Abstract
Understanding the function of individual microRNA (miRNA) species in mice would require the production of hundreds of loss-of-function strains. To accelerate analysis of miRNA biology in mammals, we combined recombinant adeno-associated virus (rAAV) vectors with miRNA `Tough Decoys' (TuDs) to inhibit specific miRNAs. Intravenous injection of rAAV9 expressing anti-miR-122 or anti-let-7 TuD depleted the corresponding miRNA and increased its mRNA targets. rAAV producing anti-miR-122—but not anti-let-7—TuD reduced serum cholesterol by >30% for 25 weeks in wild-type mice. High throughput sequencing of liver miRNAs from the treated mice confirmed that the targeted miRNAs were depleted and revealed that TuD RNAs induce miRNA tailing and trimming in vivo. rAAV-mediated miRNA inhibition thus provides a simple way to study miRNA function in adult mammals and a potential therapy for dyslipidemia and other diseases caused by miRNA deregulation.
MicroRNAs (miRNAs) repress the expression of mRNAs with which they can partially base pair; miRNAs are predicted to regulate more than half of all protein-coding genes in mammals1,2, but few miRNA-target interactions have been experimentally validated, especially in vivo.
Genetic disruption of a miRNA gene provides a powerful strategy to study miRNA function, but many miRNA genes share the same seed sequence—the 6–8 nt miRNA region that defines its target repertoire—and therefore one member of a miRNA family may compensate for loss of another. Creating an animal model in which all members of a miRNA family are deleted is daunting. Moreover, humans and mice share more than 276 miRNAs, requiring hundreds of conditional knockout strains to assess the function and contribution to disease of each conserved miRNA in adult mice. Currently, chemically modified oligonucleotides (AMOs) complementary to mature miRNAs are widely used to inhibit miRNAs in vitro and in vivo3–5. Effective AMOs typically employ expensive or proprietary modifications such as 2′-O-methyl, 2′-O-methoxyethyl, or 2′,4′-methylene (LNA) and require repeated administration to suppress expression of the cognate miRNA5–8. Current chemistries and formulations do not permit delivery of AMOs to many tissues or organs.
Plasmid DNA vectors that express miRNA “sponges”—multiple, tandem miRNA binding sites designed to competitively inhibit miRNA function—have been used in cultured cells9 and in flies10. Depletion of miR-223 using a sponge-expressing lentiviral vector to stably modify hematopoietic stem cells ex vivo, followed by bone marrow reconstitution in mice, produced a phenotype similar to that observed in a genetic miRNA knockout11. Lentiviral vectors delivered by stereotaxic injection into the brain have been used to express miRNA-inhibiting sponges to study miRNA function in neurons in vivo12. However, the risk of insertional mutagenesis and the requirement for ex vivo manipulation may limit the use of lentiviral vector-based miRNA inhibition for functional genomics studies and human therapy.
Delivery of miRNA inhibitors using the 4.7 kb single-stranded DNA parvovirus Adeno-associated virus (AAV) promises to circumvent many of the risks associated with lentiviral vectors13. In the past decade, new recombinant AAV (rAAV) vectors have been created from natural AAV serotypes, providing efficient gene transfer vehicles that target diverse tissues in mice and non-human primates14. Local delivery by direct injection of AAV2-expressed miRNA sponges into the eye allowed Filipowicz and coworkers to inhibit miR-96, miR-182, and miR-183 in the mouse retina15. The same miRNA cluster was inhibited by expressing sponges from a transgene specifically transcribed in the mouse retina16. More recently, compact, RNA polymerase III-driven miRNA decoys have been reported, including “Tough Decoy” (TuD) RNAs and miRZips, both of which inhibit miRNA function in cultured cells and in vivo17,18. The relative potencies of these inhibitors and their ability to stably and efficiently inhibit miRNAs over the long-term in adult mammals have not yet been tested.
Here, we report proof-of-concept studies using TuDs delivered systemically in mice via rAAV vectors. Each TuD depleted the targeted miRNA and increased the expression of the corresponding miRNA target genes. miRNA depletion in vivo was accompanied by the 3′ addition of non-templated nucleotides as well as 3′-to-5′ shortening of the miRNA, a degradation pathway previously observed in flies and transformed, cultured human cells19. Our data suggest that rAAV-expressing TuD RNAs could enable the study of miRNA functions in adult mammals and perhaps even form the basis for stable therapy for hypercholesterolemia and other disorders caused by aberrant miRNA expression.
