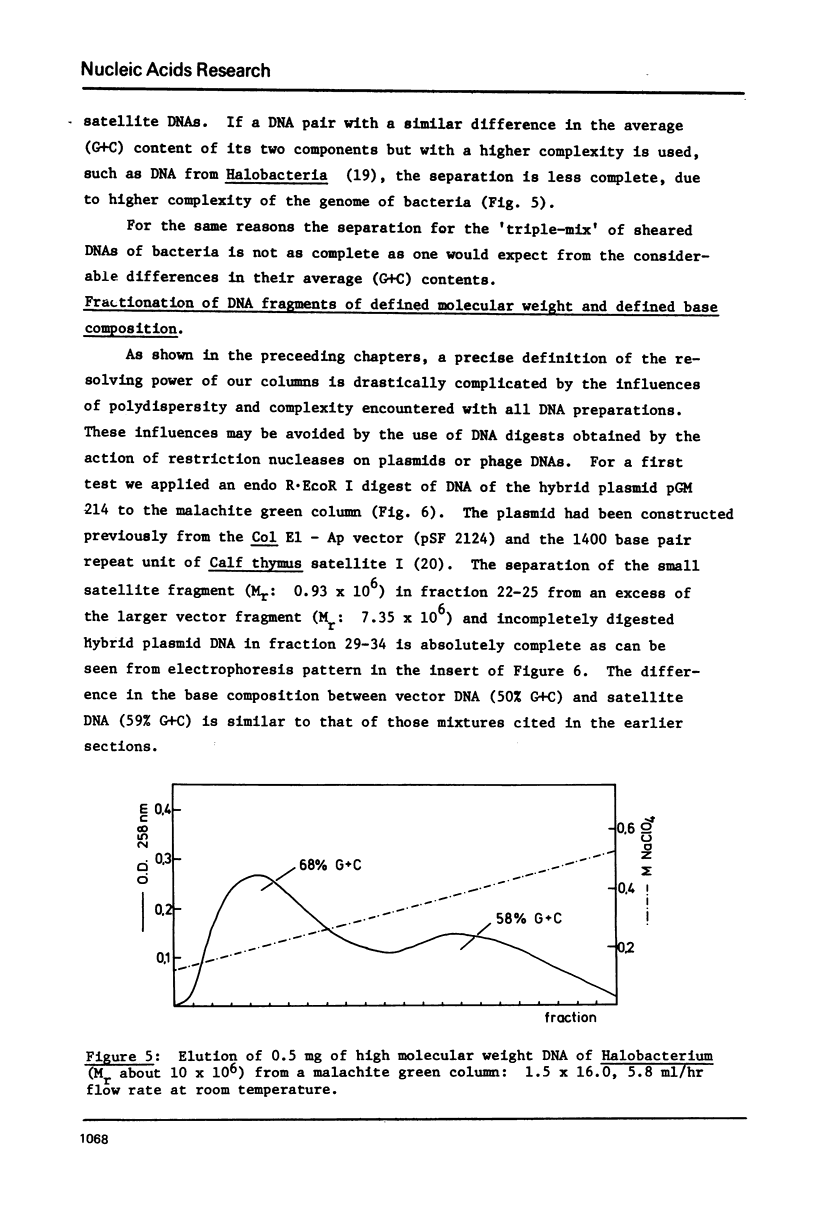
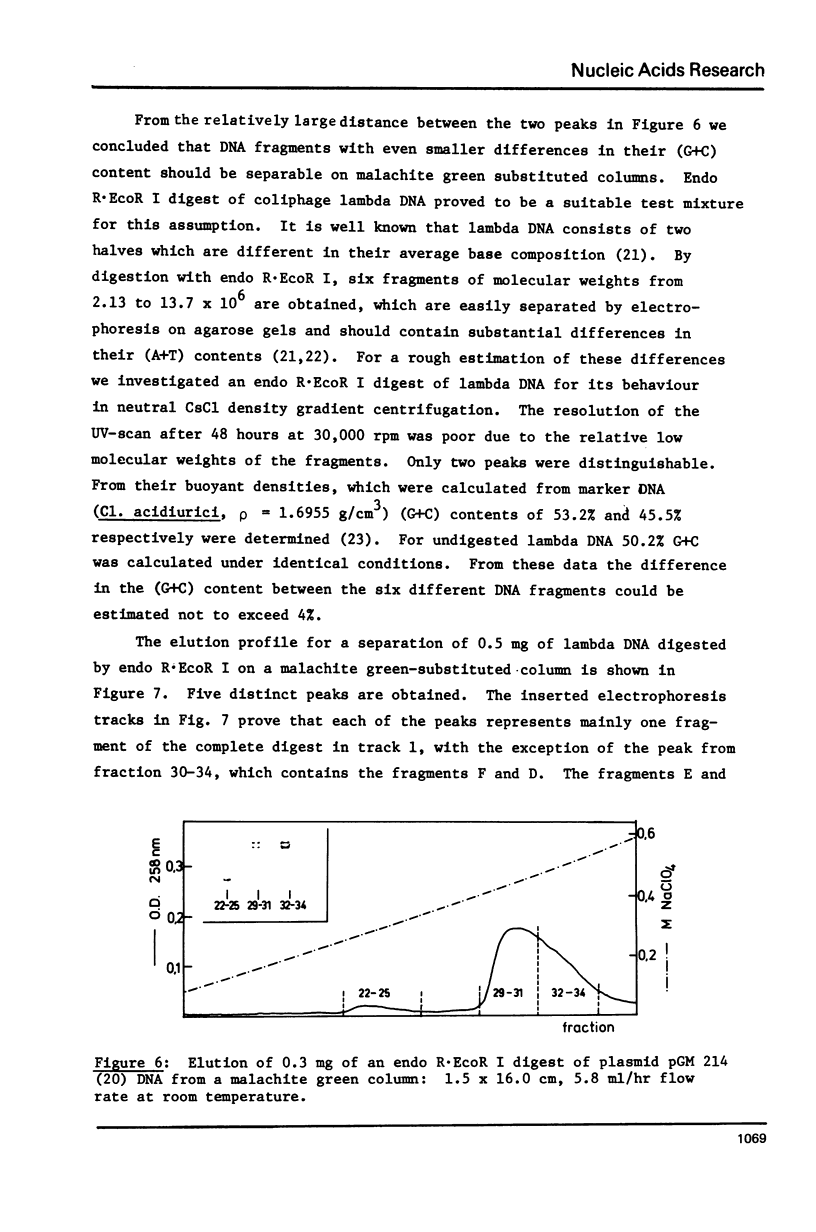
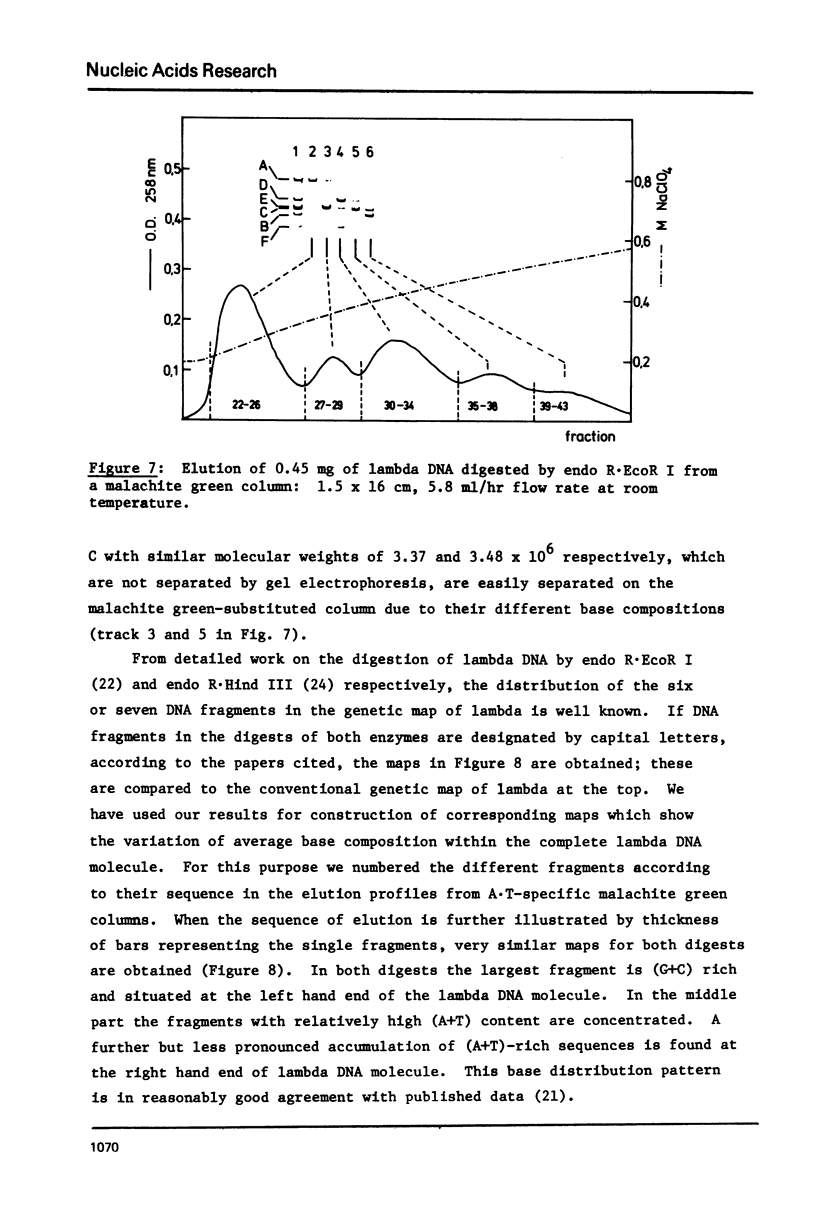
Abstract
A material suitable for base-pair-specific "affinity chromatography" of double stranded DNA is described. The synthesis of this material involves two successive polymerization reactions yielding solid particles of cross-linked bisacrylamide to which base-pair-specific dyes are covalently attached by spacers of polyacrylamide chains of different length. Materials with immobilized A.T-specific malachite green or G.C-specific phenyl neutral red were used for base-pair-specific fractionation of sheared DNA from bacterial sources, as well as of higher molecular weight Calf thymus DNA or defined fragments of coliphage lambda DNA. The excellent resolving power of this method of "affinity chromatography" of nucleic acids is comparable only to DNA fractionation in buoyant density gradients in the presence of base-pair-specific ligands. A great advantage of this material is its simple handling and its chemical stability, which allows repeated re-use of the same column.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernardi G. Chromatography of nucleic acids on hydroxyapatite. II. Chromatography of denatured DNA. Biochim Biophys Acta. 1969 Feb 18;174(2):435–448. doi: 10.1016/0005-2787(69)90274-3. [DOI] [PubMed] [Google Scholar]
- Birnstiel M., Telford J., Weinberg E., Stafford D. Isolation and some properties of the genes coding for histone proteins. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2900–2904. doi: 10.1073/pnas.71.7.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchan M. R. Bovine satellite I DNA consists of repetitive units 1,400 base pairs in length. Nature. 1974 Sep 27;251(5473):288–292. doi: 10.1038/251288a0. [DOI] [PubMed] [Google Scholar]
