Summary
The T cell receptor (TCR) serves a critical function in the immune system and represents one of the most complex receptor structures. A striking feature is the presence of nine highly conserved, potentially charged residues in the transmembrane helices. Previous models have attempted to explain assembly based on pairwise interactions of these residues. Using a novel method for the isolation of intact radiolabeled protein complexes, we demonstrate that one basic and two acidic transmembrane residues are required for the assembly of each of the three signaling dimers with the TCR. This remarkable three-helix arrangement applies to all three assembly steps and represents the organizing principle for the formation of this intricate receptor structure.
Introduction
The T cell receptor-CD3 complex (TCR-CD3) serves a critical role in the differentiation, survival, and function of T cells, and receptor triggering elicits a complex set of biological responses that serve to protect the organism from infectious agents. The receptor is composed of six different chains that form the TCR heterodimer responsible for ligand recognition, as well as the CD3γε, CD3δε, and ζζ signaling modules (reviewed in Exley et al., 1991; Garboczi et al., 1996; Sun et al., 2001). A number of competing models for receptor triggering have been proposed, driven in part by persisting uncertainties about the stoichiometry of the complex and the arrangement of its components.
A remarkable feature of the TCR-CD3 transmembrane (TM) domains may represent a key to approaching this problem: three basic residues are found in the TM domains of the TCR heterodimer while a pair of acidic residues is present in each of the three associated signaling dimers (Figure 1B). Mutagenesis experiments on some of these polar residues suggested that they play a key role in receptor assembly (Alcover et al., 1990; Blumberg et al., 1990a), but due to the complexity of the receptor it had not been possible to define the function of each polar residue or the resulting arrangement. The working hypothesis has been that these basic and acidic residues form pairwise interactions in the membrane, analogous to the aqueous environment where neighboring basic and acidic residues can form salt bridges. Experimentally, this concept was supported by transfection experiments with two-chain combinations in COS cells (Hall et al., 1991; Cosson et al., 1991). However, a number of different two-chain combinations could be observed in these experiments. In addition, a TCRα mutant in which either one of the two basic residues was mutated could still support an interaction with CD3δ, raising issues of specificity. Most importantly, the proposed pairwise interactions of basic and acidic residues resulted in two mutually exclusive proposals of TCR-CD3 stoichiometry. Three basic and six acidic residues are present in the TM domains if a single TCR heterodimer is present (Punt et al., 1994), and models with two TCR heterodimers have been proposed to account for this perceived charge imbalance (Fernandez-Miguel et al., 1999; reviewed in Jacobs, 1997). Each of these models has significant implications for the mechanism of receptor triggering.
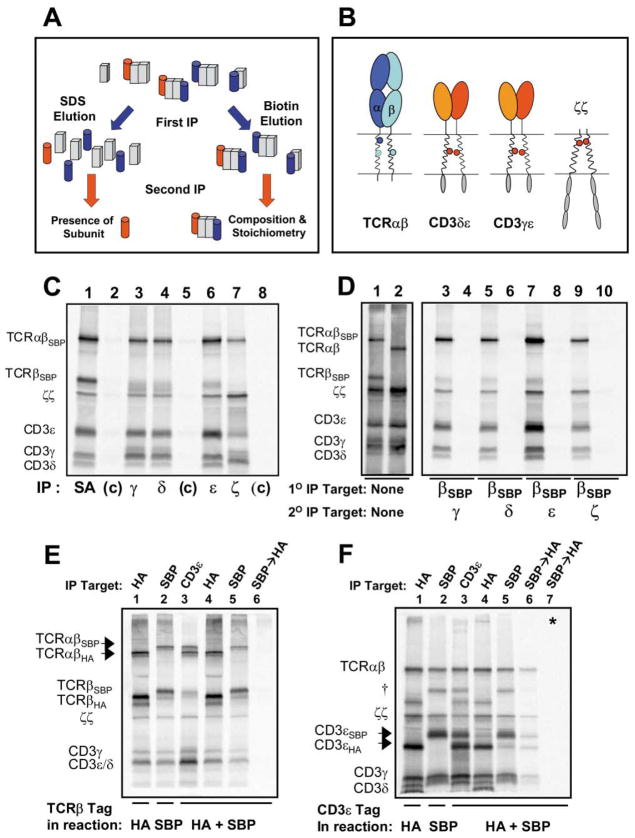
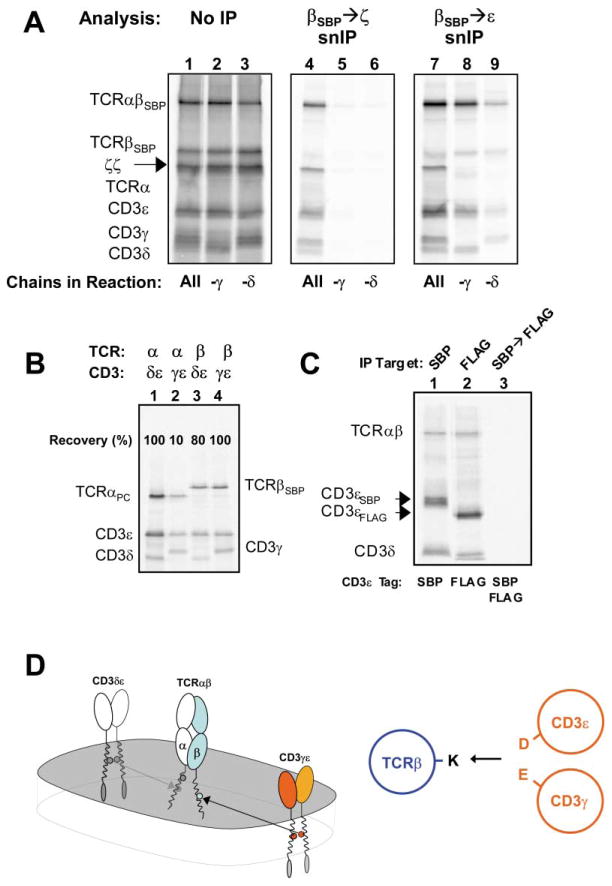
Figure 1. The TCR-CD3 Complex Assembled in the ER Contains One TCRαβ Heterodimer.
(A) Isolation of intact radiolabeled protein complexes by sequential non-denaturing immunoprecipitation (snIP). A conceptual illustration of denaturing (left side) versus non-denaturing (right side) sequential immunoprecipitation (IP) for analysis of non-covalent protein complexes.
(B) Potentially charged TM residues in the TCR and associated signaling dimers. Three conserved basic residues are present in the TM domains of the TCR (arginine and lysine in TCRα—dark and light blue, respectively—and lysine in TCRβ), while six acidic residues are located in the TM domains of the CD3δε, CD3γε, and ζζ dimers. Note that the TCRα arginine and the two acidic residues of ζζ are located in the upper third of the putative TM domains, while the lysine residues of TCRα and β as well as the acidic residues of CD3δε and γε are located approximately in the center.
(C and D) Comparison of single-step IP and two-step snIP analyses of in vitro assembled TCR-CD3 complexes.
(C) A single translation/assembly reaction containing mRNAs encoding human TCRα, TCRβ tagged with a streptavidin binding peptide (TCRβSBP), and CD3 γ, δ, ε, and ζ chains was carried out as described in Experimental Procedures. The reaction was split 8 ways and cleared digitonin lysates were analyzed by single-step IP using antibodies specific for the indicated targets (lanes 1, 3, 4, 6, and 7) or control (c) reagents (lane 2 agarose beads; lane 5 control goat polyclonal; lane 8 control mIgG1 mAb).
