabrication stage was repositioned to create a new layer. The thickness of each 2D layer can be precisely controlled by placing a glass coverslip above each layer. Consequently, 3D structures were created by sequential development of the hydrogel accordingly to the cross-sectional images generated on the DLP chip (Fig. 1B). The bottom surface of the coverslip was coated with a nonstick layer formed by Krytox 157 FSH oil (DuPont, Wilmington, DE), which readily releases the cured microstructure. Following the fabrication procedure, scaffolds were removed from the stage and gently washed using phosphate-buffered saline (PBS). The control 2D slab samples were also made following similar procedure. The photomask used for fabricating these 2D slabs were just a plain mask with no specific pattern.
Figure 1.
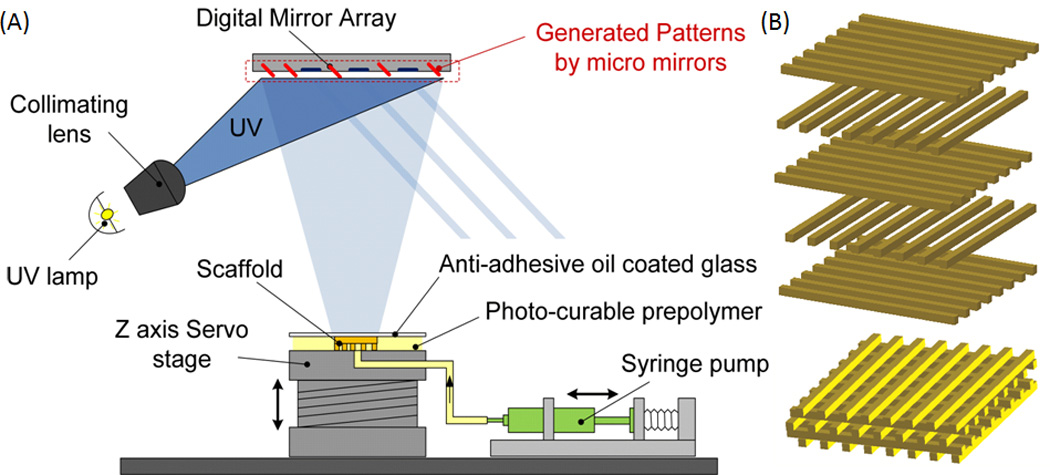
(A) Schematic representation of the digital micromirror based projection printing (DMD-PP). DMD-PP produces precise features using a system of dynamic masks generated by a digital light controlling chip. CAD software is used to design and fabricate log-pile scaffolds in photocurable PEG biomaterial. (B) Schematic of the assembled structures; the layers of crosslinked photopolymer can be stacked to form scaffolds having log-pile micro-architecture. SolidWorks files showing the structure at each layer. The layers of crosslinked PEG can be stacked to form scaffolds having log-pile micro-architecture.
Mechanical testing
Scaffold samples of the PEG polymer were stored in a solution of Isopropyl alcohol (IPA). The samples were decanted and their length and width were measured. The samples were nominally 3mm × 3mm × 1mm tall, in the shape of right angle prisms. The samples were mounted in a Perkin Elmer Thermomechanical analyzer (TMA 7) instrument. The TMA was setup to perform a static force scan, using two 20 mm circular parallel plates. The sample was then loaded with an initial force of 10 mN, and the sample height was determined by Linear Voltage Displacement Transducers (LVDT) of the instrument. During the test, the force was increased on the sample from the initial force (usually 10 mN) to the final force (usually 2500 mN) at a constant rate of 120 mN/min. The initial loading portion of each stress-strain curve was used to determine the modulus of the samples. Typically 3 samples were tested for each sample type to generate reproducible results. Compressive strain moduli were defined as the slope of the linear portion of the stress-strain curve (Fig. 2A). Stress-strain curves were plotted and analyzed to facilitate the calculation of the testing parameters. Statistical significance was determined with ANOVA (MINITAB V 14.2, Minitab Inc.) using a standard p<0.001.
Figure 2.
