Abstract
Midbrain dopaminergic neurons are implicated in various neurological and psychiatric diseases as well as drug addiction. Thus, the study of their generation and maintenance is pivotal to further our understanding of these disease-underlying mechanisms and development of novel therapeutics. Here, using an embryonic stem cell in vitro differentiation system and mutant dreher mouse, we showed that Lmx1a, an early regulator of midbrain dopamine neural progenitor phenotype specification, is also involved in the regulation of midbrain dopaminergic maturation by regulating gene expression of the dopamine transporter. Forced expression of Lmx1a induced dopamine transporter expression precociously in immature dopaminergic neurons, accompanied by significant increase in specific dopamine uptake. Lmx1a binds to well-conserved sequences in the dopamine transporter promoter region, and this binding sequence directs Lmx1a-dependent activation of reporter gene expression. Furthermore, during mouse embryonic development, dopamine transporter was more severely affected by Lmx1a mutation compared to other dopamine marker such as tyrosine hydroxylase and dopa decarboxylase, again supporting the role of Lmx1a in midbrain dopaminergic maturation in vivo. Thus, this study demonstrates that dopamine transporter is a direct target of Lmx1a and emphasizes a novel role of Lmx1a as one of regulators of mature midbrain dopaminergic neurotransmitter phenotypes.
Keywords: dopamine transporter, ES cell, neuron, Lmx1a, dreher mice, differentiation
Introduction
Dysfunction of midbrain dopamine (mDA) neurons has been implicated in various brain diseases such as neurodegenerative and psychiatric disorders, for which there are no effective treatments. A better understanding of how mDA neurons are generated and reach maturation will help develop novel therapeutic approaches for these diseases. During early brain development, mDA neurons originate from the ventral midline of the mesencephalon. The initial event of mDA neuron development depends on signaling molecules (e.g., Sonic hedgehog (Shh), fibroblast growth factor 8 (FGF8), and Wnt1) released from neighboring organizers such as Notochord and Isthmus, setting up the initial field for mDA progenitors (Smidt & Burbach 2007, Ang 2006, Prakash et al. 2006). These extrinsic signals initiate regulatory cascades leading to mDA development by inducing key transcription factors, including FoxA1/A2, Otx2, and Lmx1a/1b in mDA neural progenitor (NP) domains, then later Nurr1 and Pitx3 in mDA neuronal domains (Smidt & Burbach 2007, Ang 2006, Prakash et al. 2006, Chung et al. 2009). Early transcription factors play critical roles in inducing mDA neuronal precursor (NP) phenotype, as evidenced by their ability to induce mDA neurons when ectopically expressed in some non-DA locations (Andersson et al. 2006, Ono et al. 2007, Lin et al. 2009). Our recent work also showed that coexpression of these early factors (Otx2, FoxA2 and Lmx1a) robustly and synergistically increase mDA induction in mouse Embryonic stem cells (ESCs) (Chung et al. 2009).
Following cell fate choices of mDA NP identity by early mDA NP-inducing factors, expression of late transcription factors such as Nurr1 and Pitx3 start during or following cell cycle exit to regulate mature DA neuronal identity and survival (Smidt & Burbach 2007, Ang 2006). Especially, the dopamine transporter (DAT) is one of essential markers of mature mDA neurons, and is responsible for reuptake and homeostasis of DA (Bannon 2005). Recent studies showed that the late transcription factors Nurr1 and Pitx3 regulate DAT gene expression (Hwang et al. 2009, Martinat et al. 2006). Notably, the early transcription factor Lmx1a continues to be expressed in maturing and mature mDA neurons, thus suggesting that Lmx1a regulates maturation of mDA neurons in addition to its role in mDA NP specification (Chung et al. 2009). Indeed, we found that Lmx1a can regulate DAT expression during ESC differentiation by directly binding to conserved element(s) of DAT’s promoter region. In addition, analysis of the dreher mouse, harboring a mutation in the Lmx1a locus, showed that Lmx1a deficiency leads to a significant decrease in DAT expression. Thus, our study provides both in vitro and in vivo evidence that Lmx1a regulates DAT gene expression, emphasizing a novel role of Lmx1a in mDA maturation, in addition to its early role in mDA NP specification.