(D) Isolation of intact TCR-CD3 complexes by sequential non-denaturing IP (snIP). A single translation/assembly reaction was performed as in (C) and split 5 ways. Membrane fractions were either solubilized in SDS and run directly as loading controls (lane 1) or solubilized in digitonin and subjected to SA capture. Intact protein complexes were eluted from SA beads with free biotin and reprecipitated using antibodies to the indicated (2° IP) targets (lanes 3, 5, 7, and 9). In a control reaction (lanes 2, 4, 6, 8, and 10), the TCRβ mRNA did not encode the SBP affinity tag. Loading controls here and elsewhere represent approx. 10% of the starting material used for an IP.
(E) The core TCR-CD3 complex contains a single TCRαβ heterodimer. Assembly reactions were carried out as in (C) and included either TCRβHA (lane 1), TCRβSBP (lane 2), or both (lanes 3–6). TCRαβHA and TCRαβSBP (arrows) could be resolved on a 12% SDS gel due to the size of the SBP tag (lanes 1–3). IP for one β chain coprecipitated associated CD3 polypeptides, but not the alternative β chain (lanes 4 and 5), and TCRβSBP→TCRβHA snIP failed to recover any labeled proteins (lane 6). In a positive control experiment (F), the SBP/HA tag pair was used to isolate complexes containing two different CD3ε chains. Formation of disulfide linked CD3ε dimers (†) is a competing reaction with CD3δε and CD3γε formation and represents a side-product. This band is marked with the same symbol where it appears in other experiments. Lane 7 is a control reaction (*) in which full complexes containing each tagged CD3ε chain were assembled separately and mixed prior to solubilization.
Energetic considerations suggest that all single-spanning TM domains adopt a α-helical structure, a notion that is supported by the NMR structure of the glycophorin A (GpA) homodimer (MacKenzie et al., 1997). If TM domains indeed play a critical role in receptor assembly, it is important to explain how specificity is achieved despite such limited structural diversity and the preponderance of hydrophobic amino acids. Recent studies with model TM helices have indicated that polar interactions can play a significant role in oligomerization of membrane proteins. Two groups reported that placement of a single acidic (Glu, Asp) or carboxamide (Gln, Asn) residue in polyleucine TM helices resulted in the formation of dimers or trimers (Gratkowski et al., 2001; Zhou et al., 2001); basic residues did not induce oligomerization (Gratkowski et al., 2001) and interactions between basic and acidic residues were not examined. The observation that dimers or trimers can be formed through interaction of acidic residues is surprising and suggests that the membrane may provide a unique environment for biologically relevant protein-protein interactions.
Given the questionable specificity of two-chain interactions, we postulated that it would be critical to investigate receptor assembly at higher levels of complexity. For that purpose, we developed a novel approach for the purification of intact radiolabeled protein complexes that allowed us to dissect key assembly intermediates. Each major assembly step was found to require the formation of a three-chain interface in the membrane. The assembly of each of the three signaling dimers with the TCR thus involves creation of a trimeric interface between one basic and two acidic TM residues, such that proper placement of each of these nine polar TM residues is required for assembly of the entire structure.
Results
A Novel Method for the Isolation of Intact Membrane Protein Complexes
Given the complexity of the receptor, immunoprecipitation (IP) targeting one of the chains not only isolates the full complex, but also unassembled chains/assembly intermediates. The approach we have developed for the isolation of defined complexes is based on specialized affinity tags that permit quantitative release of complexes captured in a first IP step under non-denaturing conditions and re-IP using antibodies to other components (Figure 1A). We performed two-step and three-step sequential non-denaturing IP (snIP) experiments using the 47 amino acid streptavidin binding peptide (SBP; Wilson et al., 2001), which can be eluted from streptavidin (SA) by competition with free biotin, and the 12 amino acid protein-C derived peptide (PC), which can be eluted from the calcium-dependent anti-PC mAb using EDTA.
We tested this approach using an in vitro translation system with ER-derived microsomes. This cell-free system has been shown to yield the same specific protein interactions observed in cells (Bijlmakers et al., 1994, 1993), and resembles cellular metabolic labeling experiments in that radiolabeled proteins are translocated into ER membranes. The feasibility of assembling a complete TCR-CD3 complex by in vitro translation in the presence of canine pancreatic ER microsomes has been demonstrated (Huppa and Ploegh, 1997). Comparison of one-step IP to two-step snIP experiments (Figures 1C and 1D) demonstrates the distinct advantages of this technique. All components of the TCR-CD3 complex could be visualized with both approaches, but one-step IP resulted in an over-representation of the IP target and higher background. We utilized the TCRα and β chains from the human A6 TCR since the crystal structure of the ectodomain has been determined (Garboczi et al., 1996) and cotranslated mRNAs for TCRα, TCRβSBP, and CD3 γ, δ, ε, and ζ chains in a rabbit reticulocyte lysate system with ER microsomes and 35S-labeled methionine. When separated by SDS-PAGE under non-reducing conditions, the disulfide-linked TCRαβ heterodimer and ζζ homodimer, as well as the CD3γ, δ, and ε chains could be readily visualized. Maximum yields of fully assembled TCR-CD3 complexes were observed after an initial 15 min translation period under reducing conditions and a four-hour assembly period under oxidizing conditions. Earlier time points revealed qualitatively similar results with a lower overall yield of assembled complexes. Antibodies to CD3γ, δ, ε, or ζ precipitated all TCR-CD3 components when used in the second IP step following biotin elution of TCRβSBP (Figure 1D). The products of a TCRβSBP→snIP (lane 9) derived exclusively from fully assembled complexes, since ζζ association is the final step in TCR-CD3 assembly (Sussman et al., 1988). The two-step snIP was highly specific since proteins were not precipitated when the TCRβ chain lacked the SBP tag (lanes 4, 6, 8, and 10). The affinity tags did not interfere with assembly based on the use of four different tags, placement of tags on any chain of the complex, and comparison of complexes isolated with antibodies to ectodomains or affinity tags (not shown). All experiments shown have been performed at least twice as described and are representative of a larger body of experimental results.
These experiments required a renewable source of ER microsomes with high assembly activity. The availability of such ER microsomes was initially a critical limiting factor, and commercially available canine preparations were unsatisfactory. Due to variations in the source tissue, only one of several canine pancreatic preparations obtained from experienced laboratories was suitable for these experiments. We therefore developed a method for isolating ER microsomes with high assembly activity from established lymphoid cell lines that have a well-developed secretory apparatus (Supplemental Figure S1 available at http://www.cell.com/cgi/content/full/111/7/967/DC1). Such microsomes could be isolated from human plasmacytoma (RPMI 8226) and murine B cell hybridoma (IVD12) cells on self-generating continuous iodixanol gradients, which permit fractionation based on density, and these membranes exhibited higher assembly activity than canine microsomes. Both exocrine pancreas and plasma cells are specialized for protein secretion, but the pancreas primarily produces single-chain proteolytic enzymes while plasma cells secrete immunoglobulins and express an array of specialized surface receptors.
The Core TCR-CD3 Complex Assembled in the ER Contains One TCRαβ Heterodimer
To directly assess whether the TCR-CD3 complex assembled in the ER contains one or two TCRαβ heterodimers, we performed assembly experiments with two TCRβ chains bearing different tags (SBP and HA) (Figure 1E). Due to the size of the SBP tag, TCRαβHA and TCRαβSBP disulfide-linked heterodimers (arrows) could be readily separated by SDS-PAGE. IP for one TCRβ chain precipitated all TCR-CD3 components, but not the TCRβ chain bearing the second tag (lanes 4 and 5). A TCRβSBP→TCRβHA snIP failed to recover any products (lane 6), indicating that complexes containing both β chains were not formed. Since two CD3ε chains are present in a single complex (Blumberg et al., 1990b), we used the same approach to isolate complexes containing both SBP- and HA-tagged CD3ε chains as a positive control (Figure 1F). Experiments with another affinity tag pair on TCRα chains and with a second αβ TCR all produced similar results (M.E.C., J.P., K.W.W., unpublished data). Moreover, the outcome was not changed when ER microsomes isolated from a T cell line were used (not shown), ruling out a role for a T cell specific ER factor in forming higher-order complexes. These data clearly demonstrated that the core TCR-CD3 complex assembled in the ER contained only a single TCRαβ heterodimer. The perceived TM charge imbalance therefore could not be explained by the presence of a second TCR heterodimer.