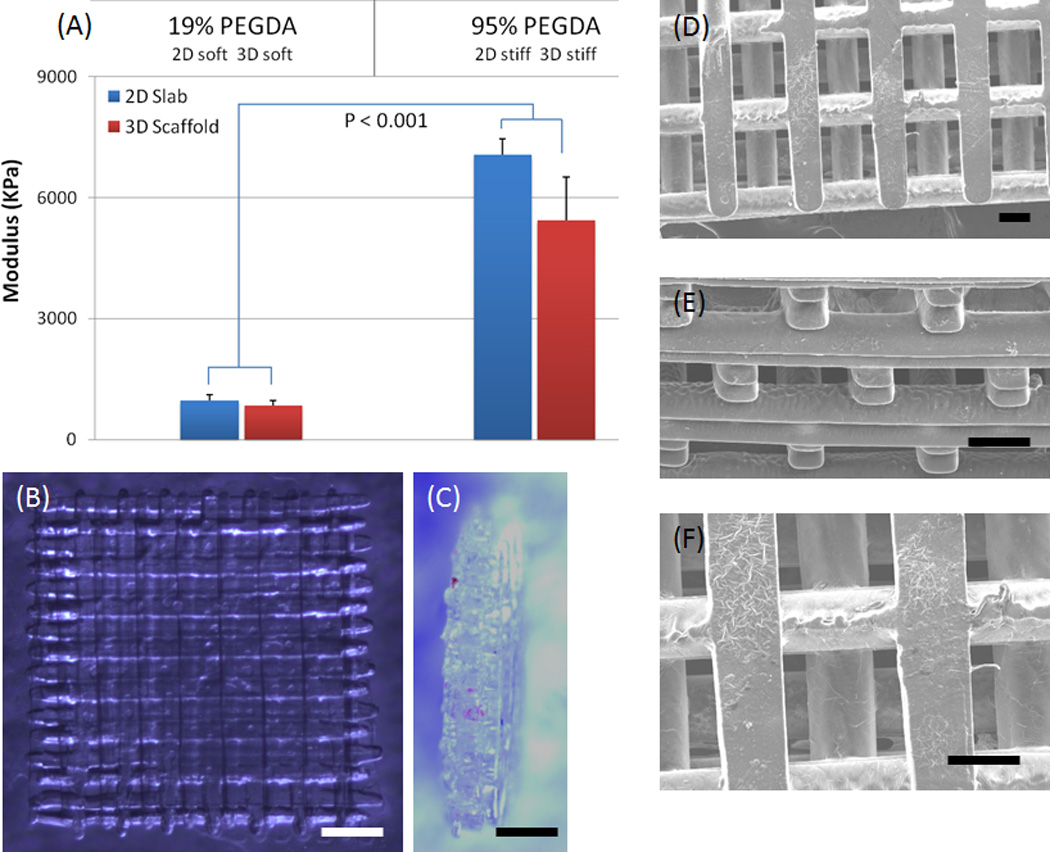
(A) Mechanical properties of DMD-PP fabricated scaffolds having 2D slab and 3D log-pile structures and fabricated using 19% (soft scaffold) and 95% (stiff scaffold) PEGDA (700 kDa molecular weight). Results showed that there is slight increase in compressive modulus for 2D stiff scaffold (~7MPa) compared to stiff 3D log-pile (~5.5MPa). However both the stiff scaffolds are about 8 times stiffer than the soft 2D and 3D scaffolds (~0.9MPa). This demonstrates that prepolymer concentration and 3D architecture can be used to tailor the mechanical properties of the PEGDA scaffolds. (B–C) Optical images show top (B) and side (C) views of the almost optically transparent 3D PEGDA scaffold and the 5-layer structure. The DMD fabrication process produced precise features using a system of dynamic masks generated by a digital light controlling chip (height ~0.5mm, each layer is ~0.1mm, scale: 500 µm). (D–F) SEM images of top (D), side (E) and magnified top view (F) of a 3D log-pile scaffold clearly shows the precise internal-geometry (scale: 100µm).
Scanning electron microscopy
Scanning electron microscopy (SEM) was used to analyze the structure of HMLE cells in the log-pile scaffolds (Figure 3A). The PEG scaffolds were fixed with glutaraldehyde and dehydrated in a series of ethanol baths. The specimens were then mounted on stubs and sputter-coated with 8nm thick Iridium using Emitech K575X. The samples were then examined in the Field Emission Environmental scanning electron microscope (FEI XL30 ESEM FEG) operated at 10 kV using high vacuum mode.
Figure 3.