Results
TuDs inhibit miRNA function more effectively than sponges or miRZips
To test the efficacy of transcribed sponges12, TuD RNAs25 (Supplementary Fig. 1) and miRZips9,17,20, we targeted the abundant miRNA miR-122, which regulates cholesterol biosynthesis, and the anti-oncogenic miRNA let-7 (Supplementary Table 1). miRNA sponges were expressed using the RNA polymerase II SV40 promoter, the liver-specific TBG promoter or the RNA polymerase III U6 promoter; the U6 promoter was used to drive TuD and miRZip expression (Fig. 1a,b).
Figure 1.
Comparison of miR-122 inhibitor strategies in cultured cells. (a) miRNA inhibitor constructs. (b) Pairing of inhibitors (black) to miR-122 (red). (c) Plasmid harboring nLacZ reporter gene with one or three sites complementary to miR-122 was co-transfected into HuH-7 cells with pTBG-driven firefly luciferase (FLuc) and either control, anti-miR-122 sponge or U6-driven anti-miR-122 TuD plasmid. The cells were stained for nLacZ expression 48 h after transfection, and the proportion of blue cells were counted and reported relative to a control reporter lacking miR-122-binding sites. n = 3 independent experiments. (d) Reporter plasmid expressing nLacZ mRNA containing 3 miR-122-binding sites was cotransfected into HuH-7 cells with a U6-driven sponge-, miRZip-, TuD-expressing, or empty (control) plasmid. n = 4 . (e) HEK 293 cells were transfected with an nLacZ reporter plasmid containing three fully complementary miR-122-binding sites together with constructs expressing anti-let-7 or anti-miR-122 TuD transcribed from a U6 promoter or anti-miR-122 sponge or anti-let-7 sponge transcribed from an SV40 promoter, as well as different amounts of a plasmid producing pri-miR-122 RNA. Percentages of nLacZ positive cells relative to the control (nLacZ without miR-122-binding sites), were determined after 48 h (c, d, and e). n = 3. (f) HuH-7 cells were transfected with plasmid expressing control Renilla reniformis luciferase (RLuc) and FLuc bearing seven miR-122-binding sites or seven mutant sites; these were either alone or in the presence of plasmid expressing anti-miR-122, anti-let-7 or a scrambled TuD RNA control. Luciferase activity was assayed after 24 h and is presented as the mean ratio of RLuc to Fluc ± s.d. (n = 3). (g and h) Evaluation of let-7 inhibitor constructs in HeLa cells. Total RNA and protein were prepared from HeLa cells transfected with plasmids expressing either anti-miR-122 or anti-let-7 TuD or anti-let-7 sponge or control plasmid. Relative Dicer mRNA abundance was measured by qRT-PCR (g) and Dicer protein abundance by western blotting (h). Values are mean ± s.d. (n = 3).
For each miRNA inhibitor, we measured its ability to de-repress a nuclear-targeted β-galactosidase (nLacZ) reporter mRNA containing one or three fully complementary miR-122-binding sites in its 3′ untranslated region (UTR). Reporter expression was reduced by ~50% when one miR-122-binding site was present in the nLacZ 3′ UTR and >80% when three sites were present (Fig.1c). We co-transfected the nLacZ reporter plasmid with each miR-122 inhibitor construct or with a control plasmid into HuH-7, a human hepatoma cell line expressing ~16,000 miR-122 molecules per cell21. Among the RNA polymerase II-driven anti-miR-122 sponges, only the strong liver-specific TBG promoter increased expression of nLacZ bearing a single miR-122-binding site, indicating that the sponge partially inhibited miR-122. However, nLacZ expression was not significantly increased by the sponge when the reporter contained three miR-122-binding sites (Fig. 1c), suggesting that the change in miR-122 activity or concentration was too small to overcome the greater repression conferred by three sites.
