Abstract
Instability of the nuclear genome is a hallmark of cancer and aging. MMS19 protein has been linked to maintenance of genomic integrity but the molecular basis of this connection is unknown. Here, we identify MMS19 as a member of the cytosolic iron-sulfur protein assembly (CIA) machinery. MMS19 functions as part of the CIA targeting complex that specifically interacts with and facilitates iron-sulfur cluster insertion into apoproteins involved in methionine biosynthesis, DNA replication, DNA repair and telomere maintenance. MMS19 thus serves as an adapter between early-acting CIA components and a subset of cellular iron-sulfur proteins. The function of MMS19 in maturation of crucial components of DNA metabolism may explain the sensitivity of MMS19 mutants to DNA damage and the presence of extended telomeres.
Maintaining genomic stability is a key cellular function and its impairment has been implicated in a variety of diseases including cancer (1–3). The process has been connected to mitochondrial function and Fe/S protein biogenesis, and may be relevant for the neurodegenerative disorder Friedreich’s ataxia (4–6). While the observation that multiple DNA replication and repair enzymes require iron-sulfur clusters for function has suggested a link between genomic stability and Fe/S protein biogenesis (7–12), in vivo evidence is lacking and the molecular basis of these connections is unclear. Synthesis of Fe/S clusters and their assembly into proteins as inorganic cofactors requires a dedicated and conserved biosynthetic pathway (13, 14). Mitochondrial Fe/S proteins are matured by the iron-sulfur cluster (ISC) assembly machinery. Extra-mitochondrial Fe/S protein biogenesis depends on both the ISC and CIA machineries. Cytosolic Fe/S clusters are first assembled on the CIA scaffold complex CFD1-NBP35 (15–18) and then transferred to apoproteins with the help of Cia1 (human CIAO1; (19)) and Nar1 (human IOP1; (20, 21)).
Mutants in eukaryotic MMS19 (also known as MET18 in yeast) show a variety of phenotypes including defects in methionine synthesis, sensitivity to genotoxic stress, and the presence of extended telomeres (22–25). A molecular function explaining these diverse cellular roles is unknown. Previous proteomic studies identified an interaction between yeast Mms19 and (putative) CIA components potentially linking MMS19 to Fe/S protein biogenesis (26, 27). Here, we show that MMS19 is a component of the CIA machinery and acts as part of the ‘CIA targeting complex’ that transfers Fe/S clusters to various DNA metabolism-associated Fe/S proteins. These findings explain the previously described MMS19 mutant phenotypes.
Yeast Mms19 is a late-acting CIA component
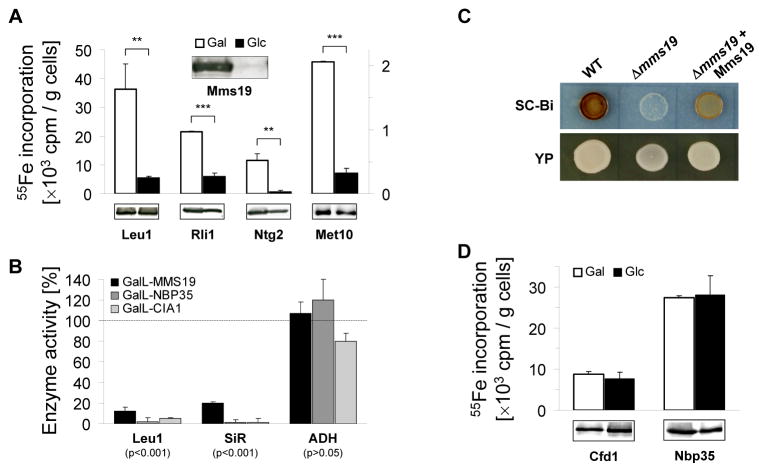