A Novel Membrane-Based Three-Helix Motif in the Assembly of CD3δε with TCRα
We postulated that each of the three basic TCR TM residues serve a definable role in the assembly process and used the in vitro translation system to examine mutations of these residues in all possible combinations. As illustrated by the helical wheel diagram in Figure 2A, the lysine and arginine residues in the TCRα TM region are predicted to fall on roughly opposite faces of the TM helix, making it unlikely that their functions are redundant. We first examined the contribution of the basic TCR TM residues to the association with the CD3δε heterodimer (Figure 2B) since assembly of TCRαβ with CD3δε occurred efficiently in the absence of CD3γ and ζ chains, consistent with experiments in Jurkat cells lacking expression of CD3γ (Dietrich et al., 1996). We used the EDTA-elutable protein-C derived peptide tag (PC) to isolate CD3δPCε heterodimers by two-step snIP, and demonstrated that the TCRα TM lysine was necessary and sufficient for association with TCRαβ. In a control (*) where wild-type TCRαβ and CD3δε were assembled separately and mixed prior to solubilization, no TCR was coprecipitated with CD3αPCε (lane 9), excluding the possibility of non-specific IP or association after solubilization. The specificity of such mixing controls is illustrated in Supplemental Figure S2 (available at above website). The same result was obtained when untagged CD3 proteins were precipitated in a single-step IP with mAb UCH-T1, proving that these results were not an artifact due to the use of an epitope tag (not shown).
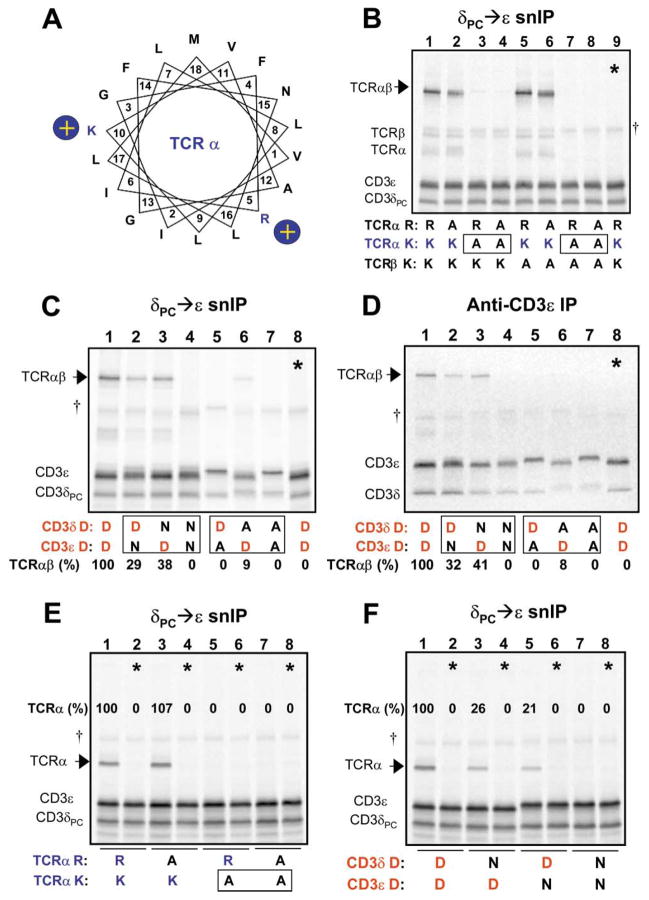
Figure 2. Interaction Among Three TM Domains that Requires One Basic and Two Acidic TM Residues.
(A) A helical wheel diagram shows the relative positions of the two basic residues in the TCRα TM region. Position 1 (V, valine) represents the first predicted TM residue at the extracellular (or ER luminal) face of the membrane. A series of experiments was performed to examine the requirements for basic and acidic TM residues in the association of CD3δε with TCR.
(B–D) analyze the four-chain TCRαβ-CD3δε complex, and (E and F) demonstrate that the same polar residues are relevant for the three-chain TCRα-CD3δε complex. CD3 heterodimers were isolated by CD3δPC→CD3ε snIP (B, C, E, and F) or single anti-CD3ε IP (D) (IP strategy indicated above each image), in order to determine the effect of mutations on assembly with TCR (arrows). In (B), the basic TM residues in the TCR chains were substituted in all possible combinations to assess their role in assembly with CD3δε (R-arginine, K-lysine, and A-alanine). The TCRα TM lysine is necessary and sufficient for this assembly step, as TCR association was lost only when this residue was substituted (boxes). The same result was obtained when TCRβ was omitted in the assembly reaction (E).
(C and D) Both acidic TM residues in CD3δε are required for association with TCR. While substitution of either aspartic acid residue with alanine [D→A] severely reduced or eliminated TCR association, conservative substitution with asparagine [D→N] was tolerated to a significant degree in either CD3 chain, but not both.
(D) The same result was obtained regardless of whether an epitope tag was used in the IP (C) or untagged complexes were precipitated with a mAb to the CD3ε extracellular domain (D).
Control lanes (*) consisted of TCR and CD3 components assembled in separate reactions and mixed prior to solubilization. Quantitation of associated TCR proteins was done by densitometry using a phosphor imager, and values are expressed as percentages relative to levels of association observed among wild-type proteins for each experiment.
This result raised the interesting question of whether one or both acidic residues in the TM regions of CD3δε are required for this interaction. Alanine substitution of either the CD3δ or ε TM aspartic acid (Figures 2C and 2D, lanes 5–7) reduced TCRαβ association to almost undetectable levels, although the mutation in CD3δ (lane 6) was slightly more tolerated than in ε (lane 5) in a reproducible fashion. Furthermore, TCR association was maintained with a conservative substitution in either CD3 subunit (D-aspartic acid to N-asparagine, lanes 2 and 3), but not when this substitution was made in both chains (lane 4). This interaction required only the lysine residue in the TM of TCRα since comparable results were obtained when the other basic TM residues were mutated to alanine (not shown).
The fact that both CD3δε TM aspartic acid residues and the TCRα TM lysine were critical for the TCRαβ–CD3δε association suggested that the TM helices from all three polypeptides form a single interface in which association is dependent on these key polar contacts. We therefore reduced the complexity of the assembly from four to three chains; this TCRα-CD3δε three-chain complex has been observed in cellular systems and therefore appears to represent a relevant assembly intermediate (Kearse et al., 1995). As observed for the four-chain complex, formation of the three-chain complex required the TCRα TM lysine (Figure 2E), and substitution of acidic residues by asparagine was tolerated in either one, but not both CD3 TM regions (Figure 2F). These results were highly reproducible, as demonstrated in Supplemental Figure 3 (available at http://www.cell.com/cgi/content/full/111/7/967/DC1). A TCRα mutant in which the lysine was the only polar TM residue (mutation of the arginine as well an asparagine and a threonine) efficiently formed the TCRα-CD3δε three-chain complex (not shown), excluding a critical role for other polar side chains. Assembly was clearly driven by TM interactions since the three-chain complex was also formed by a truncation mutant of TCRα (TCRαTM–SBP) that covered only the native TM domain with six N-terminal and five C-terminal flanking TCR residues for proper membrane insertion (Figure 3A). Importantly, assembly with this truncated TCRα chain required both acidic TM residues of the CD3δε heterodimer (lanes 3, 5, and 7).