In contrast, both the one- and three-site reporters were de-repressed by the RNA polymerase III-driven anti-miR-122 TuD RNA. For the one-site reporter, the TuD restored nLacZ expression to that observed when no miR-122 target sites were present in the reporter (Fig. 1c). To remove potential promoter differences, we compared three U6-driven miR-122 antagonist constructs—sponge, TuD, and miRZip—using the nLacZ reporter containing three miR-122-binding sites. (Sponges transcribed by RNA polymerase III were previously shown to be no more effective than those transcribed by RNA polymerase II, perhaps because the RNA polymerase III transcript accumulates in the nucleus rather than the cytoplasm9.) Again, only the TuD significantly (p-value < 0.001) de-repressed nLacZ repression by miR-122 in HuH-7 cells (Fig. 1d). The anti-miR-122 TuD expression construct but not an anti-let-7 TuD or an anti-miR-122 or anti-let-7 sponge, was similarly effective in human embryonic kidney (HEK) 293 cells artificially expressing pri-miR-122 from a co-transfected plasmid (Fig. 1e).
miRNAs that are extensively complementary to their targets direct Argonaute2 protein to cleave the mRNA, whereas less extensive complementarity generally decreases mRNA stability. To test if the TuD RNA can also inhibit repression directed by a miRNA with imperfect complementarity to its target, we designed a firefly luciferase (Fluc) reporter with seven copies of a bulged miR-122-binding site in its 3′ UTR; FLuc with seven mutant sites served as a control (Supplementary Table 1). The miR-122-responsive FLuc reporter, anti-miR-122, anti-let-7 or control TuD plasmid, and, as an internal control, a Renilla reniformis luciferase (RLuc) plasmid, were introduced into HuH-7 cells by transfection. The anti-miR-122 TuD, but not the control or anti-let-7 TuDs, fully de-repressed FLuc expression (Fig. 1f). The TuD inhibitors were also specific: the anti-let-7, but not the anti-miR-122, TuD increased expression of both Dicer mRNA and protein; Dicer is an endogenous let-7 target (Fig. 1g,h and Supplementary Fig. 2). Our in vitro data suggest that among the three miRNA antagonists we evaluated in vitro—sponge, TuD, and miRZip—a TuD transcribed from a U6 promoter is the most potent miRNA antagonist.
Monitoring miRNA function in living mice
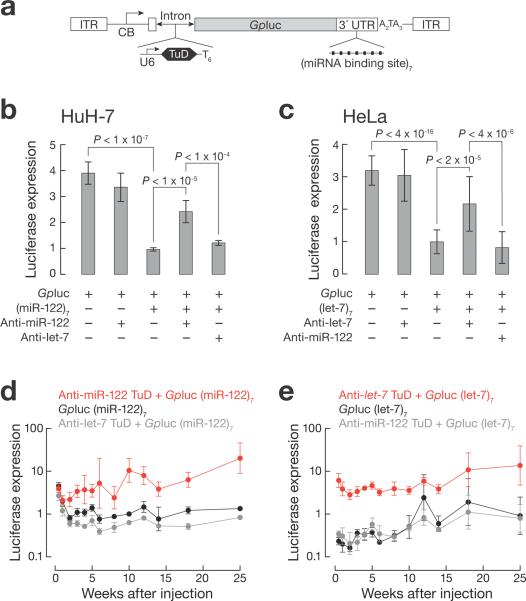
To test the ability of TuD RNAs to inhibit miRNA function in vivo, we constructed a series of rAAV vector genomes expressing a miRNA-responsive Gaussia princeps (Gp) luciferase reporter gene22 (Fig. 2a). GLuc is a secreted protein, enabling detection of the reporter in the blood or urine of live animals. We added seven bulged miR-122 or let-7 target sites to the 3′ UTR of the GLuc mRNA to render it miRNA-responsive and inserted the U6 promoter-driven TuD cassette targeting either miR-122 or let-7 into the GLuc intron. Reporters lacking either the seven miRNA-binding sites, the TuD expression cassette or both served as controls. In HuH-7 cells, the anti-miR-122, but not anti-let-7, TuD de-repressed the reporter bearing seven miR-122-binding sites (Fig. 2b). Similarly, in HeLa cells only the anti-let-7 TuD RNA de-repressed the reporter bearing seven let-7-binding sites (Fig. 2c).
Figure 2.
Real-time monitoring of endogenous miRNA activity using miRNA sensor system. (a) Schematic presentation of GLuc-expressing vectors. CB, chicken β actin promoter with CMV enhancer. AAV vector plasmids were transfected into HuH-7 (n = 6 independent trials; b) or HeLa cells (n ≥ 17; c). GLuc activity was measured after 48 h. (d, e) C57BL/6J mice were administered 1 × 1012 genome copies of scAAV9 per animal by tail vein injection. Blood was collected at the indicated times and assayed for GLuc activity, reported as mean ± s.d., relative to injected GLuc vector lacking both the TuD expression cassette and 3′ UTR miRNA-binding sites. n = 4 for each group.
