Abstract
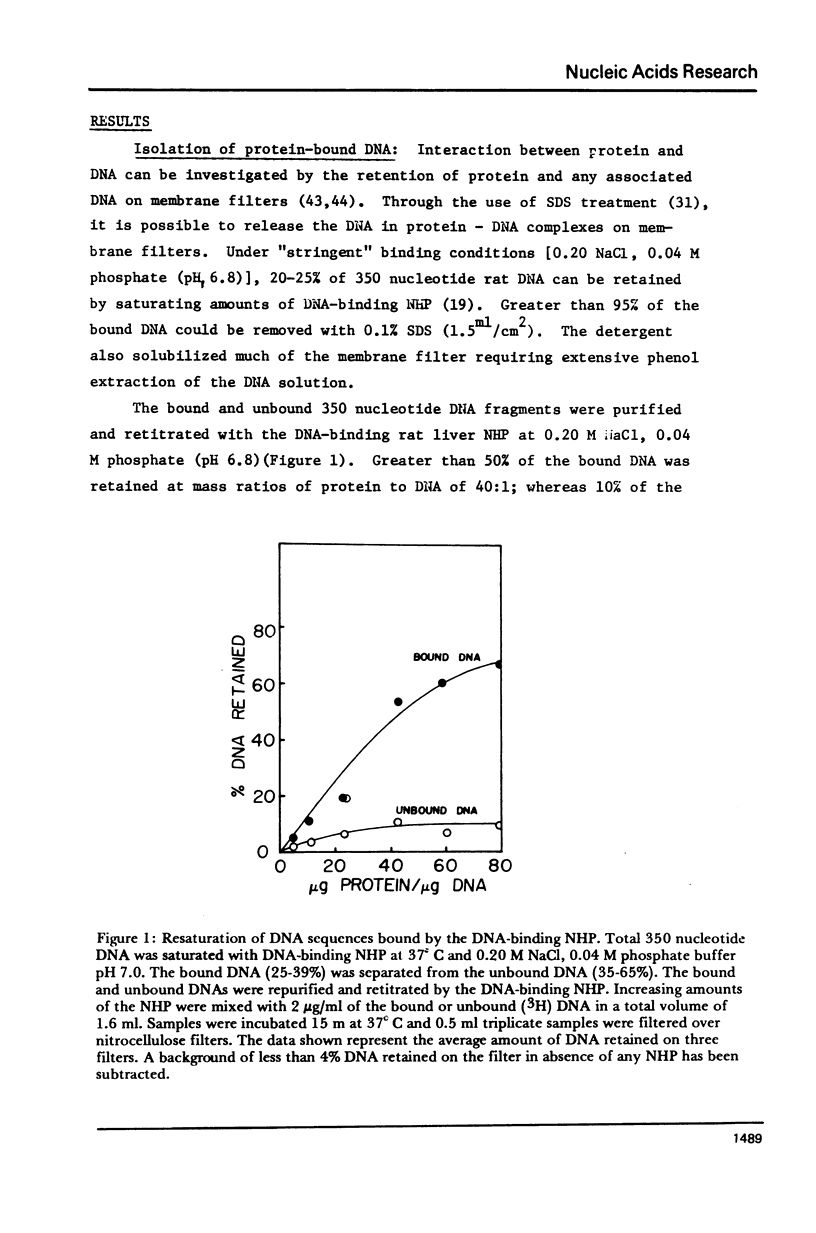
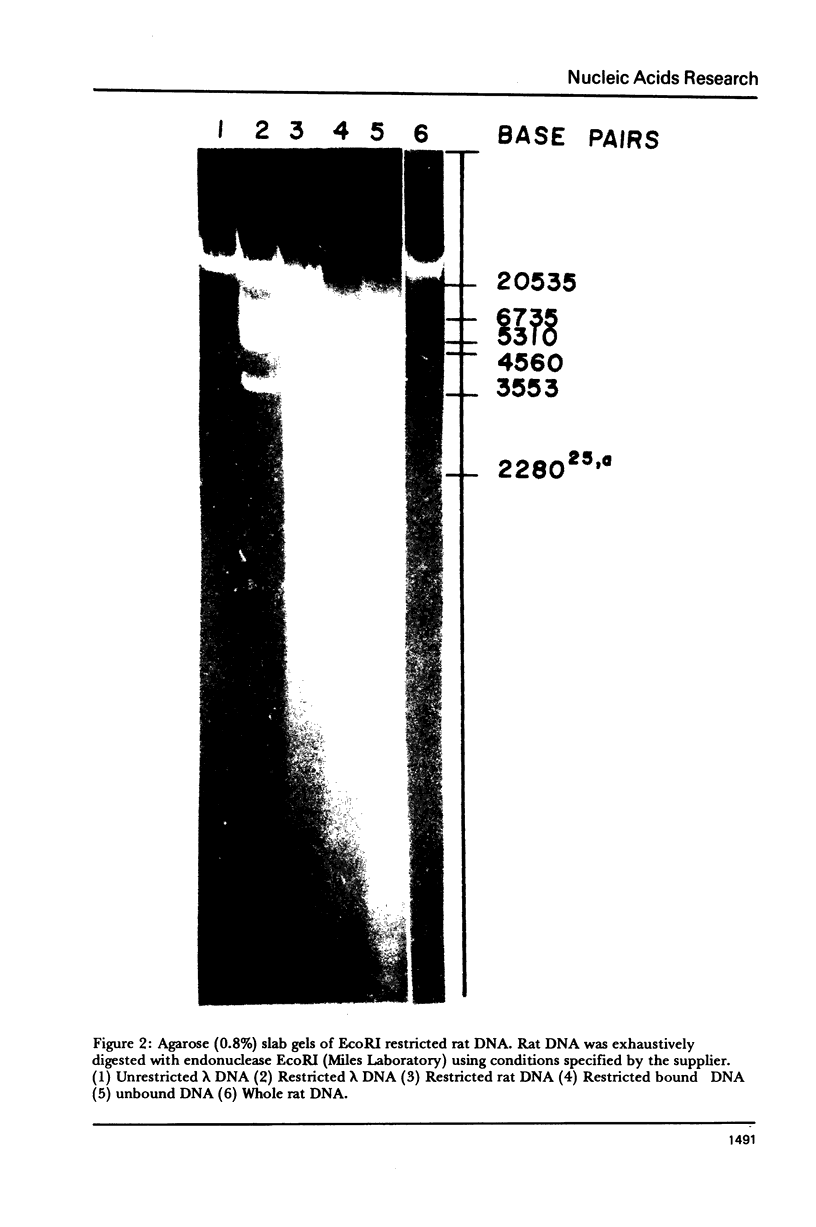
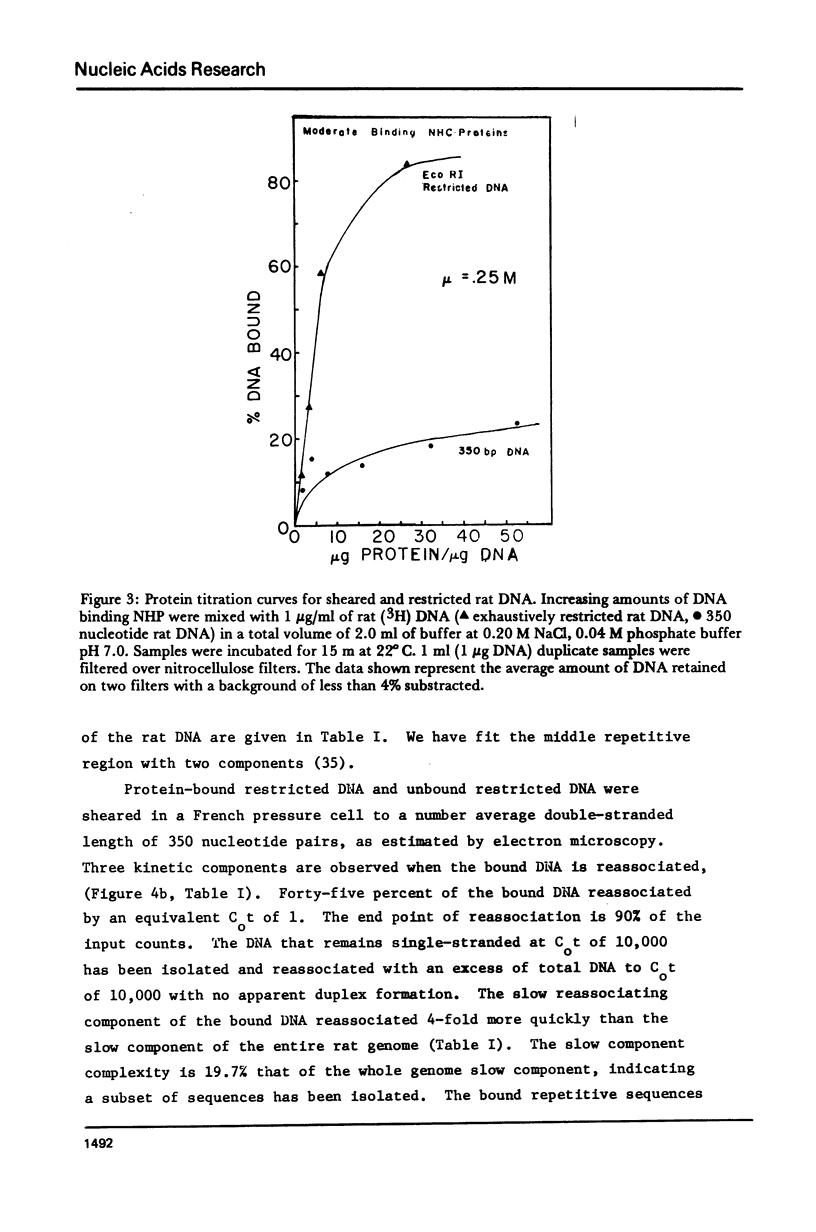
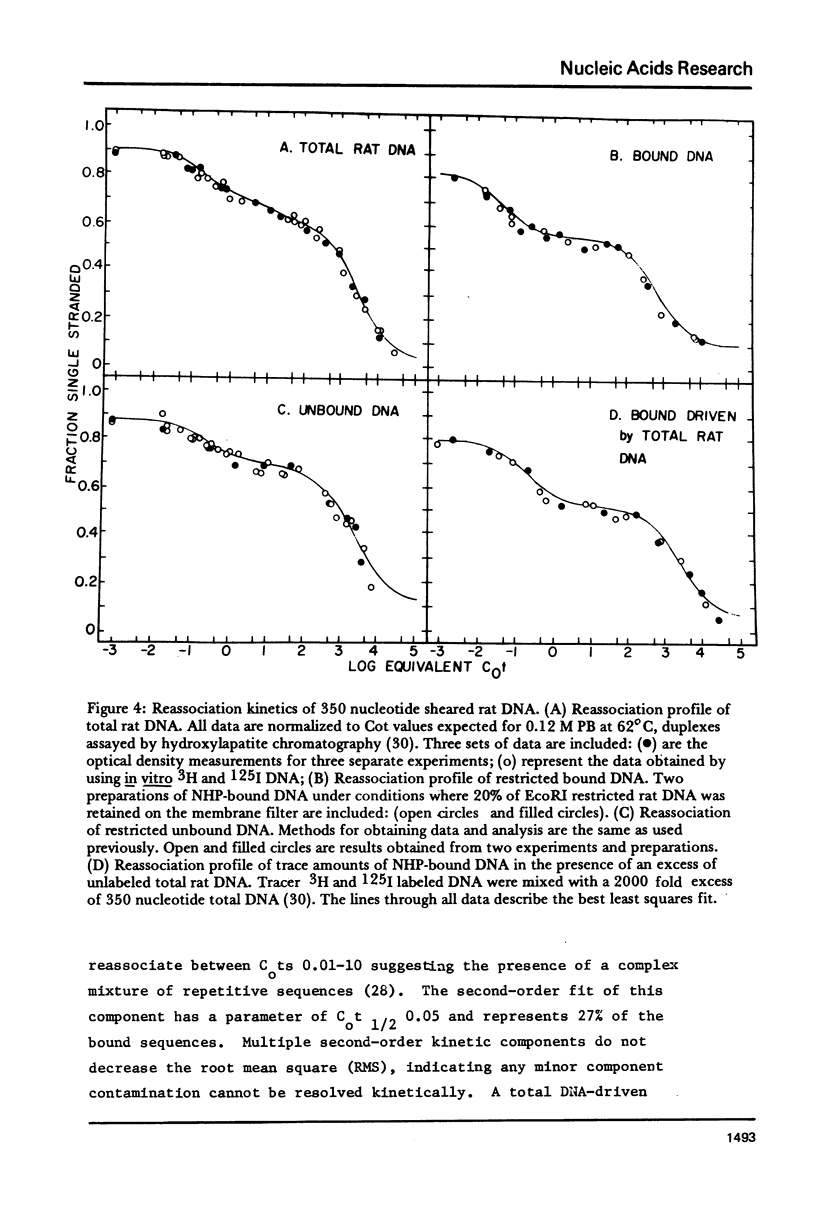
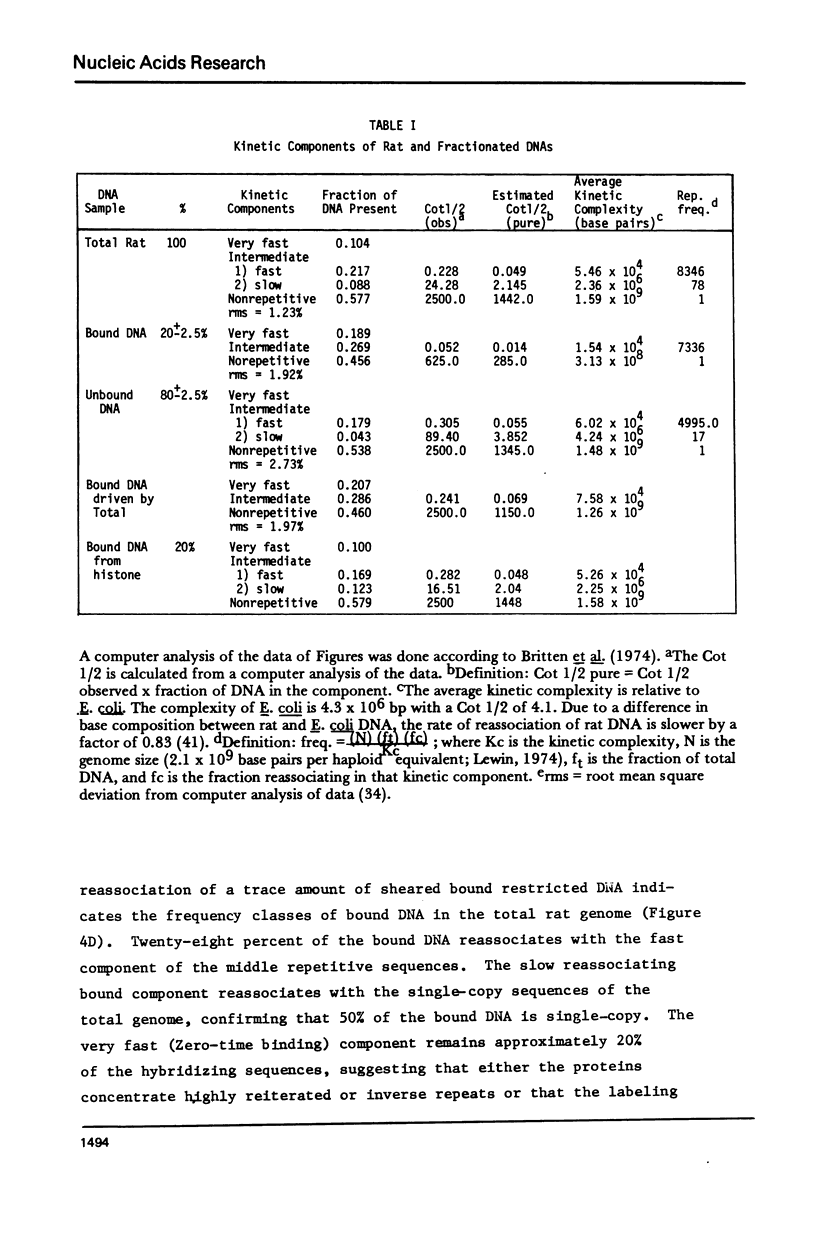
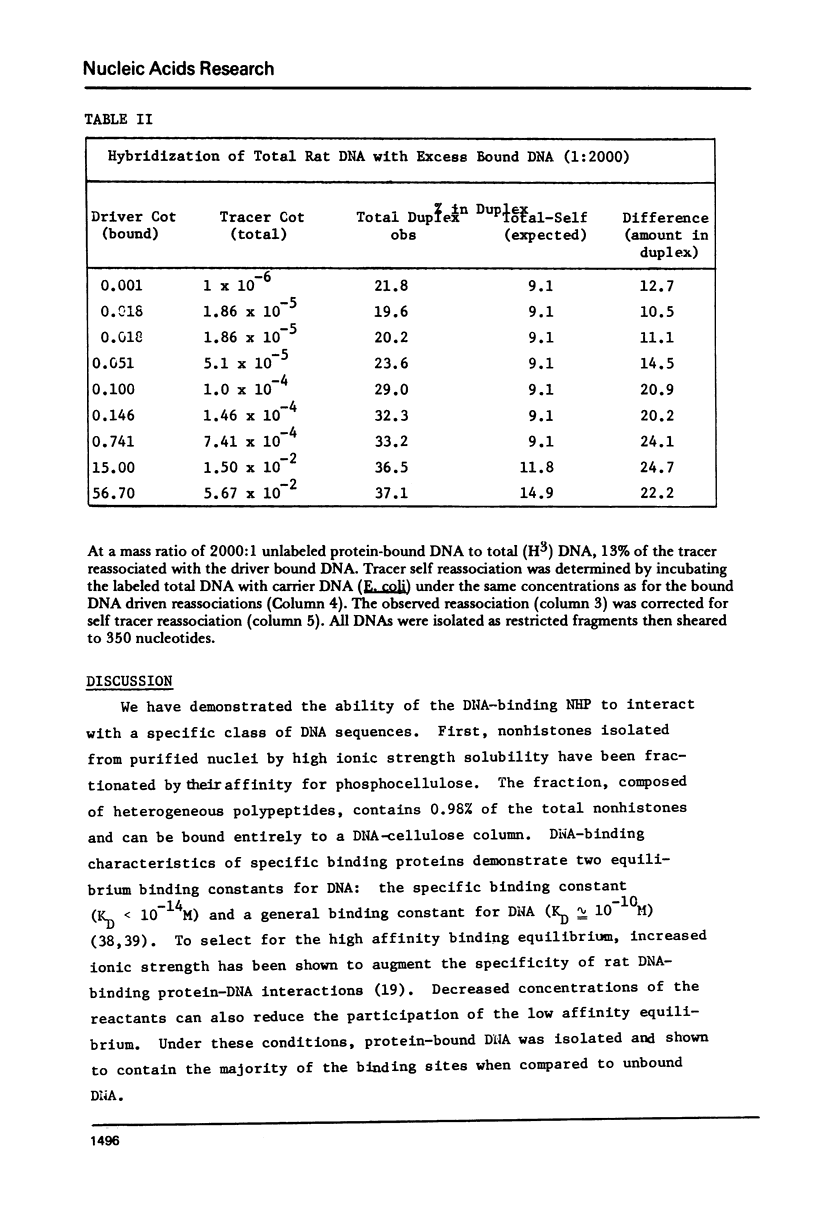
The DNA-binding nonhistone proteins (NHP) have been demonstrated to fractionate the rat genome into protein-bound and unbound DNA sequences. Twenty percent of highly sheared rat DNA [approximately 350 base pair (bp)] can be retained on membrane filters as protein complexes. When extracted from the filter and retitrated with the NHP, a 4- to 5-fold enrichment of binding sites is present in the bound DNA with few, if any, sites detected in the unbound DNA. Rat DNA restricted by EcoRI endonuclease can be fractionated by its DNA-binding NHP retention characteristics. Reassociation kinetics of the bound restricted sequences indicate that 45.6% is a subset of total single-copy sequence of the rat genome an 26.9% is repetitive sequences. Cross hybridization studies indicate the repetitive sequences of the bound DNA are not enriched as much as the slow component of the rat genome. Thus a 4-fold enrichment of a subset of the rat genome has been observed via NHP-DNA interactions.