Abstract
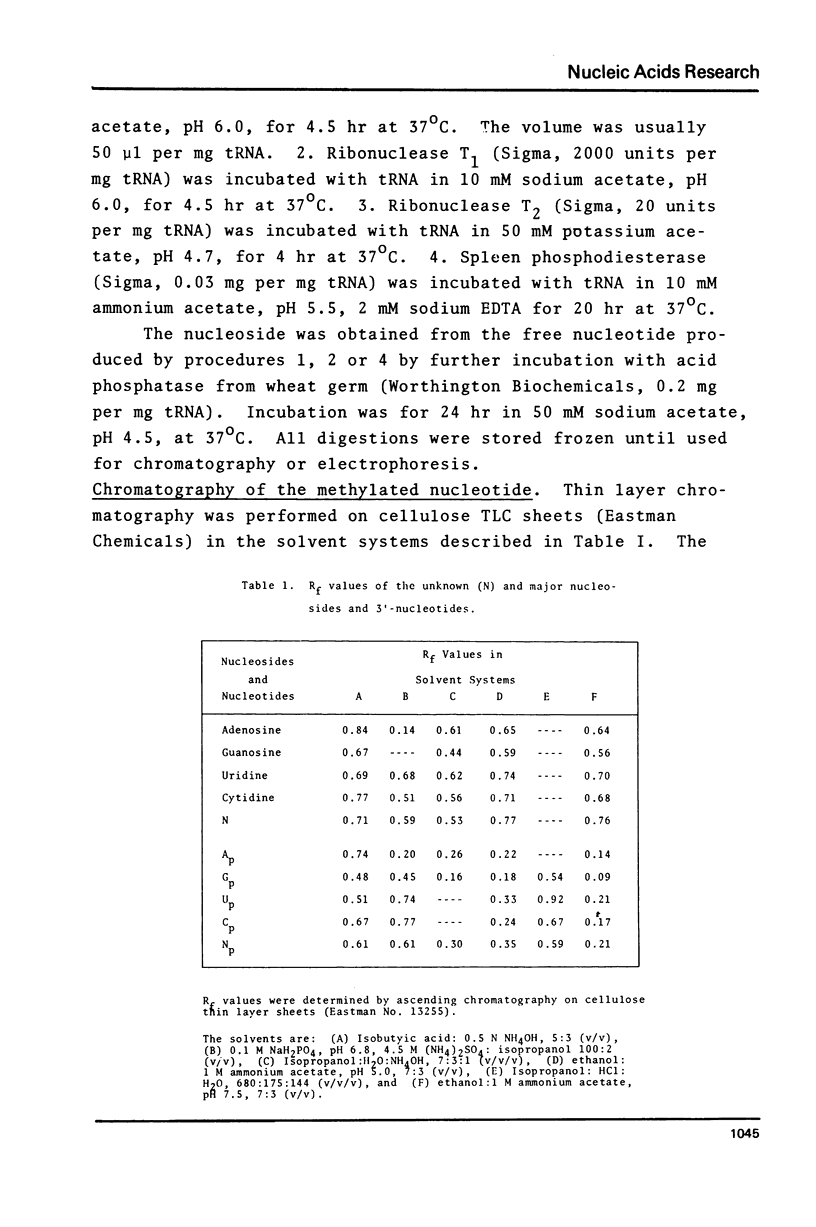
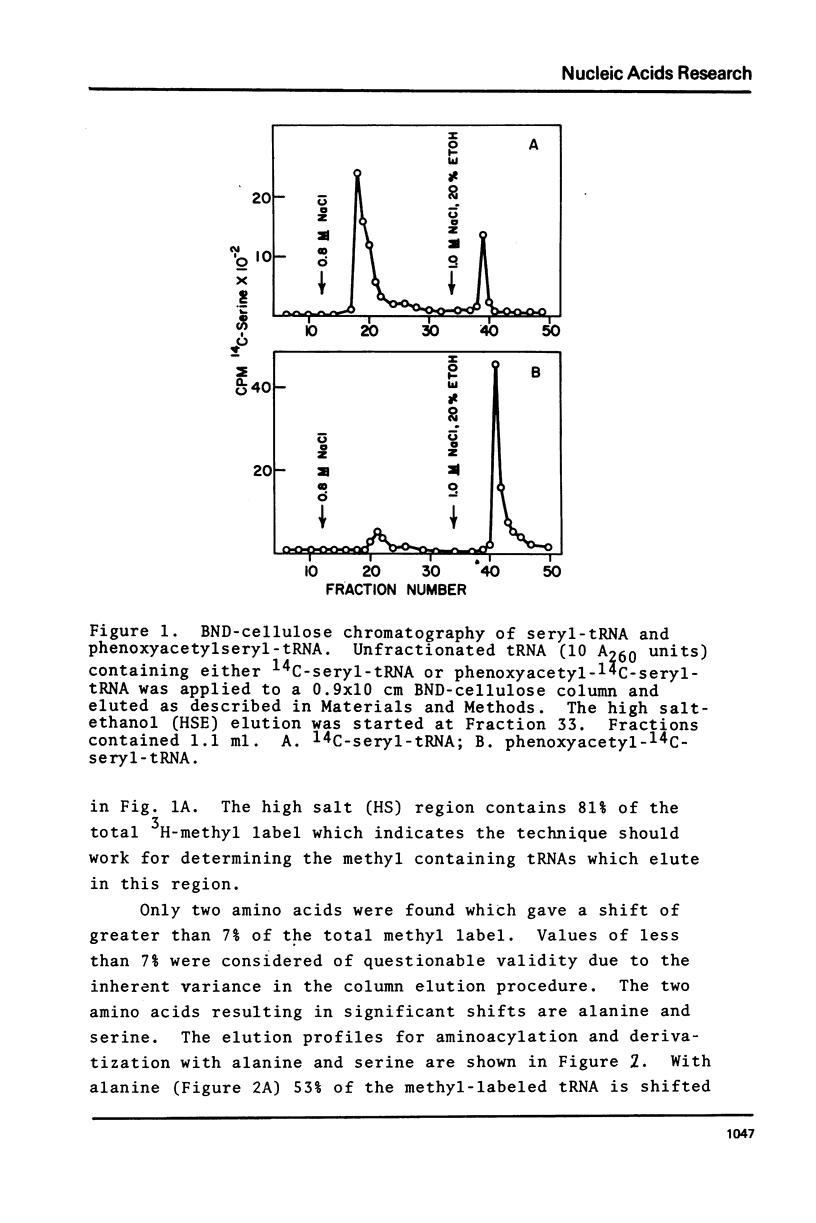
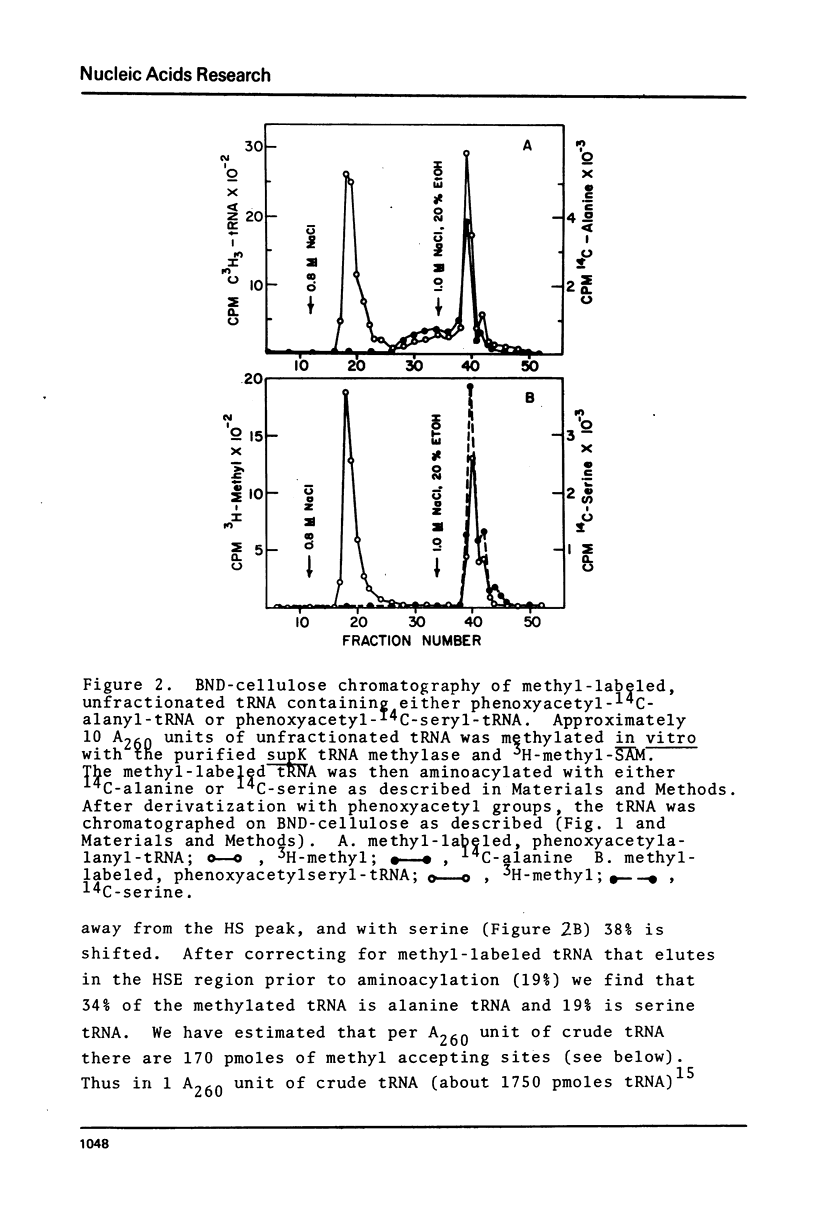
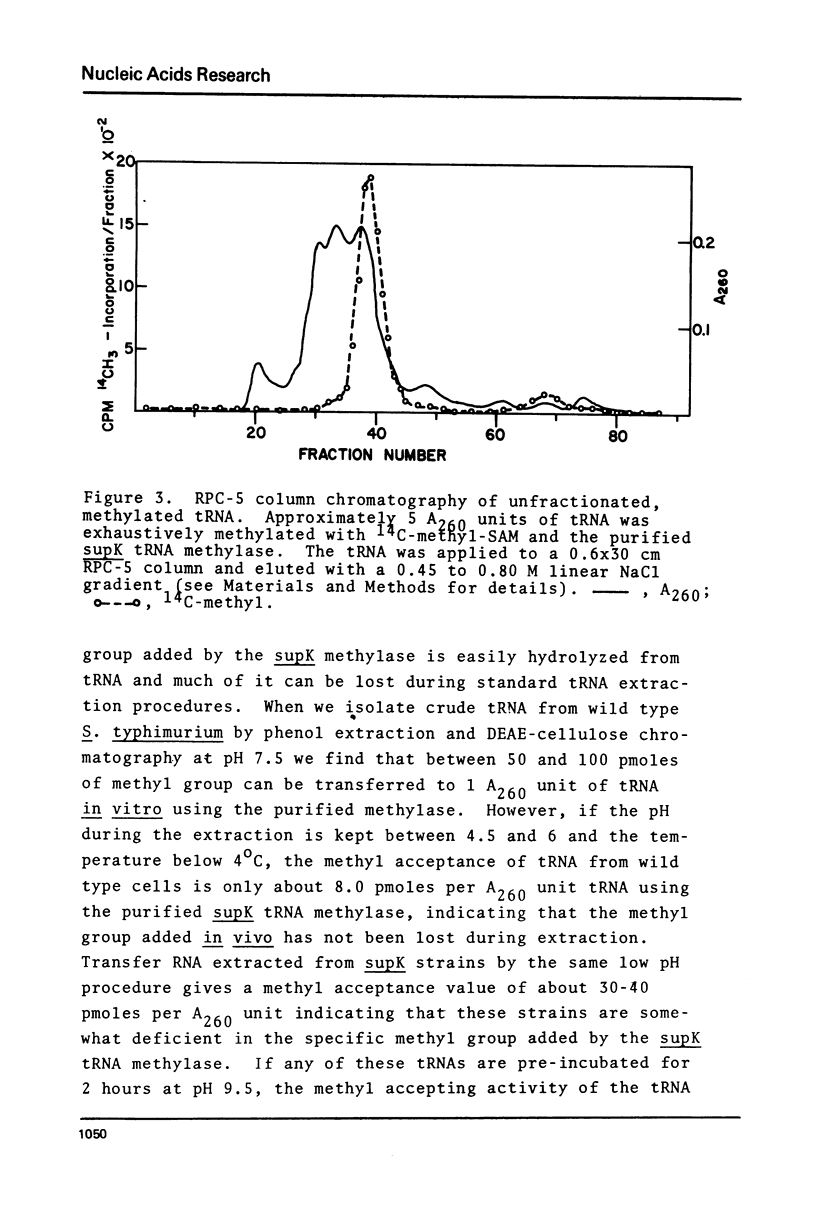
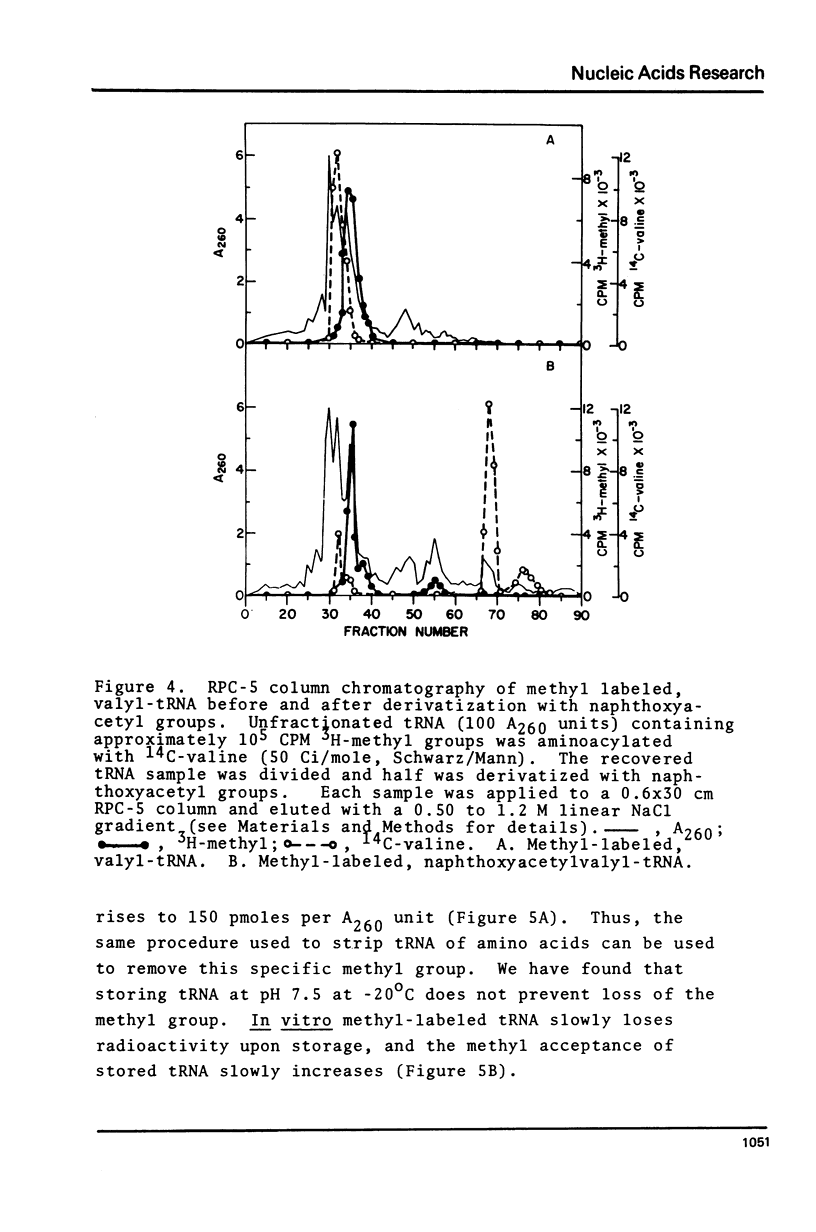
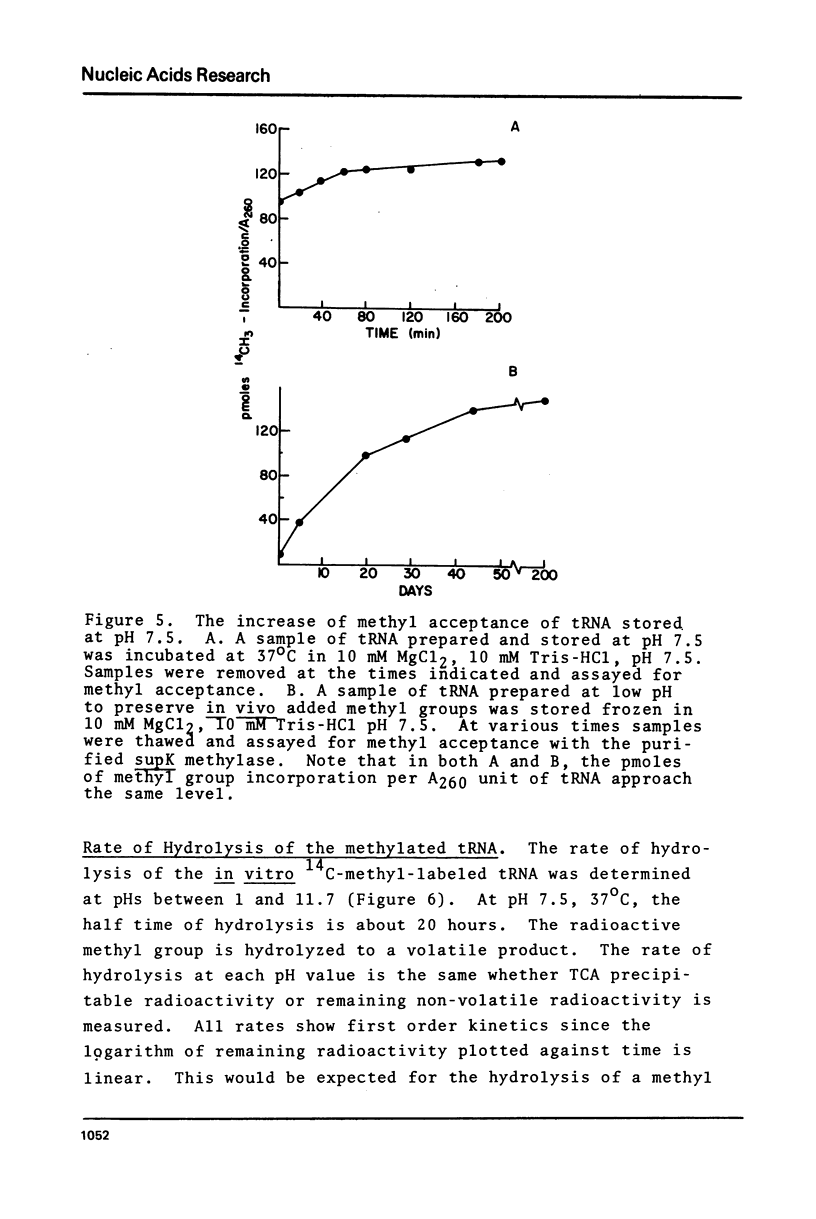
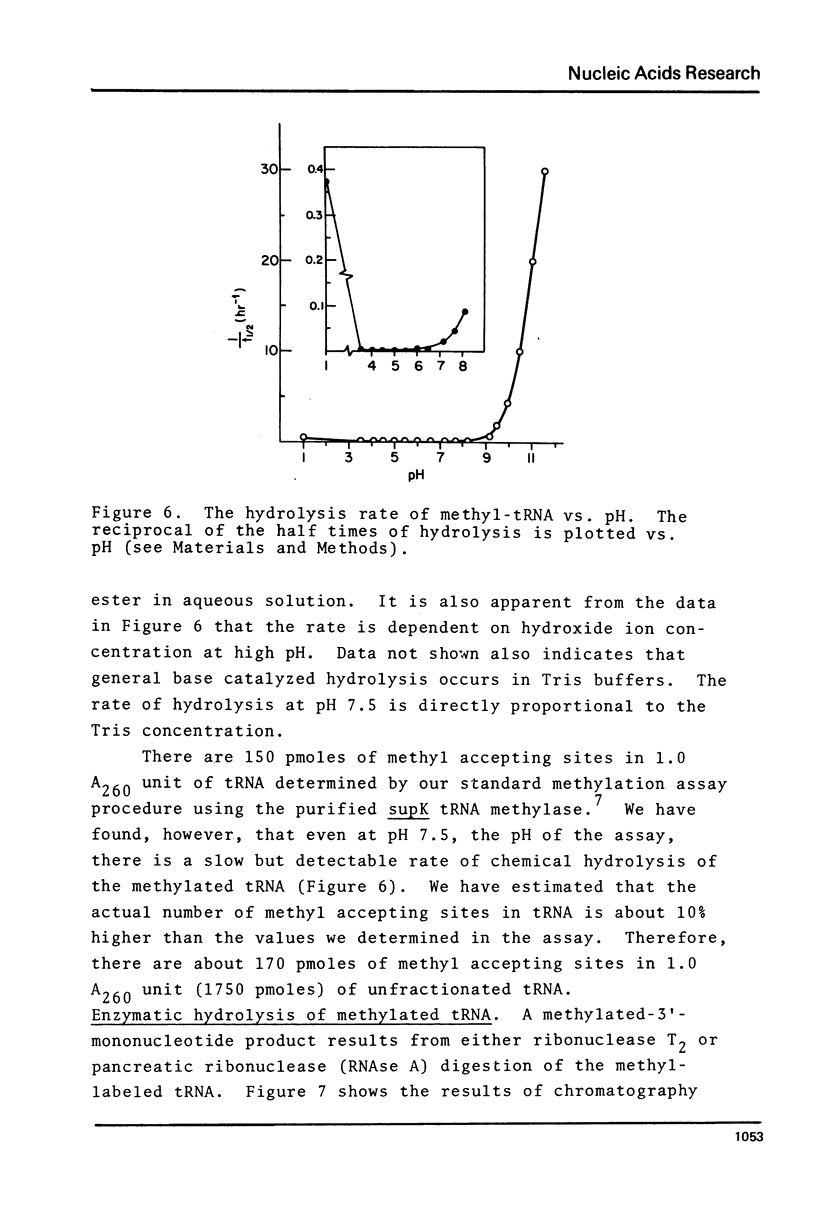
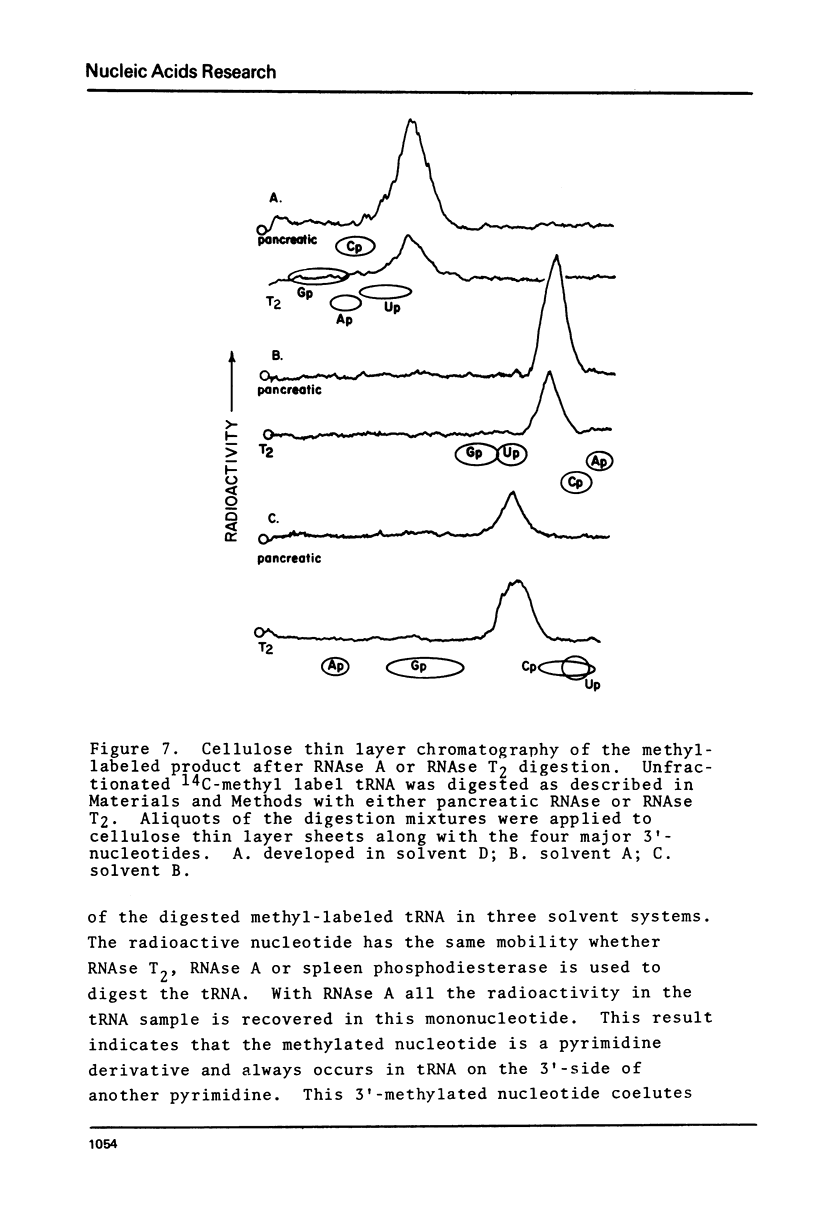
Purified preparations of the tRNA methylase deficient in supK strains of Salmonella typhimurium transfer methyl groups from S-adenosylmethionine (SAM) to at least two tRNA species, an alanine tRNA and a serine tRNA. The