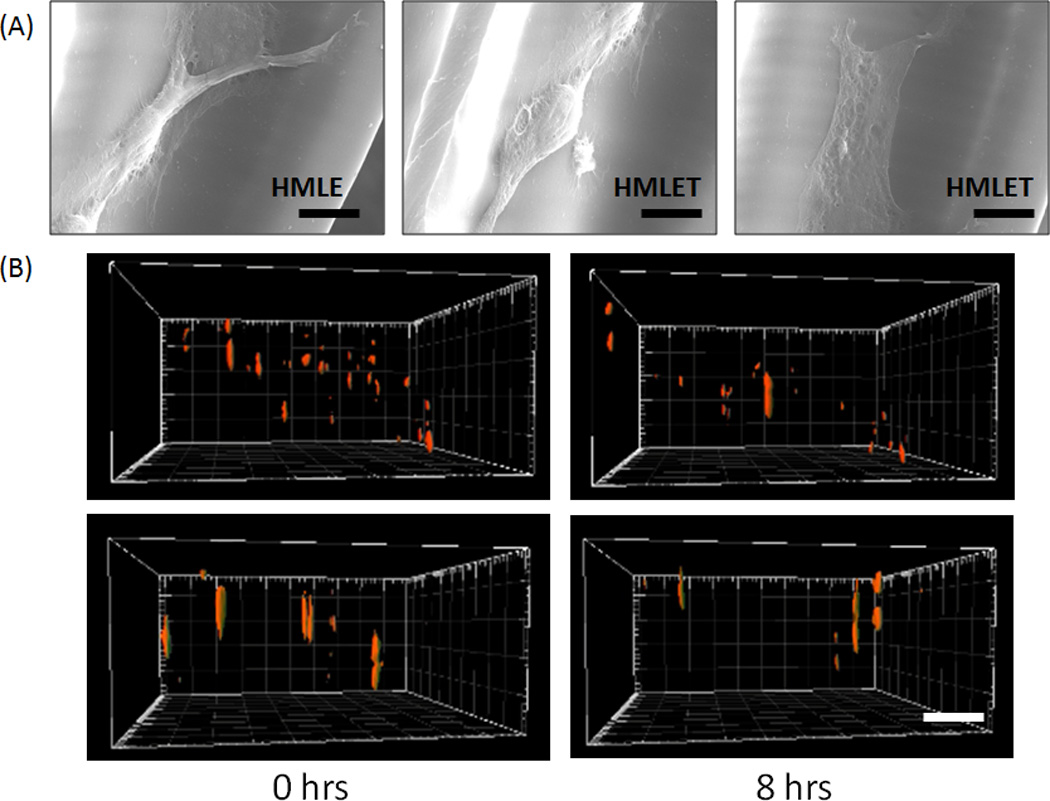
(A) Characteristic SEM images of HMLE and HMLET cells migrating inside the 3D log-pile scaffold. (Scale = 10µm). (B) Still images at 0 and 8 hours from reconstituted 3D confocal stacks of HMLE (top) and HMLET (bottom) on stiff 3D PEG log-pile scaffolds (Scale bar = 100µm).
Cell migration on PEG-based scaffolds
HMLE or HMLE-Twist cells were cultured as previously described [39, 40] in MEBM media (plus supplements) purchased from Lonza. Each scaffold was activated by incubating the scaffolds in a working solution of 0.15M 1-ethyl-3-[3–dimethylaminopropyl] carbodiimide hydrochloride (EDC) and 0.12M N-hydoxysuccinimide (NHS) in 2-[morpholino]ethanesulfonic acid (MES) buffer at pH 5 for 2 hours. Scaffolds were briefly rinsed with PBS (pH 7.4) to remove any residual NHS and EDC and immersed in a 5µg/mL human plasma fibronectin/PBS solution for 1 hour at 37°C. After rinsing non-bound fibronectin away with PBS, trypsin-dissociated and PBS-washed low-passage live cells were plated onto each scaffold in complete media in a glass-bottom chamber slide. Cells were allowed to adhere for at least 24 hours under controlled temperature and CO2 levels. Time-lapse Z-stacks were acquired on each scaffold for 10–15 hours at 20 minute intervals under buffer- and temperature-controlled conditions using a Nikon TE 2000 scanning confocal microscope. Consecutive Z-stack slices were acquired with a height step of 10 microns for a 0.2 mm-thick region of the scaffold. The stack and side-view images were constructed from 20–40 individual slices at each time point using the Imaris 3D imaging software suite (Figure 3B). Alternatively, soft and stiff 2D PEG slabs with no 3D architecture were used as described above to monitor cell migration in 2D. The object analysis function of Imaris was utilized to calculate the displacement (the distance of a straight line from initial x,y,z coordinates to final x,y,z coordinates), velocity (migration speed in microns/sec) and straightness (the quotient of displacement divided by track length – the total distance traveled by each cell) of cell migration for at least 20 cells in triplicate in both the 2D and 3D environments. Two-way or student T-test statistical analysis were performed using the Graph Pad Prism software suite.
RESULTS
Scaffold fabrication
In this study, we prepared 3D PEG scaffolds with log-pile microarchitecture and 2D PEG slabs. Fig 2B–F shows the internal log-pile pore architecture with 5 layers (~100 microns thickness of each layer). As shown in the optical images, we are able to fabricate log-pile structures according to the design. For the log-pile scaffold, the length of the gap between logs was 250~300 microns.