The interaction of yeast Mms19 (gene YIL128w) with two late-acting members of the CIA machinery, Cia1 and Cia2 (gene YHR122w; (19, 26–28)), was validated by co-immunoprecipitation (29) (fig. S1A). The potential role of Mms19 in cellular Fe/S protein biogenesis was examined by depleting the protein in a GAL promoter-regulatable MMS19 yeast strain and measuring 55Fe incorporation into known Fe/S target proteins (Leu1, Rli1, and Ntg2) by immunoprecipitation and scintillation counting (30). Mms19 depletion resulted in decreased 55Fe/S cluster binding to these proteins (Fig. 1A). The decrease in 55Fe binding by Leu1 correlated with loss of its enzymatic activity, while the activity of the non-Fe/S protein alcohol dehydrogenase was unchanged (Fig. 1B). These results were similar to those for depletion of the CIA proteins Nbp35 and Cia1(19). The defect in Leu1 activity did not result from impaired methionine biosynthesis in mms19 mutants, since other methionine synthesis mutants showed normal Leu1 activity (fig. S2A). Mms19 was also required for 55Fe/S cluster assembly and the sulfite reductase activity of the Met5-Met10 complex which supplies sulfur for methionine biosynthesis in a CIA-dependent manner (Figs. 1A–C and S2B). Together, these findings explain the methionine auxotrophy of mms19 mutants, and identify Mms19 as a CIA component. As previously found for CIA defects, both mitochondrial Fe/S protein maturation (fig. S3A-B) and cellular iron metabolism (fig. S3C) were unaffected by Mms19 deficiency. To clarify when Mms19 acted in the CIA pathway (13), we investigated its requirement for Fe/S cluster assembly on the early-acting CIA proteins Cfd1 and Nbp35. Mms19 depletion had no detrimental effect on the 55Fe/S maturation of these factors (Fig. 1D) suggesting that Mms19 functions late in cytosolic Fe/S protein assembly as a partner of Cia1 and Cia2.
Fig. 1.
Yeast Mms19 is a CIA component acting late in cytosolic Fe/S protein maturation. (A) 55Fe incorporation into cytosolic Fe/S proteins. Gal-GFP-MMS19 yeast cells producing Rli1-HA, Ntg2-HA or Met10-HA were grown in minimal medium (SC) containing galactose (Gal) or glucose (Glc). After radiolabeling with 55Fe, cell extracts were analyzed for the indicated Fe/S proteins by immunoblotting, and associated 55Fe was quantified by immunoprecipitation and scintillation counting. (B) Enzyme activities of the cytosolic Fe/S proteins isopropylmalate isomerase (Leu1) and sulfite reductase (SiR), and of alcohol dehydrogenase (ADH) were measured in extracts of indicated cells grown for 36 h in glucose-containing SC and plotted relative to wild-type cell activities. (C) SiR activity was measured in wild-type (WT) and Δmms19 cells without and with a Mms19-encoding plasmid on SC (SC-Bi) or YP glucose plates containing bismuth ammonium citrate and sodium sulfite. Sulfide produced by SiR after growth for 3 days yields Bi2S3 (brown precipitate). (D) Mms19 acts late in biogenesis. 55Fe incorporation into the indicated CIA proteins was measured as in (A) for Gal-GFP-MMS19 cells with plasmids encoding Cfd1-TAP and Nbp35-TAP.
Yeast Mms19 assembles and interacts with Fe/S proteins involved in DNA metabolism
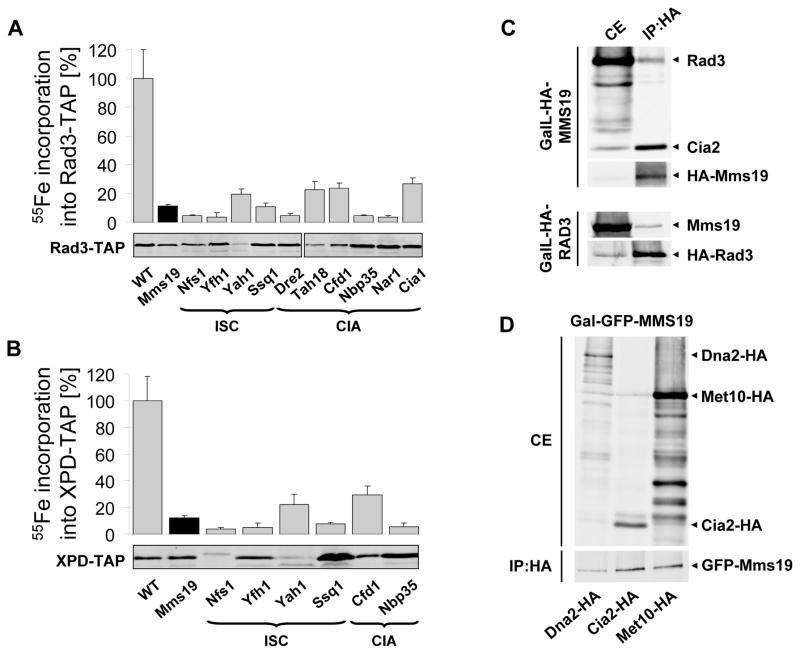