- Eigner J., Doty P. The native, denatured and renatured states of deoxyribonucleic acid. J Mol Biol. 1965 Jul;12(3):549–580. doi: 10.1016/s0022-2836(65)80312-6. [DOI] [PubMed] [Google Scholar]
- Filipski J., Thiery J. P., Bernardi G. An analysis of the bovine genome by Cs2SO4-Ag density gradient centrifugation. J Mol Biol. 1973 Oct 15;80(1):177–197. doi: 10.1016/0022-2836(73)90240-4. [DOI] [PubMed] [Google Scholar]
- Gautier F., Mayer H., Goebel W. Cloning of calf thymus satellite I DNA in Escherichia coli. Mol Gen Genet. 1976 Nov 24;149(1):23–31. doi: 10.1007/BF00275957. [DOI] [PubMed] [Google Scholar]
- Guttann T., Votavová H., Pivec L. Base composition heterogeneity of mammalian DNAs in CsCl-netropsin density gradient. Nucleic Acids Res. 1976 Mar;3(3):835–845. doi: 10.1093/nar/3.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurnit D. M., Shafit B. R., Maio J. J. Multiple satellite deoxyribonucleic acids in the calf and their relation to the sex chromosomes. J Mol Biol. 1973 Dec 15;81(3):273–284. doi: 10.1016/0022-2836(73)90141-1. [DOI] [PubMed] [Google Scholar]
- MANDELL J. D., HERSHEY A. D. A fractionating column for analysis of nucleic acids. Anal Biochem. 1960 Jun;1:66–77. doi: 10.1016/0003-2697(60)90020-8. [DOI] [PubMed] [Google Scholar]