Both miR-122 and let-7 are present in liver21, and let-7 is also found in heart23. miR-122 comprises 70% of total liver miRNA21, posing a stringent test for the ability of TuDs to inhibit the function of even the most abundant miRNA species. We packaged the rAAV genomes into the AAV9 capsid, which preferentially transduces liver and heart. To further improve transduction, all rAAVs were prepared as self-complementary (sc) genomes24. We administered the vectors intravenously to adult male C57B/6 mice and monitored GLuc activity in blood.
Initially, GLuc activity was comparable among the animals injected with vectors expressing the miR-122-regulated reporters, irrespective of the presence of a TuD RNA expression cassette (days 3 and 7). By week 2, GLuc activity declined in mice that received vectors lacking the anti-miR-122 TuD, while it increased in mice treated with the anti-miR-122 TuD-expressing vector (Fig. 2d). Similarly, GLuc activity was low in mice that received the let-7-regulated reporter and high in mice that received the same reporter containing the anti-let-7 TuD cassette (Fig. 2e). One notable difference was that the let-7-regulated reporter was silenced at the earliest time point (day 3), whereas the miR-122-regulated reporter showed an initial lag (Fig. 2d,e); we do not currently understand the source of this difference. De-repression of GLuc expression by either anti-miR-122 or anti-let-7 TuD RNA was sustained for the 25-week duration of the study, (Fig. 2d,e), during which we detected no significant decrease in AAV genome copies (Supplementary Fig. 3).
scAAV9-delivered TuDs target miRNAs for destruction in mice
Four weeks after scAAV9 administration, we analyzed miRNA expression in the liver using quantitative RT-PCR. We observed an ~80% reduction in miR-122 in mice that received the anti-miR-122 TuD vector compared to anti-let-7 TuD or control lacking a TuD (Fig. 3a). Northern hybridization confirmed the reduction of miR-122 in mice that received anti-miR-122 TuD (Fig. 3b and Supplementary Fig. 4). let-7 was similarly reduced in mice treated with anti-let-7 TuD (Fig. 3b and Supplementary Fig. 4). (Our let-7 northern probe cannot distinguish among the 9 mouse let-7 isoforms.) In contrast, no reduction was detected for miR-26a or miR-22, two abundant liver miRNAs (Fig. 3b and Supplementary Fig. 4).
Figure 3.
Analysis of miRNA expression in liver from mice administered scAAV9 expressing anti-miRNA TuD. C57BL/6J mice were injected via tail vein with 1 × 1012 genome copies of control, anti-miR-122 or anti-let-7 TuD expressing vectors. Animals were sacrificed four weeks later, and the abundance of let-7, miR-122, miR-26a, miR-22 and U6 in total liver RNA was measured by qRT-PCR (a; n = 3 per group) and northern hybridization (b; n = 4 per group). Values are mean ± s.d. U6 RNA provided a loading control. (c, d) High-throughput sequencing of total liver small RNA was used to determine the length and abundance of genome-matching (c) or prefix-matching (d) miR-122 isoforms four weeks after injection. The most abundant non-genome matching nucleotides added to the 3′ end of miR-122 fragments are indicated in the grey boxes.2(e) Nucleotide differences among the nine let-7 paralogs expressed in liver are indicated in black and their pairing to the TuD RNA is shown. The seed sequence, which determines miRNA target specificity, is underlined. The TuD targeting let-7 decreased the abundance of full-length let-7 and increased the abundance of prefix-matching let-7 sequence reads relative to the control. Isoforms are shown in black whose genome-matching reads decreased and whose prefix-matching reads increased. MM: total number of mismatches between each let-7 paralog and the TuD.