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrett T., Maryanka D., Hamlyn P. H., Gould H. J. Nonhistone proteins control gene expression in reconstituted chromatin. Proc Natl Acad Sci U S A. 1974 Dec;71(12):5057–5061. doi: 10.1073/pnas.71.12.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner T. I., Brenner D. J., Neufeld B. R., Britten R. J. Reduction in the rate of DNA reassociation by sequence divergence. J Mol Biol. 1973 Dec 5;81(2):123–135. doi: 10.1016/0022-2836(73)90184-8. [DOI] [PubMed] [Google Scholar]
- Botchan M., Topp W., Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. doi: 10.1016/0092-8674(76)90118-5. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Commerford S. L. Iodination of nucleic acids in vitro. Biochemistry. 1971 May 25;10(11):1993–2000. doi: 10.1021/bi00787a005. [DOI] [PubMed] [Google Scholar]
- Courtois Y., Dastugue B., Kamiyama M., Kruh J. Binding of chromosomal non histone proteins to DNA and to nucleohistones. Effect of in vitro phosphorylation. FEBS Lett. 1975 Feb 1;50(2):253–256. doi: 10.1016/0014-5793(75)80501-1. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Galau G. A., Angerer R. C., Britten R. J. Comparative aspects of DNA organization in Metazoa. Chromosoma. 