To explain the sensitivity of mms19 mutants to DNA damaging agents (22), we analyzed the requirement of yeast Mms19 for the maturation of several Fe/S cluster-containing DNA repair enzymes. 55Fe radiolabelling showed that the Rad3 DNA helicase bound a Fe/S cluster in vivo (Fig. 2A; (7)) and its maturation was dependent on Mms19 and the ISC CIA machinery. Similar findings were made for the human Rad3 orthologue XPD (Xeroderma pigmentosum protein D) when expressed in yeast (Fig. 2B). Mutations in Fe/S cluster-coordinating residues of XPD or the disease-relevant residue R112 destroyed Fe/S cluster binding (fig. S4A). Mms19 specifically co-immunoprecipitated with Rad3, Met10 (Fig. 2C–D), and several other Fe/S proteins including the DNA helicase/nuclease Dna2 (Figs. 2D and S4B), Rli1 and the DNA glycosylase Ntg2 (fig. S1B).
Fig. 2.
Yeast Mms19 interacts with and assembles Fe/S clusters into DNA helicases. (A–B) 55Fe incorporation into plasmid-encoded Rad3-TAP and human XPD-TAP was measured in yeast strains deficient in the indicated ISC and CIA proteins. The TAP-tagged proteins were visualized by immunostaining. For all depletions p<0.001. (C) MMS19 binds to Rad3 and Cia2. Extracts from overnight cultures of GalL-HA-MMS19 (top) and GalL-HA-RAD3 cells (bottom) grown in SC galactose medium were used for immunoprecipitation (IP) with anti-HA beads. Cell extracts (CE) and immunoprecipitates (IP:HA) were immunostained for indicated proteins. (D) Interaction of Mms19 with target Fe/S proteins. Extracts of Gal-GFP-MMS19 cells producing C-terminally HA-tagged Dna2, Cia2, or Met10 were used for immunoprecipitation with anti-HA beads. Bound proteins were analyzed by immunostaining (anti-HA, top; anti-GFP, bottom).
Human MMS19 matures only a subset of Fe/S proteins
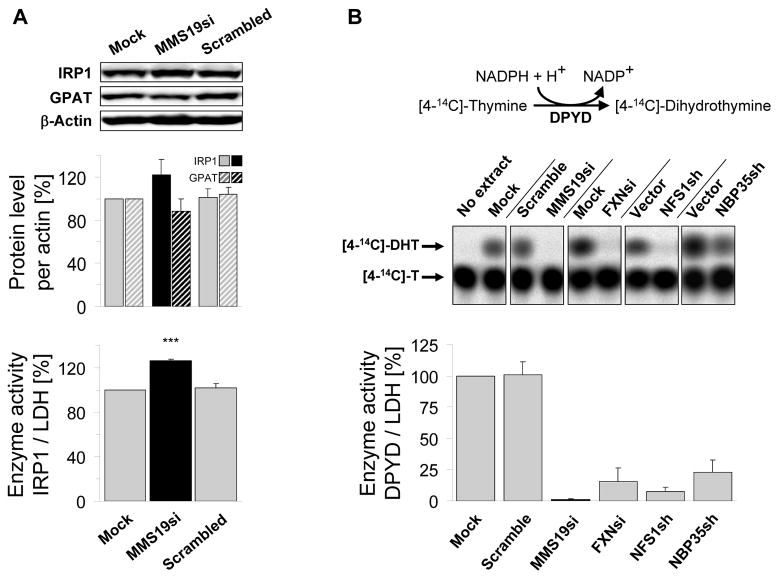
Mammalian MMS19 has been identified as part of a protein complex including XPD and two putative CIA components, CIAO1 and FAM96B (homologue of yeast Cia2). It has been functionally implicated in DNA repair, chromosome segregation, and transcription (24, 31, 32), yet no connection to Fe/S protein assembly has been proposed. To examine whether human MMS19 plays an evolutionarily conserved role in Fe/S protein biogenesis, we depleted MMS19 by RNA interference (RNAi) in HeLa cells using three different siRNA oligos either alone or as a pool (33). Efficient MMS19 silencing was achieved by three consecutive siRNA transfections performed at three day intervals (fig. S5A–B). We first tested the possible role of MMS19 in the maturation of two well-studied Fe/S proteins, cytosolic aconitase IRP1 (iron regulatory protein 1) and GPAT (glutamine phosphoribosylpyrophosphate amidotransferase; (16)). For IRP1, the enzyme activity, protein level, and binding capacity to iron-responsive RNA elements (IREs) were unchanged upon MMS19 depletion (Figs. 3A and S6A–C). Similarly, no effects on GPAT protein levels were observed (Figs. 3A and S7). Since Fe-S cluster binding is essential for GPAT stability, GPAT abundance is a reliable measure of its maturation (16).