Materials and Methods
Mouse
Heterozygous dreher mice (Millonig et al. 2000) were purchased from Jackson Laboratory (Bar Harbor, ME) and embryos were harvested after euthanasia of pregnant mice using Carbon Dioxide to minimize the discomfort. The embryos were genotyped by PCR amplification using the following primer sets followed by restriction digestion of PCR product using HpyCH4V (New England Biolabs, Ipswich, MA).
Forward primer: 5′GGAGACCACCTGCTTCTACC3′
Backward primer: 5′GCATACGGATGGACTTCCC3′
Plug date was considered as embryonic day (E) 0.5. Embryos were fixed by immersion in 4%paraformaldehyde, equilibrated in sucrose (20% in PBS), sectioned at 10 mm on a cryostat, and collected onto glass slides. For each experiment, at least three sets of littermate wt and mutant pair were used. Both male and female embryos were used for analysis.
All animal procedures were done according to National Institute of Health guidelines and were approved by the Animal Care and Use Committee (IACUC) at McLean Hospital, Harvard Medical School.
ESC and in vitro differentiation
The mouse blastocyst-derived ESC line J1 was a kind gift from Dr. Jaenisch, and was propagated and maintained as described previously (Chung et al. 2002). Briefly, undifferentiated ESCs were cultured on gelatin-coated dishes in Dulbecco’s modified Minimal Essential Medium (Invitrogen, Carlsbad, CA) supplemented with 2 mM glutamine (Invitrogen, Carlsbad, CA), 0.001% β-mercaptoethanol (Invitrogen, Carlsbad, CA), 1x non-essential amino acids (Invitrogen, Carlsbad, CA), 10% donor horse serum (Sigma, St. Louis, MO), and 2000U/ml human recombinant leukemia inhibitory factor (LIF; R & D Systems, Minneapolis, MN).
ESCs were differentiated into embryoid bodies (EBs) on nonadherent bacterial dishes (Fisher Scientific, Pittsburgh, PA) for four days in LIF-free EB medium containing 10% fetal bovine serum (Hyclone, Logan, Utah) instead of horse serum. EBs were then plated onto adhesive tissue culture surface (Fisher Scientific, Pittsburgh, PA). After 24 hrs in culture, selection of neuronal precursor cells was initiated in serum-free ITSFn medium. After 10 days of selection, cells were trypsinized and nestin+ neuronal precursors (Chung et al. 2009) were plated on polyornithine (15 μg/ml; Sigma, St. Louis, MO) and fibronectin (1 μg/ml; Sigma, St. Louis, MO) coated coverslips in N2 medium supplemented with 1μg/ml laminin (Sigma, St. Louis, MO) and 10 ng/ml bFGF (R & D Systems, Minneapolis, MN) (Neuronal precursor expansion stage: NP stage). After expansion for four days, bFGF was removed to induce differentiation to neuronal phenotypes (Neuronal differentiation stage: ND stage). Cells were eventually fixed in 4% paraformaldehyde or harvested in TriReagent (Sigma, St. Louis, MO) at stage ND3. Retrovirus was prepared using 293GPG retroviral producer cell line and used to infect NP stage cells at day 3 as described in Ory et al. (Ory et al. 1996).