1975 Jul 21;51(3):253–259. doi: 10.1007/BF00284818. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Amenson C. S., Britten R. J. General interspersion of repetitive with non-repetitive sequence elements in the DNA of Xenopus. J Mol Biol. 1973 Jun 15;77(1):1–23. doi: 10.1016/0022-2836(73)90359-8. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Klein W. H., Britten R. J. Structural genes adjacent to interspersed repetitive DNA sequences. Cell. 1975 Mar;4(3):217–238. doi: 10.1016/0092-8674(75)90170-1. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Hough B. R., Klein W. H., Britten R. J. Structural genes adjacent to interspersed repetitive DNA sequences. Cell. 1975 Mar;4(3):217–238. doi: 10.1016/0092-8674(75)90170-1. [DOI] [PubMed] [Google Scholar]
- Galau G. A., Klein W. H., Davis M. M., Wold B. J., Britten R. J., Davidson E. H. Structural gene sets active in embryos and adult tissues of the sea urchin. Cell. 1976 Apr;7(4):487–505. doi: 10.1016/0092-8674(76)90200-2. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Maizels N., Maxam A. Sequences of controlling regions of the lactose operon. Cold Spring Harb Symp Quant Biol. 1974;38:845–855. doi: 10.1101/sqb.1974.038.01.087. [DOI] [PubMed] [Google Scholar]