Fig. 3.
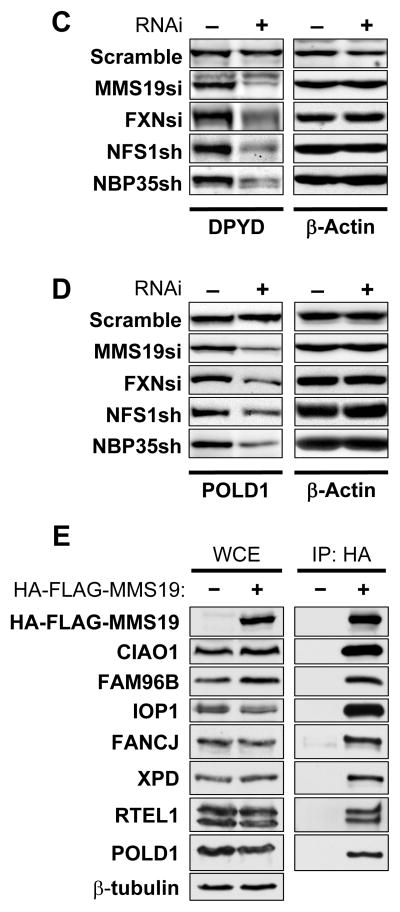
Human MMS19 interacts with and assembles Fe/S proteins involved in DNA metabolism. HeLa cells were RNAi-treated thrice at three day intervals or mock-treated. Cell extracts were prepared by digitonin lysis (cf. figs. S5–S10). (A) 9 days after starting MMS19 depletion, IRP1 (cytosolic aconitase) activities (normalized to lactate dehydrogenase; LDH) were measured and IRP1, GPAT and actin protein levels were determined by immunostaining. (B) DPYD activity was measured using thin layer chromatography and autoradiography. As controls, HeLa cells were RNAi-depleted for frataxin (FXN), NFS1, and NBP35. For all depletions p<0.001. (C) DPYD and (D) POLD1 protein levels were measured by immunostaining in cell extracts depleted for the indicated proteins. (E) Interaction of human MMS19 with Fe/S proteins and CIA components. Flp-In™TREx™-293 cells lacking or stably expressing inducible 3xHA-3xFLAG-MMS19 were induced with doxycycline (500 ng/mL) overnight. Whole cell extracts (WCE) were subjected to anti-HA immunoprecipitation (IP:HA) followed by immunostaining for indicated proteins.