Mechanical properties
Figure 2A demonstrates that PEG prepolymer concentration and 3D architecture can be used to tailor the mechanical properties of the PEGDA scaffolds. Prepolymer concentration of 19% and 95% (700 kDa molecular weight PEGDA) are referred to as soft and stiff scaffolds respectively. Results showed that there is a slight increase in compressive modulus for 2D stiff scaffold (~7MPa) compared to the stiff 3D log-pile (~5.5MPa), however both the stiff scaffolds are about 8 times stiffer than the soft 2D and 3D scaffolds (~0.9MPa).
Effect of cell transformation and substrate stiffness on 3D cell migration
Cell motility was first examined on soft and stiff 2D slabs (Supplemental Movies 1–4). Neither differences in the elastic modulus of these 2D PEG slabs or TWIST oncogene transformation of normal mammary epithelial cells (HMLE) affected cell migration parameters (Table 1, Figure 4a–b and Supplemental Movies 1–4). HMLE and Twist-transformed HMLE cells displayed similar displacements and path straightness measurements, with a slight decrease in velocity in TWIST-transformed cells on stiff substrates when compared to these cells on soft substrates. The addition of a third dimension in which the cells could migrate, induced substantial effects on nearly all aspects of cell migration. We assessed cell motility when cells were allowed to migrate in three dimensions using a 3D log-pile structure (Table 1, Figures 4c–f and Supplemental Movies 5–8). Normal HMLE cell displacement increased from 14.81 to 19.25 microns and the velocity increased from 2.96 to 6.80 microns/sec when observed on stiff 3D scaffolds in comparison to stiff 2D slabs (Table 1 and Supplemental Movies 1 and 5). TWIST-transformed HMLE cells (HMLET) displayed a decrease in displacement (15.74 to 7.91 microns) and velocity (2.71 to 1.85 microns/sec) (Table 1 and Supplemental Movies 4 and 8) when monitored on soft 3D scaffolds in comparison to soft 2D slabs. However, HMLET cells migrate further (13.05 to 27.14 microns) and faster (1.83 to 2.05 microns/sec) on stiff 3D scaffolds (13.05 microns and 1.83 microns/sec) in comparison to stiff 2D slabs (27.14 microns and 2.05 microns/sec) (Table 1 and Supplemental Movies 3 and 7). These results demonstrate that Twist transformation of HMLE cells and substrate stiffness has a notable effect on migration parameters predominantly within the 3D scaffold environments.
Table 1.
Average displacement, velocity and straightness measurements +/− standard error means for three individual replicate experiments.
| 2D Migration | ||
|---|---|---|
| HMLE | Soft | Stiff |
| Displacement (µm) | 14.14 ± 2.589 | 14.81 ± 1.865 |
| Velocity (µm/sec) | 2.954 ± 0.2898 | 2.969 ± 0.4204 |
| Straightness | 0.4841 ± 0.05145 | 0.4811 ± 0.04448 |
| HMLET | Soft | Stiff |
| Displacement (µm) | 15.74 ± 2.094 | 13.05 ± 1.062 |
| Velocity (µm/sec) | 2.707 ± 0.2975 | 1.832 ± 0.2579 |
| Straightness | 0.5112 ± 0.02415 | 0.5065 ± 0.04203 |
| 3D Migration | ||
|---|---|---|
| HMLE | Soft | Stiff |
| Displacement (µm) | 24.82 ± 1.675 | 19.25 ± 1.817 |
| Velocity (µm/sec) | 3.182 ± 0.2938 | 6.802 ± 0.3416 |
| Straightness | 0.5020 ± 0.03201 | 0.6205 ± 0.02626 |
| HMLET | Soft | Stiff |
| Displacement (µm) | 7.91 ± 0.9703 | 27.14 ± 1.785 |
| Velocity (µm/sec) | 1.844 ± 0.2793 | 2.056 ± 0.1678 |
| Straightness | 0.4498 ± 0.03760 | 0.7040 ± 0.03030 |
Figure 4.
