Abstract
An extensive analysis of dental plaque samples over the years has led to the identification of “red” complex oral bacteria that have a strong association with each other and with disease. Consequently, these bacteria have been labeled ‘periopathogens’. Studies with one of these bacteria, Porphyromonas gingivalis, have revealed that it contains several different mechanisms which either impede or modulate periodontal protective mechanisms. In a mouse model of periodontitis, it has been shown that modulation of complement function by P. gingivalis facilitates a significant change in both the amount and composition of the normal oral microbiotia. This altered oral commensal microbiota is responsible for pathologic bone loss in the mouse. Thus, P. gingivalis creates a dysbiosis between the host and dental plaque, and this may represent one mechanism by which periodontitis can be initiated. We have therefore termed P. gingivalis a keystone pathogen.
Keywords: bacteria, innate immunity, microbial ecology, microbiology, periodontal disease(s), periodontitis
Periodontitis has a rich history of proposed microbial etiologies, eloquently described by Socransky (Socransky and Haffajee, 1994). The varied hypotheses presented over the years have implicated possible etiological agents drawn from almost the complete range of the animal kingdom. This history underscores the difficulty in understanding the complex interactions between diverse microbial communities and the host. However, because of the relative ease and non-invasive nature of sampling the oral cavity, it has been possible to conduct comprehensive analyses of the oral microbiota in both health and disease (Socransky et al., 1998). Among other findings, these studies led to the identification of a “red” complex: 3 species of oral bacteria – Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola – whose detection was strongly associated with each other and with diseased sites. This landmark work and other contemporaneous studies (e.g., Curtis et al., 2011) naturally led to the investigation of potential virulence factors for these bacteria, for a fuller understanding of their association with disease. Since P. gingivalis has long been associated with periodontal disease, has a well-characterized population structure, and, critically, is the easiest of these 3 bacteria to grow and genetically manipulate, it consequently became the most well-studied (Lamont and Jenkinson, 1998; Curtis et al., 2001).
However, studies with P. gingivalis presented an apparent paradox in which a bacterium that was strongly associated with an inflammatory disease was not a potent inducer of inflammation. For example, the lipopolysaccharide (LPS) of P. gingivalis revealed an unusually low inflammatory potency, and, in fact, a P. gingivalis lipid A structure acting as TLR4 antagonist that inhibits inflammation has been discovered and characterized (Darveau et al., 1995). These surprising results stand in contrast to the well-characterized, highly inflammatory LPS obtained from Escherichia coli and many other Gram-negative bacteria (Munford and Varley, 2006). Furthermore, P. gingivalis was unusual in that it did not induce IL-8 secretion by gingival epithelial cells, unlike a variety of other oral bacteria and, in fact, inhibited the secretion of this potent chemokine for neutrophil recruitment (Darveau et al., 1998). This phenomenon was termed ‘local chemokine paralysis’ and reinforced the emerging view that P. gingivalis did not demonstrate characteristics normally associated with a bacterium contributing to an inflammatory disease. More recently, continued studies with P. gingivalis have revealed that it is an excellent immune manipulator, in that it is able to selectively induce only a limited repertoire of inflammatory responses from leukocytes through receptor cross-talk mechanisms (Hajishengallis et al., 2008; Liang et al., 2011). Specifically, it can inhibit leukocyte-mediated bacteria-killing mechanisms (Wang et al., 2010). These apparent paradoxes can be explained, however, if periodontitis is viewed as a community disease reliant upon an entire dysfunctional microbiota, as opposed to the traditional view of a conventional infectious disease caused by a single or even multiple select periopathogens (Hajishengallis et al., 2011).
Initially, evidence that the commensal microbial community may significantly contribute to periodontitis was obtained when the disease experience of germ-free (GF) mice, which are not colonized with any bacteria, was compared with that of normal specific-pathogen-free (SPF) mice in the well-established P. gingivalis gavage model (Baker et al., 2000). Although both SPF and GF mice were colonized in the oral cavity with P. gingivalis to the same extent, only SPF mice developed bone loss. Remarkably, though only approximately 100 P. gingivalis cells per maxilla (ranges from 50-350, representing < 0.01% of the total bacterial cells)/SPF mouse could be detected on the basis of quantitative real-time PCR, there was a very significant change in the commensal microbiota. First, the total microbial load measured on the basis of colony-forming units in oral swabs of P. gingivalis-infected SPF mice rose by approximately 2 log10 units. Second, there was a qualitative shift in the population structure of this commensal microbiota, leading to loss of detection of some organisms and the appearance of others. The demonstration that P. gingivalis colonized both GF and SPF mice, yet only SPF mice developed bone loss, coupled with the significant changes in the oral commensal community indicated that P. gingivalis induced dysbiosis, or a microbial shift in the commensal composition. Furthermore, the lack of disease in GF mice indicated that the commensal bacteria themselves are necessary for and directly contribute to the bone loss observed in this model.