These negative results prompted us to hypothesize that MMS19 plays a specialized role in Fe/S cluster maturation of a subset of target proteins. We therefore developed two additional assays for cytosolic Fe/S protein biogenesis. First, we examined the enzymatic activity of the Fe/S protein dihydropyrimidine dehydrogenase (DPYD; (34)) following the conversion of [4-14C]-thymine into [4-14C]-dihydrothymine by thin layer chromatography and autoradiography (Figs. 3B and S8A–C). In contrast to IRP1 and GPAT, DPYD activity was severely impaired upon MMS19 depletion. This decrease was also observed upon depletion of the ISC proteins Nfs1 and frataxin and the CIA component Nbp35, consistent with this effect being a Fe/S cluster assembly defect. Although DPYD protein levels were also depleted (Figs. 3C and S8D–E), the respective decreases were less pronounced compared to enzyme activities. Likely, the lower DPYD levels were indirect effects of apoprotein degradation. Next, we measured the amounts of the POLD1 subunit of DNA polymerase δ, the homologue of yeast Fe/S protein Pol3 (10). As impaired Fe/S protein assembly frequently results in apoprotein degradation (cf. Fig. 3C; (16)), Fe/S protein levels can be used to estimate their biogenesis. POLD1 levels were strongly decreased in MMS19-depleted cells (Figs. 3D and S9). Similar effects were observed during RNAi-mediated depletion of Nfs1, frataxin, and Nbp35. Since CIA depletion should not affect mitochondrial Fe/S proteins (cf. fig. S3; (16, 21)), we measured the levels and activities of mitochondrial aconitase (mtAco) and succinate dehydrogenase (SDH). They remained unchanged upon MMS19 depletion (fig. S10). Collectively, these results suggest that MMS19 is required for Fe/S cluster assembly of DPYD and POLD1, and may act as a specialized CIA factor with specificity for a subset of cytosolic-nuclear Fe/S proteins.
Human MMS19 is part of the CIA targeting complex
To define those Fe/S proteins that require MMS19 function, we used a proteomic approach to identify MMS19 interaction partners. MMS19-associated protein complexes were affinity-purified from a HEK293 cell line stably expressing HA-FLAG-MMS19, and then analyzed using Multidimensional Protein Identification Technology (MudPIT; (35). We identified a wide range of putative MMS19-interacting proteins including known and putative Fe/S proteins and the characterized and putative CIA components CIAO1, IOP1, and FAM96B (Tables S4A and S4B). For some of these proteins the MMS19 interaction was confirmed by co-immunoprecipitation of HA-FLAG-MMS19 from stable HEK293 cells followed by immunoblotting with specific antibodies. Endogenous CIAO1, FAM96B, IOP1, FANCJ, XPD, RTEL1, and POLD1 co-purified only in extracts containing HA-FLAG-MMS19 (Fig. 3E). The interaction between XPD and MMS19 was confirmed for endogenous levels (fig. S11). These data validate the proteomic data and show that MMS19 associates with both CIA factors and Fe/S target proteins including many involved in DNA metabolism. Based on the strong protein interactions between MMS19, CIAO1, and FAM96B as assessed by normalized spectral abundance factors (Table S4), co-immunoprecipitation studies (Fig. 3E), and the overlap between MMS19, CIAO1, and FAM96B protein interaction profiles (not shown), these three proteins likely form a complex that we term the CIA targeting complex. The results for human MMS19 agree with our yeast data and suggest that this complex acts late in Fe/S protein biogenesis to facilitate Fe/S cluster transfer from the CIA scaffold complex Cfd1-Nbp35 to target apoproteins.