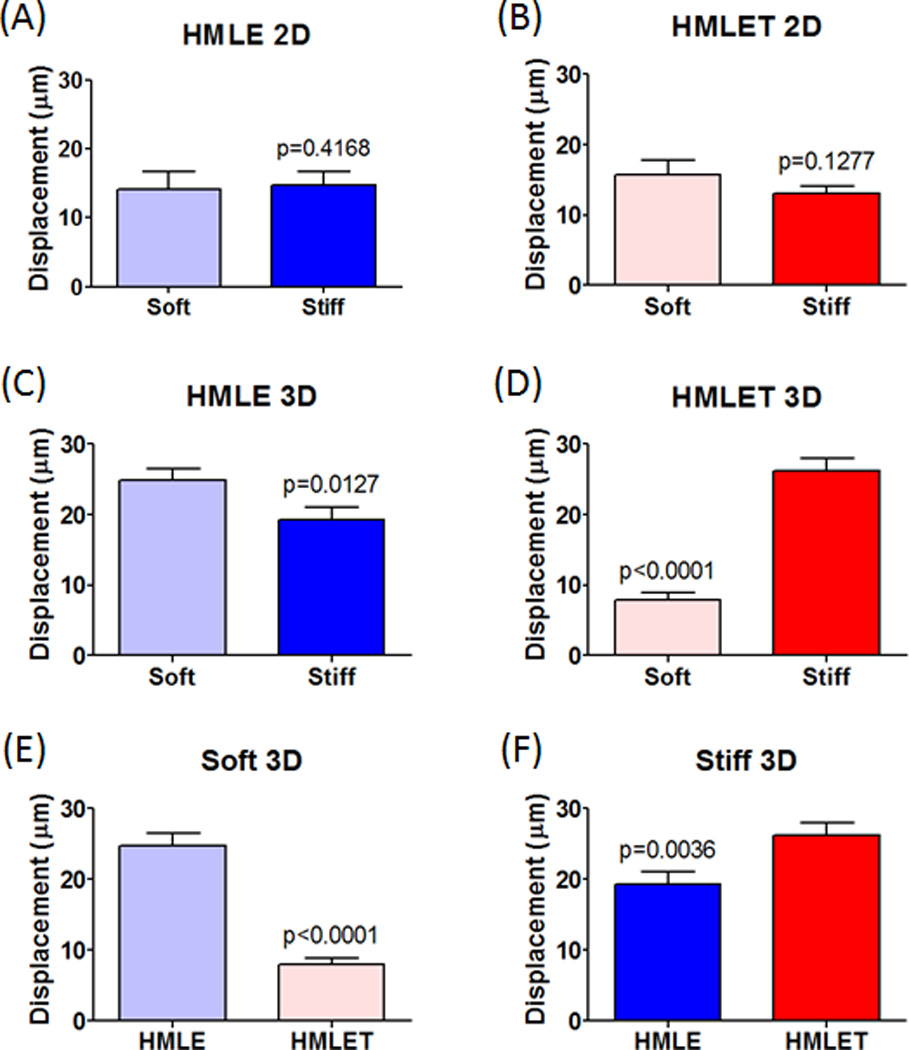
(A–B) Quantified displacement for normal mammary epithelial cells (HMLE) and TWIST modified cells (HMLET) on soft and stiff 2D slabs. (C–F) Quantified displacement for HMLE and HMLET on soft and stiff 3D scaffolds.
We also observed that differences in 3D scaffold elastic moduli had opposing effects on overall migration displacement in HMLE (soft: 24.82 microns, stiff: 19.25 microns) and HMLET (soft: 7.91 microns, stiff: 27.14 microns) cells (Table 1, Figures 4c–d and Supplemental Movies 5–8). However, cell velocity increased for both HMLE (soft: 3.182 microns/sec, stiff: 6.802 microns/sec) and HMLET (soft: 1.844 microns/sec, stiff: 2.056 microns/sec) cells on stiff versus soft 3D scaffolds (Table 1 and Supplemental Movies 5–8). Finally, while Twist transformation uniformly decreased migration velocity on both soft (HMLE: 3.182 microns/sec, HMLET: 1.844 microns/sec) and stiff (HMLE: 6.802 microns/sec, HMLET: 2.056 microns/sec) 3D scaffolds, Twist-transformed cells unexpectedly migrated a shorter distance on soft 3D scaffolds (HMLE: 24.82 microns, HMLET: 7.91 microns) while Twist transformation increased cell displacement on stiff 3D scaffolds (HMLE: 19.25 microns, HMLET: 27.14 microns) (Table 1, Figures 4e–f and Supplemental Movies 5–8). Importantly, these results demonstrate that the substrate stiffness alone within the tumor cell microenvironment may in fact be sufficient to promote or inhibit cell migration and subsequent colonization during processes such as cancer metastasis. Thus, this in vitro model may prove beneficial in predicting the tissue invasion preferences for various types of tumor cells.
