Abstract
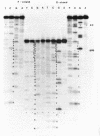
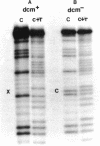
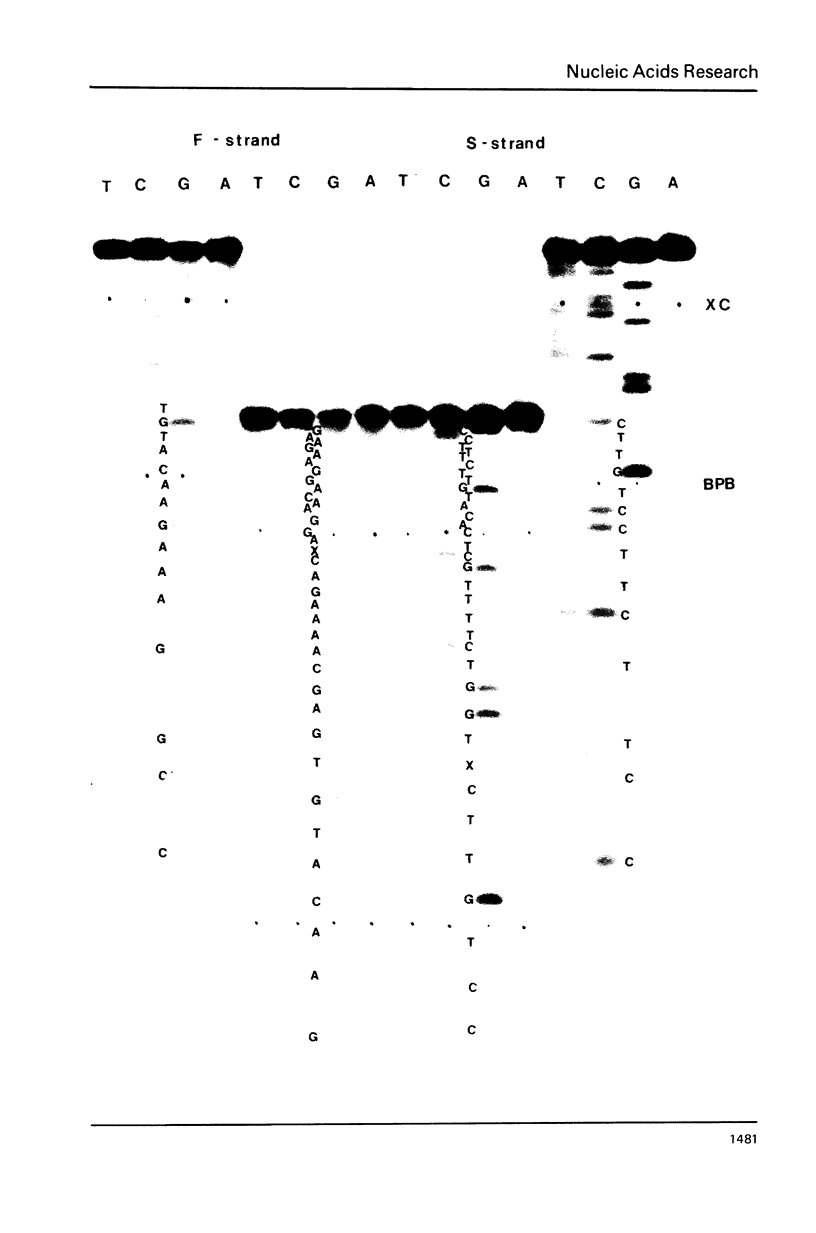
Col E1 DNA has methylated cytosine in the sequence 5'-CC*(A/T)GG-3' and methylated adenine in the sequence 5'-GA*TC-3' at the positions indicated by asterisks(*). When the Maxam-Gilbert DNA sequencing method is applied to this DNA, the methylated cytosine (5-methylcytosine) is found to be less reactive to hydrazine than are cytosine and thymine, so that a band corresponding to that base does not appear in the pyrimidine cleavage patterns. The existence of the methylated cytosine can be confirmed by analyzing the complementary strand or unmethylated DNA. In contrast, the methylated adenine (probably N6-methyladenine) cannot be distinguished from adenine with standard conditions for cleavage at adenine.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bigger C. H., Murray K., Murray N. E. Recognition sequence of a restriction enzyme. Nat New Biol. 1973 Jul 4;244(131):7–10. doi: 10.1038/newbio244007a0. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Chow L. T., Dugaiczyk A., Hedgpeth J., Goodman H. M. DNA substrate site for the EcoRII restriction endonuclease and modification methylase. Nat New Biol. 1973 Jul 11;244(132):40–43. doi: 10.1038/newbio244040a0. [DOI] [PubMed] [Google Scholar]
- Cashmore A. R., Petersen G. B. The degradation of DNA by hydrazine: a critical study of the suitability of the reaction for the quantitative determination of purine nucleotide sequences. Biochim Biophys Acta. 1969 Feb 18;174(2):591–603. doi: 10.1016/0005-2787(69)90289-5. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J., Razin A. Studies on the biological role of DNA methylation. II. Role of phiX174 DNA methylation in the process of viral progeny DNA synthesis. Nucleic Acids Res. 1976 Oct;3(10):2665–2675. doi: 10.1093/nar/3.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelinas R. E., Myers P. A., Roberts R. J. Two sequence-specific endonucleases from Moraxella bovis. J Mol Biol. 1977 Jul;114(1):169–179. doi: 10.1016/0022-2836(77)90290-x. [DOI] [PubMed] [Google Scholar]