Fe/S protein assembly defects augment sensitivity of cells to DNA damage
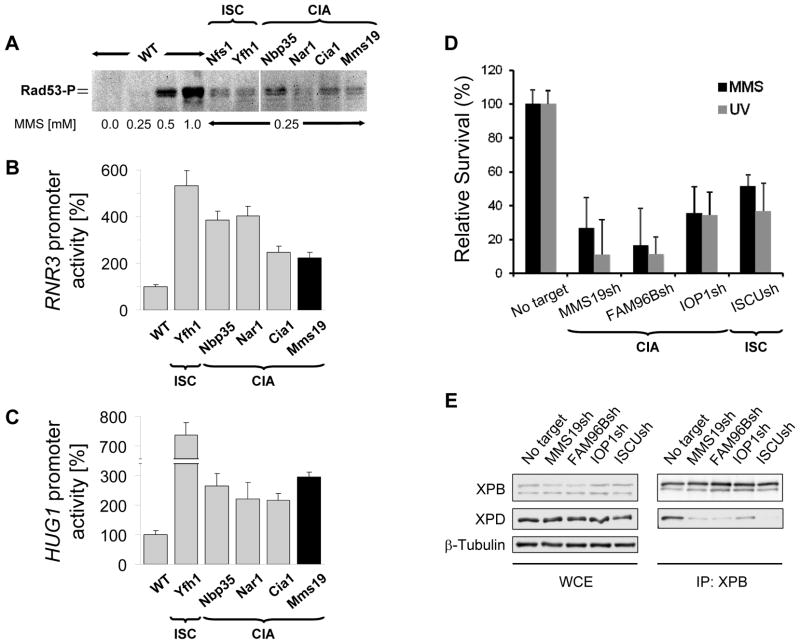
The association of MMS19 with multiple DNA replication and repair proteins led us to examine if the integrity of Fe/S protein biogenesis might be a general requirement for efficient cellular DNA damage repair. We first tested this idea in yeast and assessed whether depleting ISC or CIA components elicited effects on the DNA damage response pathway. Rad53 phosphorylation was observed in strains defective in Fe/S protein assembly after exposure to low levels of methyl methanesulfonate (MMS; (36)). These MMS levels were not sufficient to trigger Rad53 phosphorylation in wild-type yeast suggesting that the former strains accumulated higher levels of DNA damage (Fig. 4A). We then analyzed the effects of Fe/S protein deficiencies on RNR3 and HUG1 expression, two genes transcriptionally induced in response to DNA damage (37). Reporter plasmids in which the RNR3 and HUG1 promoters controlled luciferase expression were introduced into different regulatable ISC and CIA strains. A 2–6-fold increase in luciferase activity was detected in ISC- and CIA-depleted strains compared to wild-type cells (Fig. 4B–C). These results indicate that inactivation of Mms19 or other Fe/S protein assembly components leads to upregulation of the DNA damage response. We finally tested whether the requirement of intact Fe/S protein assembly for DNA damage repair was conserved in humans. HEK293 cell lines expressing shRNA constructs for silencing MMS19, FAM96B, the CIA protein IOP1, or the mitochondrial scaffold protein ISCU were generated (fig. S12). Depletion of ISC or CIA factors severely diminished survival of HEK293 cells in response to UV irradiation or MMS treatment (Fig. 4D). Together, these data suggest that the previously observed DNA repair and metabolism defects of MMS19 defective cells may be the consequence of an impaired Fe/S protein biogenesis and not related to a dedicated function of MMS19 in DNA maintenance (24, 25).
Fig. 4.
Defects in Fe/S protein assembly show increased DNA damage sensitivity. (A) Activation of Mec1-dependent DNA damage pathway in yeast. Wild-type yeast (WT) and strains deficient in the indicated proteins were treated with methyl methanesulfonate (MMS). Extracts were immunostained for phospho-Rad53 (Rad53-P). (B–C) Induction of DNA damage-inducible RNR3 or HUG1. Yeast strains depleted for indicated proteins were transformed with plasmids encoding RNR3 or HUG1 promoter-regulated luciferase, and were exposed to 0.25 mM MMS. For all depletions p<0.01. (D) Depletion of indicated proteins by shRNAs in HEK293 cells results in increased sensitivity to UV- and MMS-induced DNA damage. Stable knockdown cell lines were treated with UV (20 J/m2) or 20 μM MMS. Cell viability relative to the corresponding untreated cell line (No target) was measured using the MTS assay 7 days after treatment. Experiments are plotted as mean ± SEM (n=3). For all depletions p<0.05. (E) Fe/S protein biogenesis is required for incorporation of XPD into TFIIH. HEK293 cell extracts (WCE) from (D) were used for immunoprecipitation with XPB antibodies (IP:XPB) followed by immunoblotting for indicated proteins.