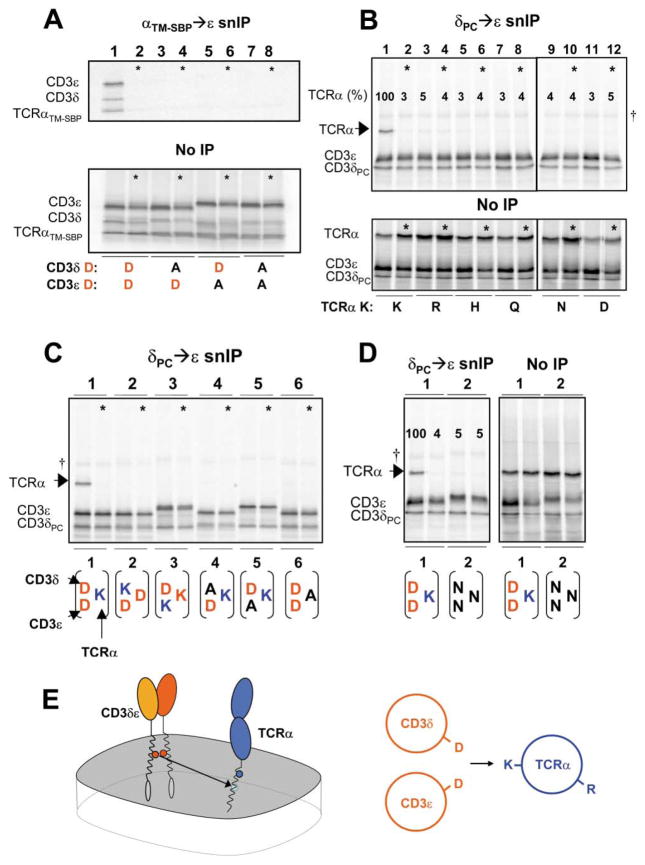
Figure 3. Highly Specific Requirements for TCRα-CD3δε TM Interactions.
(A) TCRα-CD3δε association is driven by the TM domains. An ectodomain-truncated TCRα polypeptide consisting of only the TM region and 6 flanking N-terminal and five C-terminal residues as well as a C-terminal SBP tag (TCRαTM–SBP) assembled with CD3δε (lane 1). Assembly was disrupted by D→A substitution in either CD3 chain (lanes 3, 5, and 7), as observed for full-length TCRα.
(B–D) Stringent requirement for a specific arrangement of one basic and two acidic TM residues in TCRα-CD3δε assembly. Assembly was abrogated by substituting TCRα TM lysine [K] with any other polar residues (B), or by exchanging the position of the native lysine [K] or aspartic acid [D] residues among the three TM helices (C, 1–3). Alanine substitution at the position of any potentially charged TM residue also abrogated assembly (C, 4–6), and substitution at all three positions with asparagine did not support association (D). Posttranslation mixing controls (*) were performed as before, and loading controls are included for experiments in (A), (B), and (D). IP strategy is indicated above each experiment.
(E) Illustration depicting CD3δε association with TCRα via the TM lysine [K]. TCR-CD3 components are represented as whole polypeptides (left) and as an axial view of simplified helical wheels (right).
It was relevant to determine whether the presence of polar residues at this interface was sufficient or whether a specific arrangement of basic and acidic residues was required. Experiments where the basic and acidic residues were exchanged among the three helices (Figure 3C; illustrated below gel) demonstrated that only the native arrangement supported TCRα association. Replacement of the TCRα TM lysine with arginine or other polar residues was not tolerated (Figure 3B), and an asparagine-based motif recently shown to drive homotrimerization of model TM helices via a hydrogen bonding network (Zhou et al., 2000) was not sufficient (Figure 3D). This assembly step therefore depends on the creation of a highly specific interface among three TM helices, each of which contributes a potentially charged residue (Figure 3E).
TCRβ-CD3γε Association Exhibits a Similar Requirement for One Basic and Two Acidic TM Residues
Having observed that TCRα–CD3δε assembly requires precisely one basic and two acidic TM residues, we hypothesized that the same arrangement could be relevant for CD3γε association. Alanine substitution of the TCRβ TM lysine eliminated TCR interaction with CD3γε in assembly reactions containing both CD3γ and CD3δ, as shown by a loss of TCRαβ association when CD3γε dimers were selected (Figure 4A) and by a selective loss of CD3γ with a different IP strategy (Figure 4B). Again, assembly could be reduced to three chains and formation of this TCRβ-CD3γε complex was dependent on the TCRβ TM lysine (Figure 4C).
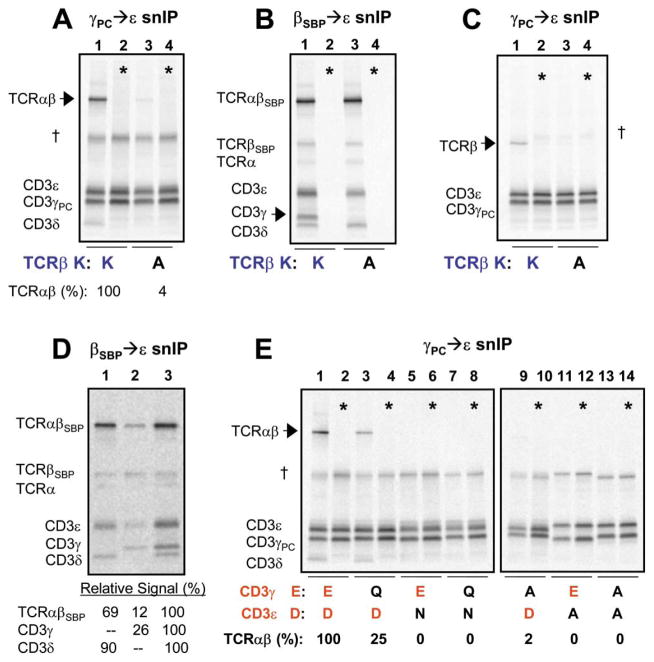
Figure 4. TCRβ TM Lysine and Both CD3γε TM Acidic Residues Are Required for TCRβ-CD3γε Association.
(A–C) TCRβ TM lysine [K] is required for CD3γε association with TCR. In reactions containing TCRα, β, and CD3γ, δ, and ε, alanine substitution of TCRβ TM K results in loss of TCR association with CD3γPCε (A) and specific loss of CD3γ from TCR-CD3 subcomplexes selected by a different IP strategy (B). The same result is obtained when the complexity is reduced to the three-chain TCRβ-CD3γε complex (C).
(D) CD3γ associates with TCR more efficiently when CD3δ is also present. TCR-CD3 complexes were assembled in reactions containing TCRαβSBP and CD3δε (lane 1), CD3γε (lane 2), or CD3γδε (lane 3). TCR-CD3 products were isolated as in (B). Relative signal recovered in the indicated bands was quantitated by densitometry using a phosphor imager.
(E) Both CD3γε TM acidic residues are required for association with TCR. Assembly reactions were performed and analyzed precisely as in (A), with conservative (aspartic acid to asparagine, D→N; glutamic acid to glutamine, E→Q) or non-conservative (alanine, A) substitutions in CD3γε TM regions.
Quantitation and mixing controls (*) were performed as above.
We observed that CD3γε associated with TCR inefficiently in the absence of CD3δ (Figure 4D) and therefore analyzed the requirement for the acidic TM residues of CD3γε in reactions containing both CD3δ and γ (Figure 4E). Alanine substitution of either acidic TM residue of CD3γε eliminated TCR association (lanes 9–14), and conservative substitution of the CD3γ TM glutamic acid to glutamine (Q) reduced TCRαβ-CD3γε association (lane 3). Asparagine (N) substitution of the CD3ε TM aspartic acid abrogated TCRαβ-CD3γε association (lane 5) since this substitution simultaneously affected both CD3δε and γε association. Comparison of TCRβ-CD3δε and TCRα-CD3δε three-chain assemblies indicated that this conservative mutation of CD3ε had a similar effect at both sites (Figure 2F and not shown).