identity of the tRNA substrates for this enzyme was determined by a change in the elution position of the methyl-labeled tRNA from BND-cellulose columns before and after aminoacylation with a specific amino acid followed by derivatization of the free primary amino group with phenoxy- or naphthoxyacetate. The radioactive methyl group enzymatically added to these tRNAs is both acid and base labile and can be hydrolyzed to a volatile product at pHs above 7.5 and also at pH 1. The methylated 3'-nucleotide isolated from digested tRNA is a pyrimidine derivative and chromatographs like a modified uridylic acid. Its identity has not been established, but it is likely that it corresponds to the methyl ester of V, uridin-5-oxyacetic acid.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alvino C. G., Remington L., Ingram V. M. Chemical studies on amino acid acceptor ribonucleic acids. 8. Degradation of purified alanine Escherichia coli B transfer ribonucleic acid by pancreatic ribonuclease. Biochemistry. 1969 Jan;8(1):282–288. doi: 10.1021/bi00829a040. [DOI] [PubMed] [Google Scholar]
- Atkins J. F., Ryce S. UGA and non-triplet suppressor reading of the genetic code. Nature. 1974 Jun 7;249(457):527–530. doi: 10.1038/249527a0. [DOI] [PubMed] [Google Scholar]
- Gillam I., Blew D., Warrington R. C., von Tigerstrom M., Tener G. M. A general procedure for the isolation of specific transfer ribonucleic acids. Biochemistry. 1968 Oct;7(10):3459–3468. doi: 10.1021/bi00850a022. [DOI] [PubMed] [Google Scholar]
- Ishikura H., Yamada Y., Nishimura S. The nucleotide sequence of a serine tRNA from Escherichia coli. FEBS Lett. 1971 Jul 15;16(1):68–70. doi: 10.1016/0014-5793(71)80688-9. [DOI] [PubMed] [Google Scholar]