High-throughput sequencing of miRNAs from the treated livers further supports the view that scAAV9-delivered TuDs effectively and specifically trigger the destruction of complementary miRNAs. The TuD targeting miR-122 (Fig. 1b) reduced the abundance of full-length, 23 nt miR-122 by 4.3-fold (Fig. 3c), consistent with our qRT-PCR results (Fig. 3a). The 21 and 22 nt miR-122 isoforms decreased less, whereas 20, 19 and 18 nt isoforms increased, suggesting that the TuD triggered 3′-to-5′ exonucleolytic trimming of miR-122 (Fig. 3c). Like antagomir-directed destruction of miRNAs in cultured human cells19, the anti-miR-122 TuD promoted the addition of non-templated nucleotides to the 3′ end of miR-122 (Fig. 3d). Prefix-matching reads—sequences that initially match the mouse genome but end with non-templated nucleotides—doubled in the anti-miR-122 TuD mouse compared to the control (Fig. 3d). The 3′ non-templated nucleotides comprised one or more adenosines. Even in the absence of the TuD, 21% of miR-122 was tailed with adenosine, suggesting that miR-122 is modified after it binds to Argonaute proteins as part of its natural turnover. Natural tailing of miR-122 is predominantly associated with the addition of a single terminal, non-templated adenosine (89% of all non-templated nucleotides in the control); TuD RNA increased the occurrence of terminal, non-templated adenosines from 3% to 11% for two adenosines and from 0.6% to 7% for three.
Mouse liver expresses nine let-7 isoforms (Supplementary Fig. 5). These differ by 1–4 nucleotides outside their common seed sequence (Fig. 3e). The TuD targeting let-7 most strongly decreased the abundance of those full-length let-7 isoforms that were fully complementary to the TuD sequence (let-7a, 12.1-fold) or contained only a single non-seed mismatch (let-7c, 5.1-fold; let-7d, 5.0-fold; let-7f, 11.0-fold; and miR-98, 6.4-fold). In contrast, the decrease was smaller for let-7b (1.6-fold) and let-7g (2.7-fold), both of which contain two 3′ mismatches to the TuD, let-7i (1.5-fold), which contains three 3′ mismatches, and let-7e (3.6-fold), which contains a purine:purine mismatch at position 9, immediately flanking the seed sequence (Fig. 3e). Relative to genome-matching reads, prefix-matching reads increased more for let-7a, c, d, and f and miR-98 (miR-98 is a let-7 isoform)—the let-7 isoforms that decreased the most in response to the anti-let-7 TuD—whereas let-7 b, e, g and i, which decreased least, showed no increase in trimmed-and-tailed species (Fig. 3e). For both the anti-miR-122 and anti-let-7 TuDs, the overall abundance of miRNAs was unaltered (Supplementary Fig. 6). (We note that qRT-PCR, northern hybridization, and high-throughput sequencing report different apparent changes in miRNA levels upon TuD expression, because each measures a different set of isoforms of the mature miRNA (Fig. 3, Supplementary Fig. 7, and Supplementary Note 1).)
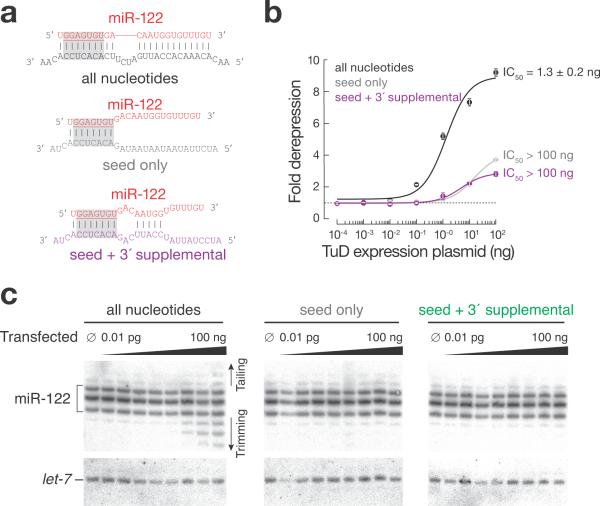
Our data suggest that TuDs act, at least in part, by targeting miRNAs for destruction by recruiting the tailing and trimming pathway to decrease their steady-state abundance. An alternative view is that the primary effect of highly abundant TuD RNA is to titrate miRNAs away from their endogenous mRNA targets, whereas miRNA decay is a secondary effect. To differentiate between these possibilities we transfected HuH-7 cells with a plasmid expressing a TuD complementary to the entire miRNA (“all nucleotides”); to only the miR-122 seed (nucleotides 2–8; “seed only”); or to the seed plus an additional region of complementarity from nucleotides 12–15, a region previously shown to enhance miRNA function25 (“seed + 3′ supplemental pairing”) (Fig. 4a). miRNA tailing and trimming requires extensive complementarity between the miRNA and its target or AMO inhibitor19. We monitored miRNA activity using an RLuc reporter bearing three miR-122 binding sites (Fig. 4b). The fully complementary TuD efficiently de-repressed the miR-122-regulated luciferase reporter achieving half-maximal inhibition (IC50) at 1.3 ± 0.2 ng transfected plasmid. In contrast, we were unable to transfect sufficient plasmid to reach half-maximal inhibition for the other TuD inhibitors; the seed only or the seed plus 3′ supplemental pairing inhibitors were >60 times less effective than the fully complementary TuD. Moreover, only the fully complementary TuD triggered the production of tailed and trimmed miR-122 isoforms (Fig. 4c). We conclude that TuDs act, at least in part, by targeting miRNAs for destruction via the tailing and trimming pathway.