TCRα-CD3δε and TCRβ-CD3γε Interactions Are Similar in the Membrane
These results indicated that the TCRα-CD3δε and TCRβ-CD3γε interactions are similar since both require a lysine in the TM region of the respective TCR chain and both acidic TM residues in the relevant CD3 heterodimer. Nevertheless, formation of fully assembled αβ TCR-CD3 complexes containing the ζ-chain strictly required both CD3γ and δ (Figure 5A), which is consistent with data showing that T cells lacking either subunit do not express TCRαβ-CD3 complexes at the cell surface (Buferne et al., 1992; Geisler, 1992). The similarity of the motifs that mediate CD3δε association with TCRα and CD3γε association with TCRβ in the membrane thus raises the question of how selectivity is achieved in the assembly process. In three-chain assembly reactions, TCRα exhibited a strong preference for interaction with CD3δε over CD3γε, but TCRβ could associate to a similar degree with either CD3 dimer in isolation (Figure 5B). The similarity of these two TM interaction sites therefore allowed the formation of alternative assembly intermediates. However, complexes with two CD3δε heterodimers bound to one TCR were not formed, as shown by snIP experiments with two different CD3ε (Figure 5C) or CD3δ chains (not shown), as well as densitometry measurements (from Figure 4D, lane 1). In the complete structure CD3δε is therefore bound only via the TCRα site, even though it can bind to either TCRα or β in isolated three-chain assembly reactions. Association of two CD3δε heterodimers with a single TCR is likely to be prevented by the EC domains rather than the TM domains, since a chimeric CD3γ chain with the TM sequence of CD3δ was incorporated into complete TCR-CD3 complexes (not shown) and rescued surface TCR-CD3 expression in a CD3γ-deficient T cell line (Wegener et al., 1995).
Figure 5. Evidence that TCRα-CD3δε and TCRβ-CD3γε Interactions Are Similar in the Membrane.
(A) Both CD3γ and δ are required for ζ-chain association. Reactions contained all TCR-CD3 components, as indicated below gels (lanes 1, 4, and 7), or singly lacked γ (lanes 2, 5, and 8), or δ (lanes 3, 6, and 9). No fully assembled complexes were recovered from reactions lacking either CD3γ or CD3δ (lanes 5 and 6; middle image), but TCR-CD3 sub-complexes not containing ζ chain were revealed by alternative snIP analysis of the same reactions (right image). Loading controls are provided as for previous experiments (left image).
(B) TCRβ can associate with either CD3 heterodimer in three-chain assembly reactions. TCRαPC or βSBP were each cotranslated with CD3δε or γε and three-chain complexes were isolated by αPC→ε (lanes 1 and 2) or βSBP→ε (lanes 3 and 4) snIP.
(C) TCRαβ-CD3δε complexes contain only one CD3δε heterodimer. A strategy similar to that employed in Figures 1E and 1F was used to assess whether two CD3δε dimers could associate with a single TCR.
(D) CD3γε associates with TCR via the TCRβ TM lysine (K) in a TM helical interaction similar to that seen for TCRδ-CD3δε. Two views are provided as in Figure 3E.
Requirements for ζζ Assembly
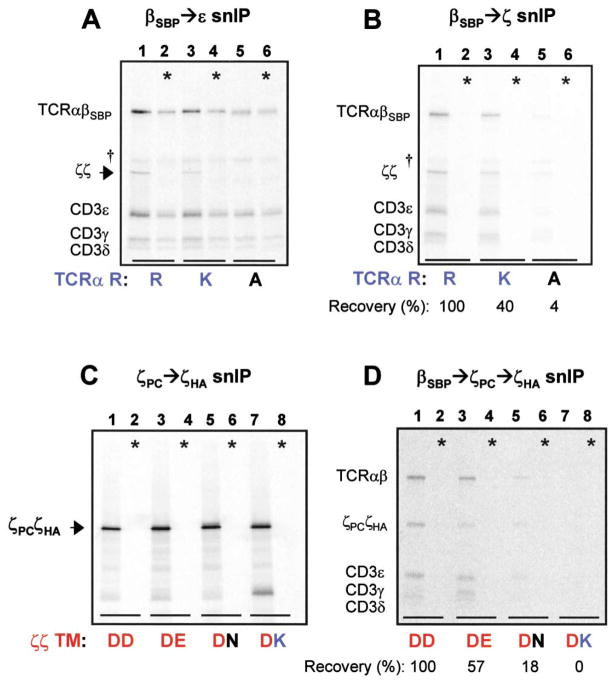
The experiments described above demonstrated that the arginine in the TCRα TM region was not required for association of CD3δε or γε heterodimers. These results raised the possibility that the arginine played a role in the last assembly step, the association of the ζζ homodimer. Substitution of the arginine by alanine (R→A) resulted in a selective loss of the ζζ dimer in complexes isolated by a TCRβSBP→CD3ε snIP (Figure 6A). Quantification of fully assembled complexes (Figure 6B) indicated that ζζ assembly with TCR was reduced to 40% of wild-type levels by the conservative lysine (K) substitution (lane 3) and to nearly undetectable levels by the alanine substitution (lane 5). Together, these data indicate that the third basic TCR TM residue, TCRα arginine, plays a key role in assembly of the ζζ homodimer with the TCR-CD3 complex.
Figure 6. The TCRα TM Arginine and Both ζ Chain TM Aspartic Acid Residues Are Important for ζζ Association.
(A and B) TCRα TM arginine is important for ζ chain association. Full assembly reactions were analyzed using two different snIP strategies to examine effects of conservative and non-conservative substitutions.
(A) Specific loss of ζζ homodimer (arrowhead) was observed when TCRα TM arginine was mutated to alanine [R→A] (lane 5).
(B) Quantitation of fully assembled complexes revealed that the R→K substitution reduced ζζ association to less than half of wild-type levels (lane 3), while the R→A substitution reduced ζζ association to almost undetectable levels (lane 5). In control reactions (*), TCR-CD3 components and ζζ homodimers were assembled separately and mixed prior to solubilization.
(C and D) Both ζ chain TM aspartic acid residues are important for ζζ association.
(C) Disulfide-linked ζζ mixed dimers containing two different ζ chains in each reaction were isolated by ζPC→ζHA snIP and separated by non-reducing SDS-PAGE. Symbols indicate the wild-type dimer (DD), and mixed dimers in which the aspartic acid of one ζ chain was substituted with glutamic acid (DE), asparagine (DN), or lysine (DK). All combinations efficiently formed covalent mixed dimers. Application of this approach to full assembly reactions (D) revealed reduced ζζ assembly with TCR when one ζ chain carried a conservative substitution (DE, lane 3; DN, lane 5), and elimination of the interaction by a non-conservative substitution (DK, lane 7). In control reactions (*), full complexes with each ζ chain were assembled separately and mixed prior to solubilization.
A role for the aspartic acid in the TM region of the ζ chain in this interaction was possible, but testing this was complicated by the fact that substitution by alanine or asparagine disrupted ζζ disulfide-linked dimer formation as well as association with TCR (Rutledge et al., 1992; data not shown). We therefore examined whether a single acidic residue was sufficient for ζζ assembly with TCR or whether both acidic residues were involved. This required an approach that would allow us to isolate ζζ dimers in which only one copy carried the desired substitution. As shown in Figure 6C, four different mixed dimers could be isolated with comparable yields by snIP when the wild-type and mutant ζ-chains carried different C-terminal peptide tags; this association did not occur during or after solubilization (* controls). We therefore used this approach to examine whether both TM aspartic acid residues were required for ζζ association with TCR (Figure 6D). A three-step snIP strategy was employed to isolate TCR-CD3 complexes containing ζζ mixed dimers. Conservative substitution by asparagine (N) in only one ζ chain (DN combination; lane 5) greatly reduced recovery of the complex, and substitution by lysine (K) in one of the two ζ chains (DK; lane 7) abrogated complex formation. Since all four mixed dimers formed with a similar efficiency (Figure 6C), these results were due to different degrees of assembly with TCR. Recovered proteins in lanes 1, 3, and 5 derived exclusively from complexes associated with ζζ mixed dimers, and not homodimers of either ζ chain, since no proteins were recovered from control reactions (*) in which full complexes were assembled with each ζ chain separately and mixed before solubilization. These data indicate that both acidic TM residues in the ζζ dimer and a basic TCR TM residue (TCRα TM arginine) contribute to this assembly step.