DISCUSSION
Cancer cells constantly encounter a variety of mechanical forces during tumor growth, invasion, and metastasis, and in turn, cells sense, process and actively exert mechanical forces on their surrounding microenvironment. Most of our current understanding about cancer cell migration comes from in vitro studies using 2D substrates, which do not take into account the true dimensionality of the physiological system. Several features of the 3D ECM microenvironment such as elastic stiffness and internal-architecture play important roles in cancer cell migration in vivo, which conventional ECM-coated 2D substrates are not able to mimic. Current fibrous scaffolds are incapable of precisely controlling internal geometry[17] and therefore the specific influence of controlled changes in 3D scaffold architecture on cell migration cannot be well understood using traditional 3D scaffolds. To better understand and model cancer migration, we need a system wherein various ECM parameters can be tuned independently.
In this report, we describe a versatile layer-by-layer microfabrication system that can not only build scaffolds of precise architecture but also tune their mechanical stiffness (Figure 2 and 3A). We have used photocrosslinkable PEGDA to produce scaffolds using a log-pile internal architecture to investigate changes in cell migration when cancer cells are presented with an option of migrating in three dimensions on either stiff or soft scaffolds. While most solid tumors have a very soft elasticity, the elasticity of the tissue microenvironment into which cancer cells invade and colonize during metastasis can vary greatly. Thus, we have chosen soft and stiff elastic moduli for these initial studies demonstrating the range of applicability for this scaffold system. The results provide a basic framework for a variety of cancer cell behavior experiments. Incorporating additional parameters to this system may allow for a more complete description of cell migration and thereby yield an in vitro model that approaches a more accurate representation of the pathophysiological behavior of tumors.
As anticipated, some changes in cell migration parameters were observed when we allowed migration of cancer cells in the third dimension and modulated the substrate stiffness. The substantial finding, however, lies in how observations from experiments conducted within a 2D system cannot be extrapolated simply to a 3D system. Specifically, whereas minimal changes in cell migration parameters were found for both HMLE and HMLET cells on 2D stiff and soft substrates, an analogous experiment conducted in a 3D log-pile scaffold system yielded significant differences in displacement, velocity, and straightness when comparing cell types or scaffold stiffness. Our findings depend directly on the biosystem we used. We acknowledge that our results are relevant to micron-scale 3D architectures and should not be directly extrapolated to scaffolds with nanoscale resolution. While scaffolds with smaller pore-sizes (Figure S1) can be fabricated using the DMD-PP system, we choose micron-scale pore-size for the log-pile scaffold to negate any steric-hindrance effects, often seen in 3D cell migration experiments using random pore-size scaffolds. Since the size of the cell is smaller than the log-dimensions, a single cell might interact with complex surface topological features in a 3D setup. Micron-scale scaffolds open the possibility to test different aspects of cancer cell migration, especially the effects of local curvatures and other complex topologies on cancer invasion characteristic. Micron-scale scaffolds are useful since the overall migration distances of cancer cells during metastasis also range to several hundred microns [41]. Moreover, epithelial cancer cells typically using mesenchymal mode of migration can be 50–200µm in length, making the micronscale scaffolds developed in this work directly applicable [42]. Micron-scale scaffolds will also be useful to investigate collective migration in highly differentiated tumors such as lobular breast cancer where collective-cell invasion has been shown to predominate [43].
Results clearly demonstrate that it is imperative to investigate cancer cell migration in a controlled 3D biosystem, with the ability to selectively tune one parameter without affecting any other parameters. PEG-based synthetic polymers used in this work, provides a clean slate to graft any number of biomolecules such as RGD peptides, growth factors, and biomolecules necessary for cell migration. This allows us to observe the cellular effects of solely adding a third dimension without concurrent changes in any other parameters. The cell migration results obtained in this work will serve as a template and a baseline to investigate concurrent effects of modulating multiple parameters simultaneously.
CONCLUSION
The complex interplay between cancer cells and the diverse micro-environments they encounter during the metastatic process remains largely unknown, particularly in terms of 3D interactions. In this work, we focused on “deconstructing” the cancer cell motility problem, by tuning the biophysical parameters including micro-architecture and mechanical stiffness that critically regulate cell migration. Results demonstrate the capability of the DMD-PP process for tuning stiffness in a 3D microenvironment, without changing other microstructural and micromechanical parameters that affect motility. We expect that the field will gain valuable insight into cancer cell motility by using the DMD-PP fabricated scaffolds, which permit orthogonal tuning of specific parameters towards the creation of complex 3D microenvironments that better reflect the native physiology.