The increased DNA damage sensitivity in cells with impaired Fe/S protein biogenesis may include the loss of nucleotide excision repair because maturation of XPD is defective. Since the Fe/S cluster of XPD is required for its DNA helicase activity in vitro (7), we investigated whether inactive apo-XPD is integrated into TFIIH complexes in ISC- or CIA-depleted cells. XPD could be detected in TFIIH immunoprecipitated with endogenous XPB from control cells but not from cells depleted of MMS19, FAM96B, IOP1, or ISCU suggesting that the inability to assemble Fe/S clusters on XPD prevented its incorporation into TFIIH complexes (Fig. 4E). The inability of cells impaired in Fe/S protein biogenesis to form functional TFIIH complexes may provide, at least in part, the mechanistic basis for the increased sensitivity of MMS19 mutants to UV- and MMS-induced DNA damage. Other Fe/S cluster-containing substrates of the CIA targeting complex such as DNA2, FANCJ and RTEL1 play key roles in maintaining genome stability and the response to other types of DNA damage (i.e. DNA double-strand breaks and DNA interstrand crosslinks; (3)). Hence, inactivation of Fe/S protein biogenesis may promote genomic instability by simultaneous inactivation of multiple DNA repair pathways.
Our study clarifies the functional role of MMS19 in DNA maintenance and provides novel insights into the mechanism of cytosolic Fe/S protein biogenesis (11, 13). MMS19 exerts its function as part of a CIA targeting complex involved in the maturation of a subset of Fe/S proteins including those with functions in DNA replication, DNA repair, and telomere stability (fig. S13). By undergoing direct interaction with target Fe/S proteins, MMS19 (and its putative functional partners CIAO1 and FAM96B) may serve an adapter function between Fe/S cluster synthesis and insertion into apoproteins. This conserved function of MMS19 can explain many, if not all, of the previously described phenotypes associated with MMS19 defects. Mitochondria perform an essential role in cellular Fe/S protein biogenesis and, as shown here, in nuclear DNA metabolism. These functions, and not ATP production, may explain the maintenance of these endosymbiontic organelles even in anaerobic eukaryotes (38). Moreover, the crucial role of mitochondria in DNA metabolism and genome maintenance may be relevant to neurodegenerative phenotypes associated with mitochondrial diseases including Friedreich’s ataxia.
Acknowledgments
We thank D.R. Dean and D.E. Gottschling for fruitful discussion, C. Doré, S.A. Freibert, M. Funke, G. Köpf, B. Niggemeyer, R. Rösser, M. Stümpfig, and H. Webert for experimental help. Our work was generously supported by funds of Deutsche Forschungsgemeinschaft (SFB 593, Gottfried-Wilhelm Leibniz program, and GRK 1216), von Behring-Röntgen Stiftung, Max-Planck Gesellschaft, Feldberg Foundation, Fonds der chemischen Industrie (all to RL), Rhön Klinikum AG (AJP and RL), National Institute of Health GM089778 (JAW), University of California Cancer Research Coordinating Committee (JAW), and the Jonsson Cancer Center at UCLA (JAW).
References
- 1.McMurray MA, Gottschling DE. Aging and genetic instability in yeast. Current opinion in microbiology. 2004;7:673. doi: 10.1016/j.mib.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Hoeijmakers JH. DNA damage, aging, and cancer. The New England journal of medicine. 2009;361:1475. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 3.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40:179. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karthikeyan G, Lewis LK, Resnick MA. The mitochondrial protein frataxin prevents nuclear damage. Hum Mol Genet. 2002;11:1351. doi: 10.1093/hmg/11.11.1351. [DOI] [PubMed] [Google Scholar]
- 6.Thierbach R, et al. The Friedreich’s ataxia protein frataxin modulates DNA base excision repair in prokaryotes and mammals. Biochem J. 2010;432:165. doi: 10.1042/BJ20101116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudolf J, Makrantoni V, Ingledew WJ, Stark MJ, White MF. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol Cell. 2006;23:801. doi: 10.1016/j.molcel.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 8.Klinge S, Hirst J, Maman JD, Krude T, Pellegrini L. An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat Struct Mol Biol. 2007;14:875. doi: 10.1038/nsmb1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeeles JT, Cammack R, Dillingham MS. An iron-sulfur cluster is essential for the binding of broken DNA by AddAB-type helicase-nucleases. J Biol Chem. 2009;284:7746. doi: 10.1074/jbc.M808526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Netz DJ, et al. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat Chem Biol. 2012;8:125. doi: 10.1038/nchembio.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheftel A, Stehling O, Lill R. Iron-sulfur proteins in health and disease. Trends Endocrinol Metab. 