These results raised the question of whether both acidic residues in the ζζ homodimer were required for an interaction with the TCRα TM arginine, or whether one is required for a polar contact with another TM helix. Depending on the TM prediction algorithm used, the TCRβ TM domain may contain a glutamic acid residue that, in contrast to the three basic TCR TM residues, is not conserved across all receptor types (αβ, γδ, and pre-TCR). Alanine substitution of this glutamic acid as well as two other polar residues in the N-terminal segment of the putative TCRβ TM (tyrosine and threonine) had no discernable effect on ζζ association, indicating that no polar TCRβ TM residue forms a critical interaction with the ζζ dimer (not shown).
Discussion
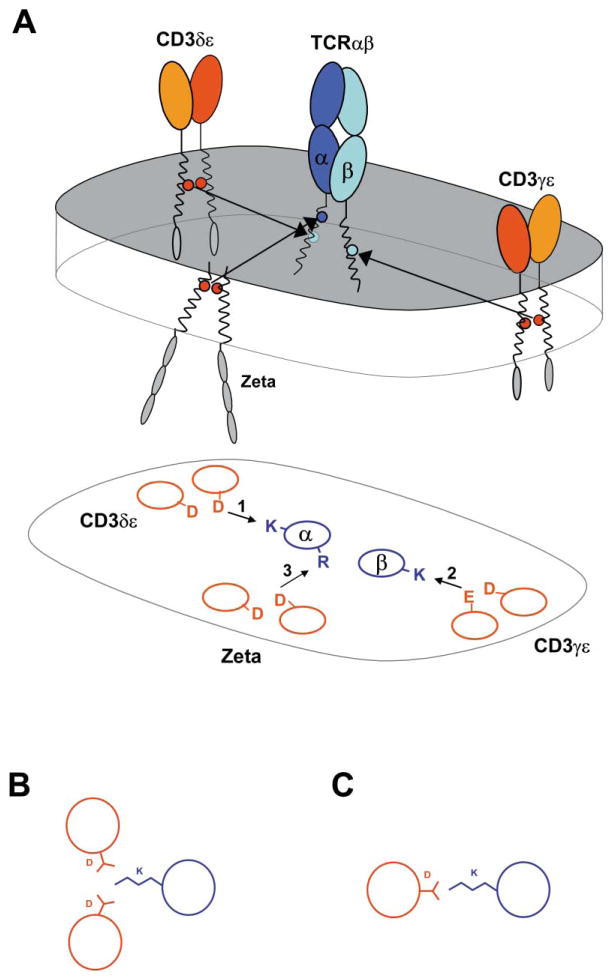
These data reveal the principal mechanism by which assembly of this complex receptor is organized. Assembly is disrupted by mutation of any one of the nine conserved basic/acidic TM residues, highlighting their essential role in the three major assembly steps that result in the creation of the complete structure (Figure 7). These results are relevant for several critical issues of TCR biology, in particular the stoichiometry of the complex, the assembly process that results in the formation of this structure, and the mechanisms of receptor triggering.
Figure 7. Assembly of the TCR-CD3 Complex Is Organized by Three Major Assembly Steps in the Membrane, Each Involving One Basic and Two Acidic Residues.
(A) Each of the three basic residues in the TCR TM regions serves as a critical contact for one of the three signaling dimers that associate with TCRαβ. TCR-CD3 polypeptides are shown in a representation of the lipid bi-layer (upper image). Lengths of TM domains are in appropriate proportion to extracellular Ig domains. Simplified helical wheel projection depicting interactions among TM domains in the same relative positions as above (lower image).
(B) Three-helix association may be favorable when a lysine (or arginine) is at the contact interface. Although lysine can adopt many conformations, close apposition of helices in the lipid may be achieved more effectively in a three-helix assembly than at a two-helix interface due to the length of lysine side chain (C).
TCR Stoichiometry and Receptor Triggering
The issue of stoichiometry is critical for defining the mechanisms of receptor triggering (Germain and Stefanova, 1999), since the two-TCR model implies that receptor activation results from a conformational change in a bivalent receptor complex. A recent study using transgenic mice expressing two different TCRβ chains reported that both were present in a single complex using an IP-Western blot approach (Fernandez-Miguel et al., 1999). However, the majority of “associated” TCRβ chains were not part of disulfide-linked TCRαβ heterodimers and the specificity of the association must therefore be questioned. In addition, the resulting dual-TCR model proposed that each TCR heterodimer associates directly with only one CD3 heterodimer, while our data clearly demonstrate that a single TCR heterodimer associates directly with both CD3δε and γε dimers. Using a similar transgenic approach and analysis of surface-labeled complexes, another group concluded that the core TCR-CD3 complex contains only a single TCR heterodimer (Punt et al., 1994). Higher-order structures were also proposed based on molecular weight estimations of detergent-solubilized complexes by fractionation on sucrose gradients (Exley et al., 1995), but very different results were in fact obtained with each of three detergents used for solubilization. Our experiments clearly demonstrate that the core TCRαβ-CD3 complex contains only a single TCR heterodimer, based on (1) sequential IP with two tags placed on TCRβ (or TCRα), (2) visualization of the two differentially tagged TCRβ chains based on an increase in MW resulting from the introduced SBP tag, and (3) quantification of components by densitometry. Most importantly, our study demonstrates that two acidic residues pair with each basic TCR residue. This solves the “charge imbalance” issue that represented the main theoretical foundation for all two-TCR models.
Organization of Receptor Assembly
Assembly of signaling dimers with the TCR occurs in a preferential sequence (CD3δε, CD3γε, and ζζ), and these higher-order assembly steps are primarily dependent on protein interactions in the membrane (Figure 7). Interactions among the extracellular domains are important in the formation of individual TCR, CD3γε, and δε dimers, and both TCR and CD3γε have been expressed as soluble dimers without their TM domains (Garboczi et al., 1996; Sun et al., 2001). In the case of TCRα-CD3δε assembly, we show directly that interactions among extra-cellular immunoglobulin domains are not required for formation of this trimeric complex (Figure 3A). Given the similarities among the TCRα-CD3δε and TCRβ-CD3γε interaction sites in the membrane, the ectodomains may contribute to complex formation by preventing association of two CD3δε or γε heterodimers with TCR (Figure 5A).
Due to the limited structural diversity and length of TM domains, assembly based on interaction of polar residues raises the issue of specificity. Fidelity in receptor assembly is critical for the TCR-CD3 complex since the engagement of a small number of receptors can result in T cell activation. In the TCR-CD3 complex, specificity is based on the requirement for three rather than two potentially charged residues in each assembly step. As illustrated in Figures 7B and 7C, the presence of a long basic side chain may favor a three-helix interface where close packing facilitates extensive contacts among neighboring amino acid side chains. The three-helix model implies that the interactions among these basic and acidic TM residues promote assembly, while additional contacts contribute stability and specificity to the binding interface. This view is consistent with the observation that low-level associations were maintained when certain basic or acidic residues were substituted by alanine. In transfection experiments, interactions between individual TCR and CD3 chains were observed in many combinations, and the two basic residues in the TCRα TM domain appeared to be functionally redundant (Cosson et al., 1991). In contrast, the lysine in the TM of TCRα was essential for the three-chain TCRα-CD3δε interaction described here and its function could not be replaced by the arginine in the TM of TCRα.