- Goldberg R. B., Crain W. R., Ruderman J. V., Moore G. P., Barnett T. R., Higgins R. C., Gelfand R. A., Galau G. A., Britten R. J., Davidson E. H. DNA sequence organization in the genomes of five marine invertebrates. Chromosoma. 1975 Jul 21;51(3):225–251. doi: 10.1007/BF00284817. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Bagi G., Berg B., Bonner J. Sequence composition of the template-active fraction of rat liver chromatin. Biochemistry. 1976 Jun 1;15(11):2472–2483. doi: 10.1021/bi00656a034. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Bagi G., Berg B., Bonner J. Sequence composition of the template-active fraction of rat liver chromatin. Biochemistry. 1976 Jun 1;15(11):2472–2483. doi: 10.1021/bi00656a034. [DOI] [PubMed] [Google Scholar]
- Hinkle D. C., Chamberlin M. J. Studies of the binding of Escherichia coli RNA polymerase to DNA. I. The role of sigma subunit in site selection. J Mol Biol. 1972 Sep 28;70(2):157–185. doi: 10.1016/0022-2836(72)90531-1. [DOI] [PubMed] [Google Scholar]
- Ide T., Nakane M., Anzai K., Ando T. Supercoiled DNA folded by non-histone proteins in cultured mammalian cells. Nature. 1975 Dec 4;258(5534):445–447. doi: 10.1038/258445a0. [DOI] [PubMed] [Google Scholar]