- Kelmers A. D., Heatherly D. E. Columns for rapid chromatographic separation of small amounts of tracer-labeled transfer ribonucleic acids. Anal Biochem. 1971 Dec;44(2):486–495. doi: 10.1016/0003-2697(71)90236-3. [DOI] [PubMed] [Google Scholar]
- Reeves R. H., Cantor C. R., Chambers R. W. Effect of magnesium ions on the conformation of two highly purified yeast alanine transfer ribonucleic acids. Biochemistry. 1970 Sep 29;9(20):3993–4002. doi: 10.1021/bi00822a019. [DOI] [PubMed] [Google Scholar]
- Reeves R. H., Roth J. R. A recessive UGA suppressor. J Mol Biol. 1971 Mar 28;56(3):523–533. doi: 10.1016/0022-2836(71)90399-8. [DOI] [PubMed] [Google Scholar]
- Reeves R. H., Roth J. R. Transfer ribonucleic acid methylase deficiency found in UGA supressor strains. J Bacteriol. 1975 Oct;124(1):332–340. doi: 10.1128/jb.124.1.332-340.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle D. L., Roth J. R. Frameshift suppressors. 3. Effects of suppressor mutations on transfer RNA. J Mol Biol. 1972 May 28;66(3):495–506. doi: 10.1016/0022-2836(72)90429-9. [DOI] [PubMed] [Google Scholar]
- White B. N., Tener G. M. Small analytical BD-cellulose columns for rapid chromatography of aminoacyl-tRNAs. Anal Biochem. 1973 Oct;55(2):394–398. doi: 10.1016/0003-2697(73)90128-0. [DOI] [PubMed] [Google Scholar]
- Williams R. J., Nagel W., Roe B., Dudock B. Primary structure of E. coli alanine transfer RNA: relation to the yeast phenylalanyl tRNA synthetase recognition site. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1215–1221. doi: 10.1016/0006-291x(74)90328-3. [DOI] [PubMed] [Google Scholar]