The data clearly demonstrate that assembly of CD3δε with TCRα is dependent on one basic and two acidic TM residues. Several lines of evidence indicate that these three potentially charged TM residues form a single functional interface: (1) all three residues are located at a similar position within the predicted TM helices, (2) alanine substitution of either aspartic acid disrupts the association, (3) conservative substitution of either aspartic acid by asparagine has a similar intermediate effect on complex formation, while asparagine substitution of both residues abrogates the interaction, and (4) substitution of all other polar TCRα TM side chains does not impair assembly. A similar interaction occurs at the interface of TCRβ and CD3γε, since this assembly step is also dependent on a lysine in the TM of the TCR and two acidic residues on the interacting CD3 dimer. Packing of two or three acidic or carboxamide side chains at interfaces between TM helices can be energetically favorable in the membrane environment (Gratkowski et al., 2001; Zhou et al., 2001). This raises the interesting possibility that the two acidic residues of CD3δε (as well as γε and ζζ) interact, and that the basic TCR residue binds to this interacting pair of acidic TM residues. The aspartic acid of ζ is located four residues C-terminal to the cysteine that forms the interchain disulfide bond and is therefore likely to be located close to the interface between the two ζ chains. Direct interaction between one basic and two acidic residues has been observed in crystal structures of solvent exposed domains, even though formation of salt bridges is more common (Palczewski et al., 2000). Our functional data demonstrate that the polar TM residues of TCRα and CD3δε shape the interface between the three chains in the membrane, but do not necessarily imply a direct interaction of all three side chains, since interactions through coordinated water and/or additional contacts to the helix backbone are possible. The high-resolution structure of bacteriorhodopsin (1.55 Å) demonstrated an interaction between an arginine (R82) and multiple acidic residues (D85, D212, and E194) through coordinated water molecules (Luecke et al., 1999).
Are the Basic and/or Acidic TCR-CD3 TM Residues Ionized?
Two distinct scenarios may account for the formation of these contacts: (1) the transmembrane helices fully equilibrate into the lipid and association of the chains occurs as a second step, and (2) unassembled chains are retained at a water-lipid interface in the vicinity of the Sec61p channel through which they enter the membrane. Since movement of charged residues into the highly hydrophobic interior of the membrane is energetically unfavorable, the first scenario implies that these polar residues may become uncharged (deprotonation of basic residues and protonation of acidic residues). This model does not explain the experimental finding that conservative substitution of both TM aspartic acid residues in the CD3δε dimer abrogates assembly since the asparagine side chain has properties similar to a protonated aspartic acid: it has a similar size and can serve as hydrogen bond donor or acceptor. The results rather indicate that the charge of the aspartic acid head groups is a critical feature of the observed assembly process. The hypothesis that receptor assembly occurs at a water-lipid interface is supported by two lines of evidence. First, unassembled TCRα and β both have a short half-life in cells due to the basic TM residues. When these residues are substituted, a longer half-life and transport out of the ER are observed (Bonifacino et al., 1990). Second, model proteins with one or two basic residues in the TM domain remain associated with the Sec61p channel rather than diffusing into the lipid (Heinrich et al., 2000).
In summary, these experiments reveal a remarkable arrangement of the transmembrane domains that is responsible for the formation of this intricate receptor structure. The unique structural arrangement of the eight transmembrane helices may be relevant for TCR triggering, and recent work in the integrin field has demonstrated that the “inside-out” signal is transmitted through the cell membrane by a conformational change of the TM domains (Takagi et al., 2002; Vinogradova et al., 2002).
Experimental Procedures
Antibodies and Reagents
Anti-CD3δ and anti-CD3γ affinity-purified goat polyclonal antibodies, anti-ζ (6B10.2) mAb, and anti-CD3ε (UCH-T1) mAb were obtained from Santa-Cruz; anti-HA mAb and calcium-dependent anti-PC mAb from Roche; and anti-FLAG mAb and streptavidin coupled to agarose beads from Sigma. Digitonin was obtained from Wako.
cDNA Constructs and In Vitro Transcription
Human CD3 γ, δ, ε, and ζ sequences were amplified by RT-PCR from peripheral blood, and A6 TCR α and β sequences from the A6 T cell clone. All sequences carried the murine H-2Kb signal sequence and were cloned into a pSP64 vector (modified by M. Kozak). Peptide tags were added as C-terminal in-frame fusions, typically with a three amino acid flexible linker. The streptavidin binding peptide (SBP) was based on sequence C4 (Wilson et al., 2001), and four methionines and one cysteine were changed to serine to simplify densitometry measurements based on incorporated 35S methionine or cysteine. Peptide tag sequences (single letter code) were: SBP (SDEKTTGWRGGHVVEGLAGELEQLRARLEHHPQGQREPSSSGGSKLG); PC peptide (EDQVDPRLIDGK); HA peptide (YPYDVPDYA); and FLAG peptide (DYKDDDDK). Mutations were introduced by PCR using overlapping primers. In vitro transcription was performed from linearized cDNA constructs using RiboMax T7 Large-Scale RNA Production kit and methyl-7G cap analog (Promega).
ER Microsome Preparation
ER microsomes for in vitro translation reactions were prepared from RPMI 8226 human plasmacytoma cells (ATCC) as described in Supplemental Data (available at http://www.cell.com/cgi/content/full/111/7/967/DC1).
In Vitro Translation and Assembly
All in vitro translation and assembly reactions were performed at 30°C. Each 25 μl reaction contained 17.5 μl nuclease-treated rabbit reticulocyte lysate (RRL) (Promega), 0.5 μl amino acid mixture minus methionine (Promega), 0.5 μl SUPERase-In RNase inhibitor (Ambion), 1.0 μl 35S-labeled methionine (Amersham), 2.0 μl human plasmacytoma ER microsomes, and equimolar amounts of each RNA (60–130 ng each). Reactions were performed with an initial translation period of 15 min at 30°C under reducing conditions (RRL contains DTT), followed by a four-hour assembly period after addition of oxidized glutathione to 4 mM. Reaction volume was adjusted as necessary for optimal signal recovery after IP procedures (typically 25 μl for single-step IP and 50 μl for two- or three-step snIP).
Immunoprecipitation and Analysis
Assembly reactions were diluted with 1 ml ice-cold PBS containing 10 mM iodoacetamide, and membrane fractions were pelleted by centrifugation for 10 min at 20,800 g at 4°C. Membrane pellets were solubilized in 400 μl solubilization/IP buffer (PBS + 1% digitonin, 10 mM iodoacetamide, 0.1% BSA, 5 μg/ml leupeptin, 1 mM PMSF; with 1 mM CaCl2 for anti-PC mAb binding) rotating for 30 min at 4°C. Lysates were filtered using 0.22 μm Spin-X microcentrifuge filter columns (Corning), and precleared for 1 hr with Tris/BSA-blocked Sepharose 4 beads at 4°C. Primary IP was performed overnight at 4°C, and beads were washed twice (0.5 ml PBS/1% digitonin, 10 mM iodoacetamide; with 1 mM CaCl2 for anti-PC mAb). Non-denaturing elution of SA- or anti-PC mAb-captured complexes was performed by incubation in 400 μl solubilization/IP buffer containing 100 μM free biotin or 5 mM EDTA, respectively, for 1 hr at 4°C. Eluted complexes were incubated with subsequent antibodies and Protein G-Sepharose 4 beads (Pharmacia) for 2 hr at 4°C and washed as before. Final precipitates were eluted by boiling in 1% SDS under reducing (three-chain assemblies) or non-reducing (other assemblies) conditions, and most samples were digested for 1 hr at 37°C with 500 U Endoglycosidase H (New England Biolabs). Labeled proteins were separated on 12% SDS-PAGE gels and transferred to PVDF membranes for exposure to phosphor imager plates. Densitometry was performed using ImageQuant software (Molecular Dynamics). Methionine (M) content of TCR-CD3 chains were: TCRα 4M, TCRβ 6M, CD3γ 3M, CD3δ 1M, CD3ε 4M, and ζ-chain 4M.