Two independent experimental approaches confirmed that oral commensal bacteria directly contribute to bone loss in the mouse. Initially, it was observed that GF mice have significantly more alveolar bone when compared with strain- and age-matched SPF mice. These measurements, which determine the gap between the alveolar crest and the cement-enamel junction, were less in GF mice. The first experimental evidence that commensals cause bone loss was the demonstration that as the SPF, but not the GF, mouse ages, the distance between the alveolar crest and the cement-enamel junction increases. This “natural” bone loss was associated with an increase in numerous inflammatory mediators in the oral tissues of SPF compared with those of GF mice, indicating that commensals naturally stimulate periodontal tissue, and this results in some non-pathologic bone loss. The second approach demonstrated that the acquisition of oral commensals by GF that have been co-caged with SPF mice resulted in bone loss. In these experiments, it was determined that after 2 wks of co-caging, GF mice had acquired an oral microbiota identical to that of SPF mice, and after 16 wks, the GF mice had lost bone to levels similar to those of their strain-matched SPF cage-mates. Analysis of these data demonstrated that bone loss in the mouse is a natural result of commensal colonization and represents a manifestation of the homeostatic relationship between the host and its oral microbial community.
Next, it was found that the natural bone loss induced by commensal colonization required complement. SPF mice deficient in either the C3a or C5a receptor (C3aR-/- or C5aR-/-) appeared similar to the GF mice in that they had more alveolar bone than their strain- and age-matched wild-type controls. Furthermore, it was found that one mechanism by which P. gingivalis induces accelerated bone loss in this mouse model was by disrupting the homeostatic relationship between the commensal oral microbiota and the complement system. C3aR-/- or C5aR-/- mice neither lost bone nor displayed a significant change in the number of commensal oral bacteria after gavage with P. gingivalis. Consistent with earlier observations that P. gingivalis can manipulate the host complement system through the gingipain-dependent proteolytic cleavage of complement and subsequent cross-talk on the leukocyte cell surface (Liang et al., 2011), a gingipain-deficient P. gingivalis strain (rgpA-/-, rgpB-/-, kgp-/-) also failed to induce bone loss or increase the oral commensal microbiota. Therefore, it appears that P. gingivalis can modulate the commensal-host homeostasis dialogue by altering complement function. This modulation increases inflammation and bone destruction. It was estimated that the bone loss seen in young SPF mice after 6-week colonization by P. gingivalis is equivalent to the bone loss seen in untreated SPF mice (i.e., not inoculated with P. gingivalis) at 18 mos of age.
Finally, these studies (Hajishengallis et al., 2011) demonstrated that P. gingivalis was not necessary to induce bone loss when immune deficiencies in the host altered the homeostatic relationship with the oral commensal microbiota. For example, mice deficient in neutrophil homing to the junctional epithelium, because of either the lack of the neutrophil leukocyte integrin, LFA-1, or the absence of the chemokine receptor, CXCR2, displayed an increase in the oral commensal microbiota and developed significant bone loss when compared with their wild-type control strains. In fact, the bone loss in LFA-1-/- mice was correlated with an increase in the numbers of oral commensal bacteria in these knock-out animals, and this bone loss did not occur when antibiotics were administered. This is similar to a much earlier publication in P- and E-selectin double-knockout mice which showed the same effect of an increased oral microbial load and increased bone loss, compared with wild-type mice, which was abrogated upon the administration of antibiotics (Niederman et al., 2001). Analysis of these data demonstrated that disruption of periodontal homeostasis by several different mechanisms can result in bone loss, but the contribution of the commensal microbiota appears to be a common factor.