Supplementary Material
ACKNOWLEDGEMENTS
The project described was supported in part by Award Number R01EB012597 from the National Institute of Biomedical Imaging And Bioengineering to S.C. This work was supported by the NIH-IRACDA (National Institutes of Health - Institutional Research and Academic Career Development Award) Postdoctoral Fellowship GM06852 (to J.A.K.) and National Institutes of Health Grants CA097022 and CA129231 (to R.L.K.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: Integrating signals from front to back. Science. 2003;302(5651):1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 3.Lauffenburger DA, Horwitz AF. Cell Migration: A physically integrated molecular process. Cell. 1996;84(3):359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg Robert A. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Sanz-Moreno V, Marshall CJ. The plasticity of cytoskeletal dynamics underlying neoplastic cell migration. Curr Opin Cell Biol. 2010;22(5):690–696. doi: 10.1016/j.ceb.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 6.Stoletov K, Kato H, Zardouzian E, Kelber J, Yang J, Shattil S, et al. Visualizing extravasation dynamics of metastatic tumor cells. J Cell Sci. 2010;123(13):2332–2341. doi: 10.1242/jcs.069443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoletov K, Montel V, Lester RD, Gonias SL, Klemke R. High-resolution imaging of the dynamic tumor cell vascular interface in transparent zebrafish. Proc Natl Acad Sci U S A. 2007;104(44):17406–17411. doi: 10.1073/pnas.0703446104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Indra I, Beningo KA. An in vitro correlation of metastatic capacity, substrate rigidity, and ECM composition. J Cell Biochem. 2011;112(11):3151–3158. doi: 10.1002/jcb.23241. [DOI] [PubMed] [Google Scholar]
- 9.Ho M-H, Kuo P-Y, Hsieh H-J, Hsien T-Y, Hou L-T, Lai J-Y, et al. Preparation of porous scaffolds by using freeze-extraction and freeze-gelation methods. Biomaterials. 2004;25(1):129–138. doi: 10.1016/s0142-9612(03)00483-6. [DOI] [PubMed] [Google Scholar]
- 10.Ovsianikov A, Deiwick A, Van Vlierberghe S, Pflaum M, Wilhelmi M, Dubruel P, et al. Laser fabrication of 3D gelatin scaffolds for the generation of bioartificial tissues. Materials. 2011;4(1):288–299. doi: 10.3390/ma4010288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker EL, Srivastava J, Yu D, Bonnecaze RT, Zaman MH. Cancer cell migration: Integrated roles of matrix mechanics and transforming potential. PLoS ONE. 2011;6(5):20355. doi: 10.1371/journal.pone.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaman MH, Trapani LM, Sieminski AL, MacKellar D, Gong H, Kamm RD, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci U S A. 2006;103(29):10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willerth SM, Arendas KJ, Gottlieb DI, Sakiyama-Elbert SE. Optimization of fibrin scaffolds for differentiation of murine embryonic stem cells into neural lineage cells. Biomaterials. 2006;27(36):5990–6003. doi: 10.1016/j.biomaterials.2006.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raeber GP, Lutolf MP, Hubbell JA. Molecularly Engineered PEG Hydrogels: A novel model system for proteolytically mediated cell migration. Biophys J. 2005;89(2):1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma PX, Choi J-W. Biodegradable polymer scaffolds with well-defined interconnected spherical pore network. Tissue Eng. 2001;7(1):23–33. doi: 10.1089/107632701300003269. [DOI] [PubMed] [Google Scholar]
- 16.Kuntz RM, Saltzman WM. Neutrophil motility in extracellular matrix gels: mesh size and adhesion affect speed of migration. Biophys J. 1997;72(3):1472–1480. doi: 10.1016/S0006-3495(97)78793-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi Y, Kang T, Lee LP. Plasmon resonance energy transfer (PRET)-based molecular imaging of cytochrome c in living cells. Nano Lett. 2008;9(1):85–90. doi: 10.1021/nl802511z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nonlinear elasticity in biological gels. Nature. 2005;435(7039):191–194. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- 19.Chandran PLBV. Affine versus non-affine fibril kinematics in collagen networks: theoretical studies of network behavior. J Biomech Eng. 2006;128(2):259–270. doi: 10.1115/1.2165699. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen J, Swartz M. Mechanobiology in the third dimension. Ann Biomed Eng. 2005;33(11):1469–1490. doi: 10.1007/s10439-005-8159-4. [DOI] [PubMed] [Google Scholar]