- Kostraba N. C., Wang T. Y. Inhibition of transcription in vitro by a non-histone protein isolated from Ehrlich ascites tumor chromatin. J Biol Chem. 1975 Dec 10;250(23):8938–8942. [PubMed] [Google Scholar]
- Lin S., Riggs A. D. The general affinity of lac repressor for E. coli DNA: implications for gene regulation in procaryotes and eucaryotes. Cell. 1975 Feb;4(2):107–111. doi: 10.1016/0092-8674(75)90116-6. [DOI] [PubMed] [Google Scholar]
- Orosz J. M., Wetmur J. G. In vitro iodination of DNA. Maximizing iodination while minimizing degradation; use of buoyant density shifts for DNA-DNA hybrid isolation. Biochemistry. 1974 Dec 31;13(27):5467–5473. doi: 10.1021/bi00724a003. [DOI] [PubMed] [Google Scholar]
- Park W. D., Stein J. L., Stein G. S. Activation of in vitro histone gene transcription from Hela S3 chromatin by S-phase nonhistone chromosomal proteins. Biochemistry. 1976 Jul 27;15(15):3296–3230. doi: 10.1021/bi00660a020. [DOI] [PubMed] [Google Scholar]
- Paul J., Gilmour R. S. Organ-specific restriction of transcription in mammalian chromatin. J Mol Biol. 1968 Jul 14;34(2):305–316. doi: 10.1016/0022-2836(68)90255-6. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Davidson E. H., Britten R. J. A program for least squares analysis of reassociation and hybridization data. Nucleic Acids Res. 1977 Jun;4(6):1727–1737. doi: 10.1093/nar/4.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S. On the assay, isolation and characterization of the lac repressor. J Mol Biol. 1968 Jul 14;34(2):361–364. doi: 10.1016/0022-2836(68)90260-x. [DOI] [PubMed] [Google Scholar]
- Sevall J. S., Cockburn A., Savage M., Bonner J. DNA-protein interactions of the rat liver non-histone chromosomal protein. Biochemistry. 1975 Feb 25;14(4):782–789. doi: 10.1021/bi00675a021. [DOI] [PubMed] [Google Scholar]
- Tashiro T., Kurokawa M. A contribution of nonhistone proteins to the conformation of chromatin. Eur J Biochem. 1975 Dec 15;60(2):569–577. doi: 10.1111/j.1432-1033.1975.tb21035.x. [DOI] [PubMed] [Google Scholar]
- Teng C. S., Teng C. T., Allfrey V. G. Studies of nuclear acidic proteins. Evidence for their phosphorylation, tissue specificity, selective binding to deoxyribonucleic acid, and stimulation effects on transcription. J Biol Chem. 1971 Jun 10;246(11):3597–3609. [PubMed] [Google Scholar]
- Thomas T. L., Patel G. L. Optimal conditions and specificity of interaction of a distinct class of nonhistone chromosomal proteins with DNA. Biochemistry. 1976 Apr 6;15(7):1481–1489. doi: 10.1021/bi00652a019. [DOI] [PubMed] [Google Scholar]