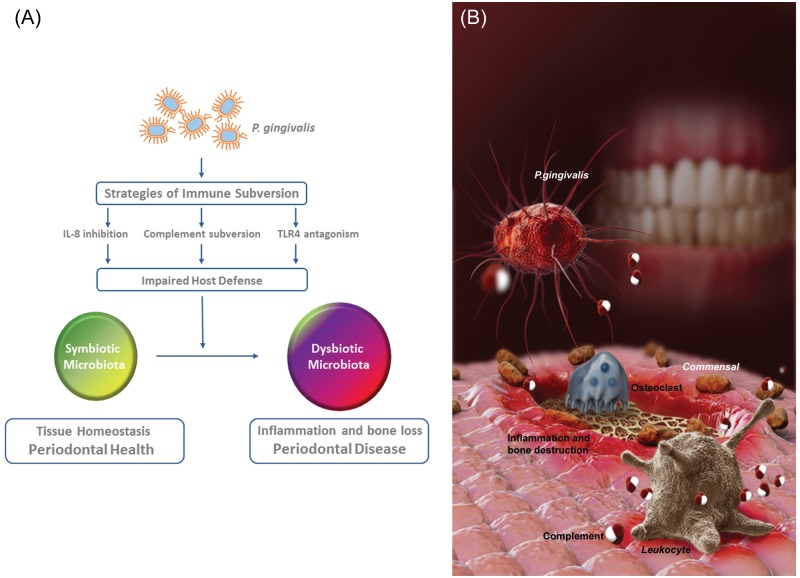
Based upon the fact that P. gingivalis was present in low abundance yet had such a profound effect on both the amount and composition of the oral microbiota, leading to periodontitis, we have designated this bacterium a “keystone” species. The concept of a keystone species derives from ecological studies and is defined as a species that is present in low abundance yet provides a major supporting role for an entire ecological community (Paine, 1969; Power et al., 1996). We were able to demonstrate that the continued presence of low numbers of P. gingivalis was required for the significant increase in mouse oral microbiota which was associated with disease. Moreover, P. gingivalis could be viewed as a ‘keystone pathogen’, that is, a keystone species which supports and remodels a microbial community in ways that also promote disease pathogenesis (Fig.).
Figure.
The red complex bacterium P. gingivalis causes inflammation and bone loss by remodeling the oral commensal microbiota. (A) Studies have shown that P. gingivalis modulates innate host defense functions that can have global effects on the oral commensal community. Immune subversion of IL-8 secretion, complement activity, or TLR4 activation can result in an impaired host defense. The inability of the host to control the oral commensal microbial community in turn results in an altered oral microbial composition and an increased microbial load. This alteration from a symbiotic to a dysbiotic microbiota is responsible for pathologic inflammation and bone loss. (B) P. gingivalis, a low-abundance oral anaerobic bacterium (shown as a fimbriated reddish rod), exploits complement (depicted in red and white) and subverts leukocytes (in gray), leading to alterations in the amount and composition of the oral commensal microbiota (brown rods). Collectively, these changes disrupt host homeostasis and lead to destructive inflammatory periodontitis (indicated by the reddened/inflamed tissue and osteoclast-mediated bone erosion). The disease requires the presence of the commensal microbiota and intact complement pathways, since P. gingivalis fails to cause periodontitis in germ-free mice or in conventionally raised mice deficient in the complement anaphylatoxin receptors.
Although P. gingivalis is not a natural periopathogen in mice, several observations indicate that P. gingivalis may act as a keystone pathogen in human disease. First, the mouse model is similar to humans in that a significant increase in the total oral microbial load (including oral commensals) is observed in disease when compared with health (Darveau et al., 1997). Second, the innate defense status, including complement and neutrophil transit, is similar in mice and humans (Page and Schroeder, 1982), allowing for the investigation of periopathogen effects on the innate defense system. Third, consistent with the keystone pathogen hypothesis in mice (Hajishengallis et al., 2011), in that oral commensal bacteria induce destructive inflammation, studies with human dental plaque obtained from either healthy or diseased sites have revealed that they are both potent inducers of inflammation through either TLR2 or TLR4 (Yoshioka et al., 2008). This observation demonstrates that human oral commensal communities in both health and disease have a similar potential to induce inflammation. Moreover, the specific targeting of P. gingivalis adversely affects the total subgingival bacterial load even in hosts (non-human primates) where P. gingivalis is a natural inhabitant of the periodontal biofilm (Page et al., 2007).