DA reuptake assay
NP stage cells (3×104 cells/well) were infected with Control or Lmx1a virus, followed by in vitro differentiation for 7 days and DA reuptake assay (at day 7 of the ND stage; ND7). Cells were washed with PBS and incubated with 50 nM [3H]DA in PBS (51 Ci/mmol, Amersham Co., Buckinghamshire, UK; 80,000 cpm per well) without or with 10 mM nomifensine (RBI, Natick, MA, USA), a dopamine transporter (DAT) blocker, to determine non-specific uptake. After incubation for 10 min at 37°C, the uptake reactions were terminated by aspiration of the reaction solution and washing twice with ice-cold PBS. Cells were lysed in 0.5 M NaOH and the radioactivity was measured by liquid scintillation counting. Specific DA uptake was calculated by subtracting non-specific uptake (with nomifensine). The samples from three independent différentiation were used for analysis
Immunocytochemistry & immunohistochemistry
For immunofluorescence staining, cells were fixed in 4% formaldehyde (Electron Microscopy Sciences, Ft. Washington, PA) for 30 minutes, rinsed with PBS and then incubated with blocking buffer (PBS, 10 % normal donkey serum; NDS) for 10 minutes. Cells were then incubated overnight at 4°C with primary antibodies diluted in PBS containing 2% NDS. The following primary antibodies were used: rabbit anti-β-tubulin (Covance; 1:2000), sheep anti-tyrosine hydroxylase (TH) (Pel-Freez, Rogers, Arkansas; 1:500) and rat anti-DAT antibody (Chemicon; 1:1,000). After additional rinsing in PBS, the samples were incubated in fluorescent-labeled secondary antibodies (Alexa 488- or Alexa 594 -labeled IgG; Invitrogen, Carlsbad, CA) in PBS with 2% NDS for 30 minutes at room temperature. After rinsing in PBS, Hoechst 33342 (4 mg/ml) was used for counterstaining, and coverslips/tissues sections were mounted onto slides in Mowiol 4–88 (Calbiochem, Gibbstown, NJ). Confocal analysis was performed using a Zeiss LSM510/Meta Station (Carl Zeiss, Thornwood, NY).
Real time PCR analysis
For all other samples, total RNA was prepared using TriReagent (Sigma, St. Louis, MO) followed by further purification using the RNeasy mini kit (Qiagen, Valencia, CA). For RT-PCR analysis, 2 μg of RNA were transcribed into cDNA with the SuperScript™ II RT (Invitrogen, Carlsbad, CA) and oligo (dT) primers. For quantitative analysis of the expression level of mRNAs, real-time PCR analyses using SYBR green I were performed using the DNA engine Opticon™ (MJ Research, Waltham, MA). To reduce non-specific signals, oligonucleotides amplifying small amplicons were designed using the MacVector software (Oxford Molecular Ltd.) and the sequences of the primers are available upon request. Amplifications were performed in 25 μl containing 0.5 mM of each primer, 0.5 × SYBR Green I (Molecular Probes, OR), and 2 μl of 5 fold diluted cDNA. Fifty cycles of PCR were performed with the temperature profile of 95°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec, and 79°C for 5 sec. The dissociation curve of each PCR product was determined to ensure that the observed fluorescent signals were only from specific PCR products. After each PCR cycle, the fluorescent signals were detected at 79°C to melt primer dimers (Tms of all primer dimers used in this study were < 76°C). A standard curve was constructed using plasmid DNAs containing the GAPDH gene (from 102 to 108 molecules). The fluorescent signals from specific PCR product were normalized against that of β-actin, and then relative values were calculated by setting the normalized value of control as 100 (or 1 in some cases). All reactions were repeated using more than three samples from independent differentiations. The sequences of the primers are available upon request.