- 21.Kaufman LJ, Brangwynne CP, Kasza KE, Filippidi E, Gordon VD, Deisboeck TS, et al. Glioma expansion in collagen I matrices: Analyzing collagen concentration-dependent growth and motility patterns. Biophys J. 2005;89(11):635–650. doi: 10.1529/biophysj.105.061994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gelse K, Pöschl E, Aigner T. Collagens-structure, function, and biosynthesis. Adv Drug Deliv Rev. 2003;55(12):1531–1546. doi: 10.1016/j.addr.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Winer JP, Oake S, Janmey PA. Non-linear elasticity of extracellular matrices enables contractile cells to communicate local position and orientation. PLoS ONE. 2009;4(7):6382. doi: 10.1371/journal.pone.0006382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pathak A, Kumar S. Biophysical regulation of tumor cell invasion: moving beyond matrix stiffness. Integr Biol. 3(4):267–278. doi: 10.1039/c0ib00095g. [DOI] [PubMed] [Google Scholar]
- 25.Hayen W, Goebeler M, Kumar S, Riessen R, Nehls V. Hyaluronan stimulates tumor cell migration by modulating the fibrin fiber architecture. J Cell Sci. 1999;112(13):2241–2251. doi: 10.1242/jcs.112.13.2241. [DOI] [PubMed] [Google Scholar]
- 26.Lannutti J, Reneker D, Ma T, Tomasko D, Farson D. Electrospinning for tissue engineering scaffolds. Mater Sci Eng C Mater Biol Appl. 2007;27(3):504–509. [Google Scholar]
- 27.Krause B, Diekmann K, van der Vegt NFA, Wessling M. Open nanoporous morphologies from polymeric blends by carbon dioxide foaming. Macromolecules. 2002;35(5):1738–1745. [Google Scholar]
- 28.da Silva J, Lautenschlager F, Sivaniah E, Guck JR. The cavity-to-cavity migration of leukaemic cells through 3D honey-combed hydrogels with adjustable internal dimension and stiffness. Biomaterials. 2009;31(8):2201–2208. doi: 10.1016/j.biomaterials.2009.11.105. [DOI] [PubMed] [Google Scholar]
- 29.Tayalia P, Mendonca CR, Baldacchini T, Mooney DJ, Mazur E. 3D cell-migration studies using two-photon engineered polymer scaffolds. Adv Mater. 2008;20(23):4494–4498. [Google Scholar]
- 30.Yung-Shin S, Shih-Wei P, Keng-Hui L, Ji-Yen C. Electrotaxis of lung cancer cells in ordered three-dimensional scaffolds. Biomicrofluidics. 2012;6(1):014102. doi: 10.1063/1.3671399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khademhosseini A, Langer R. Microengineered hydrogels for tissue engineering. Biomaterials. 2007;28(34):5087–5092. doi: 10.1016/j.biomaterials.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 32.Lee KY, Mooney DJ. Hydrogels for Tissue Engineering. Chem Rev. 2001;101(7):1869–1880. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- 33.Fozdar DY, Soman P, Lee JW, Han L-H, Chen S. Three-Dimensional Polymer Constructs Exhibiting a Tunable Negative Poisson's Ratio. Adv Funct Mater. 2011;21(14):2712–2720. doi: 10.1002/adfm.201002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han L-H, Mapili G, Chen S, Roy K. Projection microfabrication of three-dimensional scaffolds for tissue engineering. J Manuf Sci E–T. 2008;130(2):021005. [Google Scholar]
- 35.Han L-H, Suri S, Schmidt CE, Chen S. Fabrication of three-dimensional scaffolds for heterogeneous tissue engineering. Biomed Microdevices. 2010;12:721–725. doi: 10.1007/s10544-010-9425-2. [DOI] [PubMed] [Google Scholar]
- 36.Lu Y, Mapili G, Suhali G, Chen SC, Roy K. A digital micro-mirror device-based system for the microfabrication of complex, spatially patterned tissue engineering scaffolds. J Biomed Mater Res A. 2006;77(2):396–405. doi: 10.1002/jbm.a.30601. [DOI] [PubMed] [Google Scholar]
- 37.Soman P, Lee JW, Phadke A, Varghese S, Chen S. Spatial tuning of negative and positive Poisson's ratio in a multi-layer scaffold. Acta Biomater. 2012;8(7):2587–2594. doi: 10.1016/j.actbio.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soman P, Fozdar DY, Lee JW, Phadke A, Varghese S, Chen S. A three-dimensional polymer scaffolding material exhibiting a zero Poisson's ratio. Soft Matter. 2012;8(18):4946–4951. doi: 10.1039/C2SM07354D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mani SA, Guo W, Liao M-J, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Green CE, Liu T, Montel V, Hsiao G, Lester RD, Subramaniam S, et al. Chemoattractant signaling between tumor cells and macrophages regulates cancer cell migration, metastasis and neovascularization. PLoS ONE. 2009;4(8):6713. doi: 10.1371/journal.pone.0006713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3(5):362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 43.Bell C, Waizbard E. Variability of cell size in primary and metastatic human breast carcinoma. Invasion Metastasis. 1986;6(1):11–20. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