Figure 4.
Analysis of TuD-directed inhibition of miR-122 in HuH-7 cells. (a) Anti-miR-122 TuDs used. (b) Dose-response curves for inhibition of miR-122 function in HuH-7 cells. Cells were transfected with constructs expressing the TuDs, together with a psiCHECK-2 reporter plasmid bearing three sites complementary to miR-122. Values are mean ± s.d. (n = 3). (c) Northern hybridization analysis of the experiment in (b).
TuDs delivered with scAAV9 increase expression of endogenous miRNA-regulated mRNAs
TuD inhibitors delivered using scAAV9 de-repressed miR-122- and let-7-regulated mRNAs in vivo (Fig. 5). We used qRT-PCR (Fig. 5a) and western blotting (Fig. 5b and Supplementary Fig. 8) to analyze the expression of known miR-122 and let-7 targets in liver and heart four weeks after injection of the scAAV9 vectors. Mice injected with scAAV9 expressing the GLuc reporter but no TuD served as a control. For anti-miR-122 TuD-treated mice, we detected in the liver a 2.5 to 4.2-fold increase in Aldolase A (3.3 ± 0.5; p-value < 0.04), Tmed3 (4.2 ± 1.5; p-value < 0.01), Hfe2 (3.3 ± 1.0; p-value < 0.02), and Cyclin G1 (2.5 ± 0.4; p-value < 0.001) mRNAs, all previously shown to be regulated by miR-122; expression of these was unaltered in the heart, which lacks miR-122 (Fig. 5a). We found no significant change in these four miR-122-regulated mRNAs in either liver or heart from mice that received the vector expressing anti-let-7 TuD RNA (Fig. 5a). We also detected a significant increase in Hfe2 (1.5 ± 0.1; p-value < 0.02) and Tmed3 (1.6 ± 0.1; p-value < 0.008) proteins in the livers of anti-miR-122 TuD-treated mice (Fig. 5b and Supplementary Fig. 8).
Figure 5.
Expression in TuD-treated mice of previously validated regulatory targets of miR-122 and let-7. scAAV9 vectors (control, TuD targeting miR-122 or let-7) were injected by tail vein into C57BL/6J mice (1 × 1012 genome copies each). The mice were sacrificed four weeks later and the expression of representative endogenous targets of miR-122 (Aldolase A, Cyclin G1, Tmed3, Hfe2, and Cat-1 mRNA) and let-7 (Kras, Hras, Nras, and Dicer mRNA) analyzed in liver (left) or heart (right) by qRT-PCR (a) or in liver by western blotting (b). Values are mean ± s.d. comparing TuD-treated to control scAAV-treated mice; each group used four (a) or three (b) mice.
The mRNA encoding the miRNA-producing enzyme Dicer is itself repressed by let-7. We used qRT-PCR to measure Dicer mRNA abundance in mice that received scAAV9 vector expressing either anti-miR-122 or anti-let-7 TuD RNA (Fig. 5a). When let-7 was inhibited, Dicer mRNA increased in both liver (1.9 ± 0.2; p-value ≤ 0.001) and heart (2.4 ± 0.4; p-value ≤ 0.003). Similarly, Dicer protein levels increased in the liver (2.0 ± 0.6; p-value < 0.05; Fig. 5b). (We did not examine protein levels in the heart.) The RAS family genes Hras1, Nras and Kras have also been reported to be repressed by let-7. We detected increased expression of Nras in both liver (1.3 ± 0.1; p-value ≤ 0.01) and heart (1.3 ± 0.1; p-value ≤ 0.02) and of Hras1 (1.3 ± 0.1; p-value ≤ 0.04) in heart in anti-let-7-treated, but not anti-miR-122 TuD–treated mice (Fig. 5a).