Supplementary Material
Acknowledgments
We would like to thank Drs. Martin Hemler and Tim Springer for reading of the manuscript; Dr. William Biddison for providing the A6 T cell clone; and Dr. Marilyn Kozak for the modified pSP64 vector. This work was supported by grants AI45757 and NS39096 from NIH (to K.W.W.).
References
- Alcover A, Mariuzza RA, Ermonval M, Acuto O. Lysine 271 in the transmembrane domain of the T-cell antigen receptor β chain is necessary for its assembly with the CD3 complex but not for α/β dimerization. J Biol Chem. 1990;265:4131–4135. [PubMed] [Google Scholar]
- Bijlmakers MJ, Neefjes JJ, Wojcik-Jacobs EH, Ploegh HL. The assembly of H2-Kb class I molecules translated in vitro requires oxidized glutathione and peptide. Eur J Immunol. 1993;23:1305–1313. doi: 10.1002/eji.1830230618. [DOI] [PubMed] [Google Scholar]
- Bijlmakers MJ, Benaroch P, Ploegh HL. Assembly of HLA-DR1 molecules translated in vitro: binding of peptide in the endoplasmic reticulum precludes association with invariant chain. EMBO J. 1994;13:2699–2707. doi: 10.1002/j.1460-2075.1994.tb06560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg RS, Alarcon B, Sancho J, McDermott FV, Lopez P, Breitmeyer J, Terhorst C. Assembly and function of the T cell antigen receptor. Requirement of either the lysine or arginine residues in the transmembrane region of the α chain. J Biol Chem. 1990a;265:14036–14043. [PubMed] [Google Scholar]
- Blumberg RS, Ley S, Sancho J, Lonberg N, Lacy E, McDermott F, Schad V, Greenstein JL, Terhorst C. Structure of the T-cell antigen receptor: evidence for two CD3ε subunits in the T-cell receptor-CD3 complex. Proc Natl Acad Sci USA. 1990b;87:7220–7224. doi: 10.1073/pnas.87.18.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Klausner RD. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell. 1990;63:503–513. doi: 10.1016/0092-8674(90)90447-m. [DOI] [PubMed] [Google Scholar]
- Buferne M, Luton F, Letourneur F, Hoeveler A, Couez D, Barad M, Malissen B, Schmitt-Verhulst AM, Boyer C. Role of CD3δ in surface expression of the TCR/CD3 complex and in activation for killing analyzed with a CD3δ-negative cytotoxic T lymphocyte variant. J Immunol. 1992;148:657–664. [PubMed] [Google Scholar]
- Cosson P, Lankford SP, Bonifacino JS, Klausner RD. Membrane protein association by potential intramembrane charge pairs. Nature. 1991;351:414–416. doi: 10.1038/351414a0. [DOI] [PubMed] [Google Scholar]
- Dietrich J, Neisig A, Hou X, Wegener AM, Gajhede M, Geisler C. Role of CD3γ in T cell receptor assembly. J Cell Biol. 1996;132:299–310. doi: 10.1083/jcb.132.3.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exley M, Terhorst C, Wileman T. Structure, assembly and intracellular transport of the T cell receptor for antigen. Semin Immunol. 1991;3:283–297. [PubMed] [Google Scholar]
- Exley M, Wileman T, Mueller B, Terhorst C. Evidence for multivalent structure of T-cell antigen receptor complex. Mol Immunol. 1995;32:829–839. doi: 10.1016/0161-5890(95)00046-h. [DOI] [PubMed] [Google Scholar]
- Fernandez-Miguel G, Alarcon B, Iglesias A, Bluethmann H, Alvarez-Mon M, Sanz E, de la Hera A. Multivalent structure of an αβ T cell receptor. Proc Natl Acad Sci USA. 1999;96:1547–1552. doi: 10.1073/pnas.96.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- Geisler C. Failure to synthesize the CD3γ chain. Consequences for T cell antigen receptor assembly, processing, and expression. J Immunol. 1992;148:2437–2445. [PubMed] [Google Scholar]
- Germain RN, Stefanova I. The dynamics of T cell receptor signaling: complex orchestration and the key roles of tempo and cooperation. Annu Rev Immunol. 1999;17:467–522. doi: 10.1146/annurev.immunol.17.1.467. [DOI] [PubMed] [Google Scholar]
- Gratkowski H, Lear JD, DeGrado WF. Polar side chains drive the association of model transmembrane peptides. Proc Natl Acad Sci USA. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, Berkhout B, Alarcon B, Sancho J, Wileman T, Terhorst C. Requirements for cell surface expression of the human TCR/CD3 complex in non-T cells. Int Immunol. 1991;3:359–368. doi: 10.1093/intimm/3.4.359. [DOI] [PubMed] [Google Scholar]
- Heinrich SU, Mothes W, Brunner J, Rapoport TA. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- Huppa JB, Ploegh HL. In vitro translation and assembly of a complete T cell receptor-CD3 complex. J Exp Med. 1997;186:393–403. doi: 10.1084/jem.186.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H. Pre-TCR/CD3 and TCR/CD3 complexes: decamers with differential signalling properties? Immunol Today. 1997;18:565–569. [PubMed] [Google Scholar]
- Kearse KP, Roberts JL, Singer A. TCRα-CD3δε association is the initial step in αβ dimer formation in murine T cells and is limiting in immature CD4+ CD8+ thymocytes. Immunity. 1995;2:391–399. doi: 10.1016/1074-7613(95)90147-7. [DOI] [PubMed] [Google Scholar]
- Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 Å resolution. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Punt JA, Roberts JL, Kearse KP, Singer A. Stoichiometry of the T cell antigen receptor (TCR) complex: each TCR/CD3 complex contains one TCRα, one TCRβ, and two CD3ε chains. J Exp Med. 1994;180:587–593. doi: 10.1084/jem.180.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge T, Cosson P, Manolios N, Bonifacino JS, Klausner RD. Transmembrane helical interactions: ζ chain dimerization and functional association with the T cell antigen receptor. EMBO J. 1992;11:3245–3254. doi: 10.1002/j.1460-2075.1992.tb05402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZJ, Kim KS, Wagner G, Reinherz EL. Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3εγ heterodimer. Cell. 2001;105:913–923. doi: 10.1016/s0092-8674(01)00395-6. [DOI] [PubMed] [Google Scholar]
- Sussman JJ, Bonifacino JS, Lippincott-Schwartz J, Weissman AM, Saito T, Klausner RD, Ashwell JD. Failure to synthesize the T cell CD3ζ chain: structure and function of a partial T cell receptor complex. Cell. 1988;52:85–95. doi: 10.1016/0092-8674(88)90533-8. [DOI] [PubMed] [Google Scholar]
- Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas TA, Plow EF, Qin J. A structural mechanism of integrin αIIbβ3 “inside-out” activation as regulated by its cytoplasmic face. Cell. 2002;110:587–597. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- Wegener AM, Hou X, Dietrich J, Geisler C. Distinct domains of the CD3γ chain are involved in surface expression and function of the T cell antigen receptor. J Biol Chem. 1995;270:4675–4680. doi: 10.1074/jbc.270.9.4675. [DOI] [PubMed] [Google Scholar]
- Wilson DS, Keefe AD, Szostak JW. The use of mRNA display to select high-affinity protein-binding peptides. Proc Natl Acad Sci USA. 2001;98:3750–3755. doi: 10.1073/pnas.061028198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat Struct Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Polar residues drive association of polyleucine transmembrane helices. Proc Natl Acad Sci USA. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.