2010;21:302. doi: 10.1016/j.tem.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 12.White MF, Dillingham MS. Iron-sulphur clusters in nucleic acid processing enzymes. Current opinion in structural biology. 2011 doi: 10.1016/j.sbi.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Lill R. Function and biogenesis iron-sulphur proteins. Nature. 2009;460:831. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 14.Ye H, Rouault TA. Human iron-sulfur cluster assembly, cellular iron homeostasis, and disease. Biochemistry. 2010;49:4945. doi: 10.1021/bi1004798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy A, Solodovnikova N, Nicholson T, Antholine W, Walden WE. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 2003;22:4826. doi: 10.1093/emboj/cdg455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stehling O, et al. Human Nbp35 is essential for both cytosolic iron-sulfur protein assembly and iron homeostasis. Molecular and cellular biology. 2008;28:5517. doi: 10.1128/MCB.00545-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mühlenhoff U, et al. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 2010;12:373. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Netz DJ, et al. Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat Chem Biol. 2010;6:758. doi: 10.1038/nchembio.432. [DOI] [PubMed] [Google Scholar]
- 19.Balk J, Aguilar Netz DJ, Tepper K, Pierik AJ, Lill R. The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol Cell Biol. 2005;25:10833. doi: 10.1128/MCB.25.24.10833-10841.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balk J, Pierik AJ, Aguilar Netz D, Mühlenhoff U, Lill R. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 2004;23:2105. doi: 10.1038/sj.emboj.7600216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song D, Lee FS. A role for IOP1 in mammalian cytosolic iron-sulfur protein biogenesis. J Biol Chem. 2008;283:9231. doi: 10.1074/jbc.M708077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prakash L, Prakash S. Three additional genes involved in pyrimidine dimer removal in Saccharomyces cerevisiae: RAD7, RAD14 and MMS19. Molecular & general genetics: MGG. 1979;176:351. doi: 10.1007/BF00333097. [DOI] [PubMed] [Google Scholar]
- 23.Askree SH, et al. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci U S A. 2004;101:8658. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito S, et al. MMXD, a TFIIH-independent XPD-MMS19 protein complex involved in chromosome segregation. Molecular cell. 2010;39:632. doi: 10.1016/j.molcel.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 25.Kou H, Zhou Y, Gorospe RM, Wang Z. Mms19 protein functions in nucleotide excision repair by sustaining an adequate cellular concentration of the TFIIH component Rad3. Proc Natl Acad Sci U S A. 2008;105:15714. doi: 10.1073/pnas.0710736105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 27.Ben-Aroya S, et al. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. . Mol Cell. 2008;30:248. doi: 10.1016/j.molcel.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.See supporting material on Science Online.
- 30.Pierik AJ, Netz DJA, Lill R. Analysis of iron-sulfur protein maturation in eukaryotes. Nat Protoc. 2009;4:753. doi: 10.1038/nprot.2009.39. [DOI] [PubMed] [Google Scholar]
- 31.Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475:244. doi: 10.1038/nature10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seroz T, et al. Cloning of a human homolog of the yeast nucleotide excision repair gene MMS19 and interaction with transcription repair factor TFIIH via the XPB and XPD helicases. Nucleic acids research. 2000;28:4506. doi: 10.1093/nar/28.22.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stehling O, et al. In: Methods in Molecular Biology. Leister D, Herrmann JM, editors. Vol. 372. Humana Press Inc; Totowa, NJ: 2007. pp. 325–342. [DOI] [PubMed] [Google Scholar]
- 34.Schnackerz KD, Dobritzsch D, Lindqvist Y, Cook PF. Dihydropyrimidine dehydrogenase: a flavoprotein with four iron-sulfur clusters. Biochim Biophys Acta. 2004;1701:61. doi: 10.1016/j.bbapap.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 35.Vashisht AA, et al. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science. 2009;326:718. doi: 10.1126/science.1176333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pellicioli A, et al. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. The EMBO journal. 1999;18:6561. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benton MG, Glasser NR, Palecek SP. The utilization of a Saccharomyces cerevisiae HUG1P-GFP promoter-reporter construct for the selective detection of DNA damage. Mutation research. 2007;633:21. doi: 10.1016/j.mrgentox.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Goldberg AV, et al. Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature. 2008;452:624. doi: 10.1038/nature06606. [DOI] [PubMed] [Google Scholar]