Anti-miR-122 TuD RNA reduces cholesterol levels
miR-122 is required for normal cholesterol biosynthesis; inhibition of miR-122 with AMOs decreases cholesterol metabolism in adult mice5,8 and non-human primates6,7. In wild-type mice, a single intravenous injection of scAAV9 expressing anti-miR-122 TuD significantly reduced total serum cholesterol (45 ± 5%; p-value ≤ 0.001) and high-density lipoprotein (HDL; 42 ± 5%; p-value ≤ 0.001) beginning two weeks after injection, and this reduction was sustained for the 25-week duration of the study. LDL levels also decreased (88 ± 102%; p-value ≤ 0.05) by the third week and remained depressed throughout the study (Fig. 6a). Total serum cholesterol, HDL, and LDL levels were unaltered in mice that received the anti-let-7 TuD. We detected no significant weight loss (Supplementary Fig. 9) or increase in serum alanine aminotransferase (ALT) or aspartate aminotransferase (AST; Fig. 6b) over the course of 25 weeks.
Figure 6.
Change in cholesterol profiles of wild-type C57BL/6J mice after administration of scAAV expressing both the TuD targeting miR-122 and the GLuc reporter bearing miR-122-binding sites, relative to control GLuc reporter lacking the TuD and the miR-122 binding sites. (a) Four-to-six week–old male wild-type C57BL/6J mice were intravenously injected with 1 × 1012 genome copies of scAAV9 per mouse. Serum levels of total cholesterol, high-density lipoprotein (HDL) and low-density lipoprotein (LDL) were measured at different times after injection. (b) The serum transaminases aspartate, aminotransferase (ASL) and alanine aminotransferase (ALT) were assayed to assess liver toxicity. Values are mean ± s.d.; n = 4, control and anti-miR-122 TuD groups and n = 3, anti-let-7 TuD group.
DISCUSSION
Inhibitors of miRNA function promise to accelerate the understanding of miRNA functions in mammals, especially in adults. Chemically modified oligonucleotide miRNA inhibitors are effective, but are currently expensive, require repeated dosing that risks long-term toxicity, and many tissues are not currently accessible to delivery of oligonucleotides.
Transcribed miRNA-binding RNAs provide an alternative to oligonucleotides. Their small size makes them readily incorporated into a variety of gene transfer vectors. Primate AAV-derived vectors represent attractive tools for this application because of their unique tissue tropism, high efficiency of transduction, stability of in vivo gene transfer, and low toxicity.
We compared several designs of miRNA antagonists in vitro, then used the most effective design, TuDs, in vivo to inhibit miR-122 and let-7 by incorporating TuD expression cassettes into scAAV9. A single administration of rAAV9 expressing a TuD RNA efficiently depleted the targeted miRNA (Fig. 3) and increased the abundance of its endogenous target mRNAs (Fig. 1 and 5), which in turn produced the predicted phenotypic change in metabolism (Fig. 6).
Our data suggest that, in mice, TuD RNAs inhibit their miRNA targets via the target-RNA directed tailing and trimming pathway19 (Fig. 3 and 4). This work, in addition to observations in flies and cultured cells, suggests that the pathway is widely conserved among animals. Targeted miRNA destruction triggered by TuDs was surprisingly sequence-specific for the nine distinct let-7 isoforms. Compared to let-7a, for which the TuD was fully complementary, a single purine:purine mismatch between the TuD and let-7e reduced miRNA degradation by >3-fold, and four mismatches (let-7i) reduced miRNA destruction 8-fold. We are therefore optimistic that future studies will reveal the rules for designing TuD inhibitors that target individual miRNA isoforms that differ by just one or two nucleotides.
The GLuc sensor system described here provides a simple means to detect changes in specific miRNA function, such as those caused by miRNA inhibitors, in live adult mammals across time (Fig. 2). This system allows one to assess the activity of a specific miRNA in a cell line, tissue or organ, providing a quantitative measure of the effectiveness of a miRNA antagonist.
Retrospective profiling has linked aberrant miRNA expression to a variety of diseases, suggesting that miRNAs may provide new targets for therapy. Indeed, miR-122 inhibition by AMOs5–8 or scAAV-delivered TuD RNA (Fig. 6) lowers both HDL and LDL. However, the current view that HDL protects against heart attack argues that therapy for dyslipidemia should lower LDL but raise HDL levels. Recently, miR-33 was identified as a repressor of HDL biogenesis; miR-33 inhibition raises serum HDL level18. Perhaps simultaneous inhibition of miR-122 and miR-33 by a pair of TuD RNAs expressed from a single scAAV vector may achieve a more balanced and healthy cholesterol profile and provide long-lasting therapy for familial hypercholesterolemia.