- Hughes S. G. The sensitivity of bacteriophage lambda DNA to restriction endonuclease RII. J Mol Biol. 1975 Nov 5;98(3):645–647. doi: 10.1016/s0022-2836(75)80093-3. [DOI] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977 Jul;114(1):153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- Marinus M. G. Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol Gen Genet. 1973 Dec 14;127(1):47–55. doi: 10.1007/BF00267782. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M. S., Hattaman S. Deoxyribonucleic acid-cytosine methylation by host- and plasmid-controlled enzymes. J Bacteriol. 1975 Apr;122(1):129–138. doi: 10.1128/jb.122.1.129-138.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J., Myers P. A., Morrison A., Murray K. A specific endonuclease from Haemophilus haemolyticus. J Mol Biol. 1976 May 5;103(1):199–208. doi: 10.1016/0022-2836(76)90060-7. [DOI] [PubMed] [Google Scholar]
- Schlagman S., Hattman S., May M. S., Berger L. In vivo methylation by Escherichia coli K-12 mec+ deoxyribonucleic acid-cytosine methylase protects against in vitro cleavage by the RII restriction endonuclease (R. Eco RII). J Bacteriol. 1976 May;126(2):990–996. doi: 10.1128/jb.126.2.990-996.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TEMPERLI A., TUERLER H., RUEST P., DANON A., CHARGAFF E. STUDIES OF THE NUCLEOTIDE ARRANGEMENT IN DEOXYRIBONUCLEIC ACIDS. IX. SELECTIVE DEGRADATION OF PYRIMIDINE DEOXYRIBONUCLEOTIDES. Biochim Biophys Acta. 1964 Nov 15;91:462–476. doi: 10.1016/0926-6550(64)90076-3. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. I., Ohmori H., Bird R. E. Origin of replication of colicin E1 plasmid DNA. Proc Natl Acad Sci U S A. 1977 May;74(5):1865–1869. doi: 10.1073/pnas.74.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VERWOERD D. W., ZILLIG W. A specific partial hydrolysis procedure for soluble RNA. Biochim Biophys Acta. 1963 Mar 26;68:484–486. doi: 10.1016/0006-3002(63)90171-9. [DOI] [PubMed] [Google Scholar]