- Ullman J. S., McCarthy B. J. Alkali deamination of cytosine residues in DNA. Biochim Biophys Acta. 1973 Feb 4;294(1):396–404. doi: 10.1016/0005-2787(73)90094-4. [DOI] [PubMed] [Google Scholar]
- Wang S., Chiu J. F., Klyszejko-Stefanowicz L., Fujitani H., Hnilica L. S. Tissue-specific chromosomal non-histone protein interactions with DNA. J Biol Chem. 1976 Mar 10;251(5):1471–1475. [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Wu J. R., Hurn J., Bonner J. Size and distribution of the repetitive segments of the Drosophila genome. J Mol Biol. 1972 Feb 28;64(1):211–219. doi: 10.1016/0022-2836(72)90330-0. [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Alberts B. The interaction of estradiol-receptor protein with the genome: an argument for the existence of undetected specific sites. Cell. 1975 Apr;4(4):301–310. doi: 10.1016/0092-8674(75)90150-6. [DOI] [PubMed] [Google Scholar]
- van den Broek H. W., Noodén L. D., Sevall J. S., Bonner J. Isolation, purification, and fractionation of nonhistone chromosomal proteins. Biochemistry. 1973 Jan 16;12(2):229–236. doi: 10.1021/bi00726a009. [DOI] [PubMed] [Google Scholar]