Low miR-122 levels have been associated with hepatocellular carcinoma in rodents and humans26–28, although no direct causal link has been established27,28. Because AAV vector expression is stable for years in rodents and primates, animals treated with scAAV9 expressing anti-miR-122 should enable testing the safety of prolonged miR-122 inhibition in general and the increased risk of developing hepatocellular carcinoma in particular.
Supplementary Material
Acknowledgements
This work was supported in part by an EMBO long term fellowship (ALTF 522-2008) and an Erwin Schrödinger-Auslandsstipendium (Austrian Science Fund FWF, J2832-B09) to S.L.A. and by grants from the National Institutes of Health to P.D.Z. (GM62862 and GM65236), to P.D.Z. and G.G. (UL1RR031982), to the UMass Mouse Metabolic Phenotypic Center (U24-DK093000), and to T.R.F. (P01 DK58327), and from the University of Massachusetts Medical School to G.G. T.R.F., J.K, P.D.Z, and G.G. are members of the UMass DERC, which is supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases (P30 DK32520).
Footnotes
Authors' Contributions J.X. created the miRNA inhibitor constructs, performed the experiments in cultured cells and mice. J.-H.H. S.L.A., and Z.W. analyzed high throughput sequencing data. S.L.A. designed, conducted and analyzed the experiments to study how TuDs inhibit miRNAs. R.F. and J.K.K. measured cholesterol profiles and liver function in the study animals. Y.Z. contributed to most of the anti-miRNA TuD RNA studies in mice. R.H. and Q.X. contributed to vector construction as well as in vitro characterization of miRNA inhibitors. L.Z., M.L., H.L. and X.M. contributed to the miRNA inhibitor studies in mice and tissue sample analyses. Q.S. and R.H. produced the AAV vectors. H.Z. assisted in in vitro characterization. J.B. designed and cloned anti-miRNA sponges. T.R.F. contributed to the development of rAAV delivered miRNA therapeutics for treating hyperlipidemia. J.X., S.L.A., P.D.Z., and G.G. conceived the research. S.L.A., J.X., P.D.Z. and G.G. wrote the manuscript. S.L.A. and P.D.Z prepared the figures.
Competing Financial Interests P.D.Z. is a member of the scientific advisory board of Regulus Therapeutics.
References
- 1.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 2.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 3.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krutzfeldt J, et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 6.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 7.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat. Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentner B, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat. Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 12.Luikart BW, et al. miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS One. 2011;6:e19077. doi: 10.1371/journal.pone.0019077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berns KI, Giraud C. Biology of adeno-associated virus. Curr. To.p Microbiol. Immunol. 1996;218:1–23. doi: 10.1007/978-3-642-80207-2_1. [DOI] [PubMed] [Google Scholar]
- 14.Gao GP, et al. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krol J, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 16.Zhu Q, et al. Sponge transgenic mouse model reveals important roles for the microRNA-183 (miR-183)/96/182 cluster in postmitotic photoreceptors of the retina. J. Biol. Chem. 2011;286:31749–31760. doi: 10.1074/jbc.M111.259028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rayner KJ, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ameres SL, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. www.systembio.com/microrna-research/microrna-knockdown/mirzip/
- 21.Chang J, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 22.Tannous BA. Gaussia luciferase reporter assay for monitoring biological processes in culture and in vivo. Nat. Protoc. 2009;4:582–591. doi: 10.1038/nprot.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sen CK, Gordillo GM, Khanna S, Roy S. Micromanaging vascular biology: tiny microRNAs play big band. J. Vasc. Res. 2009;46:527–540. doi: 10.1159/000226221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarty DM. Self-complementary AAV vectors; advances and applications. Mol. Ther. 2008;16:1648–1656. doi: 10.1038/mt.2008.171. [DOI] [PubMed] [Google Scholar]
- 25.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutay H, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J. Cell. Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai WC, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–1582. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao G, et al. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6081–6086. doi: 10.1073/pnas.0937739100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 32.Gao G, et al. Biology of AAV serotype vectors in liver-directed gene transfer to nonhuman primates. Mol. Ther. 2006;13:77–87. doi: 10.1016/j.ymthe.2005.08.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.