Furthermore, consistent with a keystone contribution of P. gingivalis to disease, studies in humans have shown that P. gingivalis is present in low abundance when compared with the total oral microbiota in diseased sites (Kumar et al., 2006). Furthermore, both non-human primate (Page et al., 2007) and rabbit (Hasturk et al., 2007) animal models of periodontitis have shown that the introduction of P. gingivalis significantly increases the total oral microbial load and alters its composition, leading to dysbiosis (Hasturk et al., 2007). In addition, selective removal of P. gingivalis by the addition of a C5aR antagonist (Hajishengallis et al., 2011) or by immunization in non-human primates (Page et al., 2007) subsequently reduced the microbial load and attenuated bone loss. These data are all consistent with P. gingivalis being a keystone species in the dysbiosis associated with periodontitis.
Nevertheless, it is not clear why P. gingivalis, which may also be found in the periodontal microbiota of healthy individuals, is not routinely associated with disease. The most likely explanations, which are not mutually exclusive, involve changes in either the bacterium or the host. For example, the pathogenicity of P. gingivalis may depend upon strain and virulence diversity within the population structure of this bacterium, which in certain cases may have co-evolved with its host in ways that allow for its co-existence and persistence without detrimental effects on host tissue (Yilmaz, 2008). It was further posited that local changes in the environment may affect this delicate relationship. For example, P. gingivalis protease production is regulated by local environmental conditions (Curtis et al., 2001), and conditions which promote its secretion may result in the modulation of complement activity, as we have described in the mouse (Hajishengallis et al., 2011). Conversely, changes in the status of the host may also result in an increased ability of P. gingivalis to act as a keystone species. We have recently shown that aging adversely affects the ability of the host to regulate neutrophil recruitment and inflammation (Eskan et al., 2012), and these environmental changes in the periodontium may facilitate the ability of P. gingivalis to capitalize on its keystone phenotype. Moreover, it is possible that certain individuals can either resist or tolerate the conversion of the microbiota from a symbiotic to a dysbiotic one, by virtue of their intrinsic immuno-inflammatory status (e.g., hyporesponsive or lack-of-function polymorphisms that attenuate inflammation or microbial immune subversion). Indeed, there are clinical cases of individuals who are periodontally healthy despite massive accumulation of dental plaque at dento-gingival sites.
In summary, we have shown that, in a validated animal model of periodontitis, P. gingivalis contributes to disease in an indirect fashion. Rather than a direct assault on host periodontal tissue, its presence, even at low abundance, alters the total commensal microbial load and composition, which overwhelms normal host-tissue-protective mechanisms and results in disease. Ingeniously, P. gingivalis accomplishes this significant microbial change by inhibiting key features of the normal host-protective mechanisms in the periodontium. Importantly, the P. gingivalis-induced dysbiotic effects can be reversed by pharmacologic blockade of the complement receptors that P. gingivalis exploits. It should be noted, however, that complement is one target of P. gingivalis action that does not necessarily exclude other innate host defense functions whose manipulation may facilitate significant changes in the oral microbial community. Moreover, other putative periopathogens of the oral microbial community may also disrupt the homeostatic relationship between dental plaque and the host. In this regard, SPF mice with defective leukocyte recruitment (CXCR2KO and LFA-1KO) display dramatic bone loss relative to age-matched wild-type controls (Hajishengallis et al., 2011). Consequently, P. gingivalis or other bacteria that could interfere with mechanisms of leukocyte trafficking to the periodontium could thereby contribute to periodontal disease pathogenesis. The fact that healthy mice show modest bone loss (by comparison with germ-free mice; Hajishengallis et al., 2011) demonstrates that commensal bacteria have multiple opportunities to interfere with the multitude of tissue functions that maintain periodontal structure and function, especially when environmental or host genetic factors are not favorable for maintaining homeostasis. Interference with innate host protection is one mechanism; others could include modulation of collagen deposition, epithelial cell proliferation, and other tissue homeostatic mechanisms necessary for a properly functioning periodontium. Future studies will likely uncover some of these approaches used by commensal oral bacteria that have co-evolved with us, yet are not always friendly to our tissue.
Conversely, although our data are consistent with alterations in the oral commensal bacterial community being directly responsible for disease, we cannot exclude the possibility that a low-abundance pathogen is also increased in the presence of P. gingivalis and may have direct effects on host periodontal function. Furthermore, it must be considered that adult chronic periodontitis most likely has multiple etiologies, and the presence of P. gingivalis is just one of several that have yet to be elucidated. This becomes more evident when one considers that an inflammatory disease such as periodontitis fundamentally represents a disruption of tissue homeostasis; therefore, at least in principle, any factor (whether microbial or host-based) that can destabilize the homeostatic balance with the periodontal microbiota can contribute to the pathogenesis of periodontitis.