- Moore R. L., McCarthy B. J. Base sequence homology and renaturation studies of the deoxyribonucleic acid of extremely halophilic bacteria. J Bacteriol. 1969 Jul;99(1):255–262. doi: 10.1128/jb.99.1.255-262.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K., Murray N. E. Phage lambda receptor chromosomes for DNA fragments made with restriction endonuclease III of Haemophilus influenzae and restriction endonuclease I of Escherichia coli. J Mol Biol. 1975 Nov 5;98(3):551–564. doi: 10.1016/s0022-2836(75)80086-6. [DOI] [PubMed] [Google Scholar]
- Müller W., Bünemann H., Dattagupta N. Interactions of heteroaromatic compounds with nucleic acids. 2. Influence of substituents on the base and sequence specificity of intercalating ligands. Eur J Biochem. 1975 May;54(1):279–291. doi: 10.1111/j.1432-1033.1975.tb04138.x. [DOI] [PubMed] [Google Scholar]
- Müller W., Crothers D. M. Interactions of heteroaromatic compounds with nucleic acids. 1. The influence of heteroatoms and polarizability on the base specificity of intercalating ligands. Eur J Biochem. 1975 May;54(1):267–277. doi: 10.1111/j.1432-1033.1975.tb04137.x. [DOI] [PubMed] [Google Scholar]
- Müller W., Gautier F. Interactions of heteroaromatic compounds with nucleic acids. A - T-specific non-intercalating DNA ligands. Eur J Biochem. 1975 Jun;54(2):385–394. doi: 10.1111/j.1432-1033.1975.tb04149.x. [DOI] [PubMed] [Google Scholar]
- Pakroppa W., Müller W. Fractionation of DNA on hydroxyapatite with a base-specific complexing agent. Proc Natl Acad Sci U S A. 1974 Mar;71(3):699–703. doi: 10.1073/pnas.71.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHILDKRAUT C. L., MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J Mol Biol. 1962 Jun;4:430–443. doi: 10.1016/s0022-2836(62)80100-4. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]