Footnotes
This work was supported by grants from the NIH (DE015254, DE018292, and DE021580 to G.H.; DE18274 to R.P.D.) and the Medical Research Council (UK; G0900408 to M.A.C.).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Baker PJ, Dixon M, Roopenian DC. (2000). Genetic control of susceptibility to Porphyromonas gingivalis-induced alveolar bone loss in mice. Infect Immun 68:5864-5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MA, Aduse-Opoku J, Rangarajan M. (2001). Cysteine proteases of Porphyromonas gingivalis. Crit Rev Oral Biol Med 12:192-216 [DOI] [PubMed] [Google Scholar]
- Curtis MA, Zenobia C, Darveau RP. (2011). The relationship of the oral microbiotia to periodontal health and disease. Cell Host Microbe 10:302-306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP, Cunningham MD, Bailey T, Seachord C, Ratcliffe K, Bainbridge B, et al. (1995). Ability of bacteria associated with chronic inflammatory disease to stimulate E-selectin expression and promote neutrophil adhesion. Infect Immun 63:1311-1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP, Tanner A, Page RC. (1997). The microbial challenge in periodontitis. Periodontol 2000 14: 12-32 [DOI] [PubMed] [Google Scholar]
- Darveau RP, Belton CM, Reife RA, Lamont RJ. (1998). Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun 66:1660-1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskan MA, Jotwani R, Abe T, Chmelar J, Lim JH, Liang S, et al. (2012). The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nat Immunol 13:465-473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Wang M, Liang S, Triantafilou M, Triantafilou K. (2008). Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proc Natl Acad Sci USA 105:13532-13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, et al. (2011). Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10:497-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasturk H, Kantarci A, Goguet-Surmenian E, Blackwood A, Andry C, Serhan CN, et al. (2007). Resolvin E1 regulates inflammation at the cellular and tissue level and restores tissue homeostasis in vivo. J Immunol 179:7021-7029 [DOI] [PubMed] [Google Scholar]
- Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. (2006). Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol 44:3665-3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. (1998). Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev 62:1244-1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Krauss JL, Domon H, McIntosh ML, Hosur KB, Qu H, et al. (2011). The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J Immunol 186:869-877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munford RS, Varley AW. (2006). Shield as signal: lipopolysaccharides and the evolution of immunity to Gram-negative bacteria. PLoS Pathog 2:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederman R, Westernoff T, Lee C, Mark LL, Kawashima N, Ullman-Culler M, et al. (2001). Infection-mediated early-onset periodontal disease in P/E-selectin-deficient mice. J Clin Periodontol 28:569-575 [DOI] [PubMed] [Google Scholar]
- Page RC, Schroeder HE. (1982). Periodontics in man and other animals, a comparative review. Basel: S. Karger Publications [Google Scholar]
- Page RC, Lantz MS, Darveau R, Jeffcoat M, Mancl L, Houston L, et al. (2007). Immunization of Macaca fascicularis against experimental periodontitis using a vaccine containing cysteine proteases purified from Porphyromonas gingivalis. Oral Microbiol Immunol 22:162-168 [DOI] [PubMed] [Google Scholar]
- Paine RT. (1969). A note on trophic complexity and community stability. Am Nat 103:91-93 [Google Scholar]
- Power ME, Tilman D, Estes JA, Menge BA, Bond WJ, Mills S, et al. (1996). Challenges in the Quest for Keystones. BioScience 46:609-620 [Google Scholar]
- Socransky SS, Haffajee AD. (1994). Evidence of bacterial etiology: a historical perspective. Periodontol 2000 5: 7-25 [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. (1998). Microbial complexes in subgingival plaque. J Clin Periodontol 25:134-144 [DOI] [PubMed] [Google Scholar]
- Wang M, Krauss JL, Domon H, Hosur KB, Liang S, Magotti P, et al. (2010). Microbial hijacking of complement-Toll-like receptor crosstalk. Sci Signal 3:ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O. (2008). The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology 154(Pt 10):2897-2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H, Yoshimura A, Kaneko T, Golenbock DT, Hara Y. (2008). Analysis of the activity to induce Toll-like receptor (TLR)2- and TLR4-mediated stimulation of supragingival plaque. J Periodontol 79:920-928 [DOI] [PubMed] [Google Scholar]