ChIP-real time PCR analysis
For Lmx1a ChIP, HA-tagged Lmx1a retrovirus was transduced into J1 cells at the NP stage (MOI=4) and further differentiated until stage ND3. Fixing of cells and immunoprecipitation of Chromatin were performed using the ChIPAssay kit (Upstate, Billerica, MA) according to the manufacturer’s instruction. Briefly, after fixing using 1% formaldehyde for 10 minutes at 37°C, chromatin was sheared with a Sonic Dismembrator (Fisher Scientific; output setting 2.5, ten 20-sec pulses with 30-sec incubation on ice between pulses) to a size of 100bp to 700 bp as verified on a 1% agarose gel. Clarified nuclear lysates were incubated overnight at 4°C with anti-HA rabbit polyclonal antibody (Abcam, Cambridge, MA) or rabbit normal IgG as a control. Following this incubation, Protein A agarose beads were added and allowed to bind for 60 min at 4°C. Immunoprecipitates were washed and crosslinks were reversed for 4 hours at 65°C. ChIP DNA was purified by incubation with RNase A for 1 h at 37°C, with 200 μg/ml Proteinase K for 2 h at 45°C, phenol:chloroform:isoamyl alcohol extraction, and precipitation with 0.1 volumes of 3M sodium acetate, 2.5 volumes of 100% ethanol and 1 μl of glycogen as a carrier.
For Lmx1a ChIP from mouse embryonic ventral midbrain, timed pregnant mice (C57BL/6) were purchased from Charles River laboratory (Wilmington, MA). The ventral mesencephalon from E13.5 embryos was dissected, fixed, and processed for ChIP as described above except that, instead of protein A-agarose beads, protein A-dyna beads (Invitrogen, Carlsbad, CA) were used as described in (Dahl & Collas 2008). Minimal amount (10μl) of dyna beads were used for each ChIP to decrease the nonspecific background caused by small input of material. Lmx1a antibody is a kind gift from Dr. German (UCSF).
For real time PCR analysis of ChIP fragments, ECRs (evolutionally conserved region) in the promoter region of each gene were analyzed with an ECR browser (http://ecrbrowser.dcode.org) and well-conserved binding sites within ECRs were identified. PCR primer pairs were designed to amplify genomic fragments containing well-conserved binding sites using the MacVector software (Oxford Molecular Ltd.). The sequences of the primers used for ChIP are available upon request. Real-time PCR was carried out using SYBR green I as described above. Samples from three independent ChIP assays were analyzed for each candidate target sites.
Generation of reporter constructs and Luciferase assay
Double-stranded DNAs containing DAT-A (5′-CAGTTTTAATTATGCGTAT-3′ for sense and 5′-GATACGCATAATTAAAACT-3′ for antisense) and DAT-D (5′-GTAGGCATTACTCTCTTAATCAGAGCA-3′ for sense and 5′-CTGCTCTGATTAAGAGAGTAATGCCTA-3′ for antisense) were generated by annealing the sense and antisense oligonucleotides. These double-stranded DNAs were self-ligated to generate multiple copies. The multimers were blunt-ended and inserted into the upstream of TATA sequence of TATA-luciferase plasmid. Each clone was sequenced and the clones containing 14 copies of DAT-A and 15 copies of DAT-D were used for our experiments. Transfections were performed using Lipofectamine™ (Invitrogen). Plasmids for transfection were prepared using Qiagen columns (Qiagen). One day before transfection, cells were plated in individual wells of 24-well plates at a density of 1×105 cells per well. For the human neuroblastoma SK-N-BE(2)C cell-lines, each well was transfected with an equimolar amount (0.1 pmol) of each reporter construct, 0.125μg of pRSV-βgal, and pUC19 plasmid for a total of 0.625μg DNA. In cotransfection analysis, a half molar amount of the reporter construct was used for the pCMV-Lmx1A. Cells were harvested 24 to 48 h after transfection, and assayed for luciferase activities. All experiments were done in triplicate and repeated three to six times with different DNA preparations. To correct for differences in transfection efficiencies, luciferase activity was normalized to β-galactosidase activity.
Statistical analysis
For statistical analysis, we used the Statview software and performed analysis of variance (ANOVA) with an alpha level of 0.05 to determine possible statistical differences between group means. When significant differences were found, post hoc analysis was performed using Fisher’s PLSD (alpha=0.05).
Results and Discussion
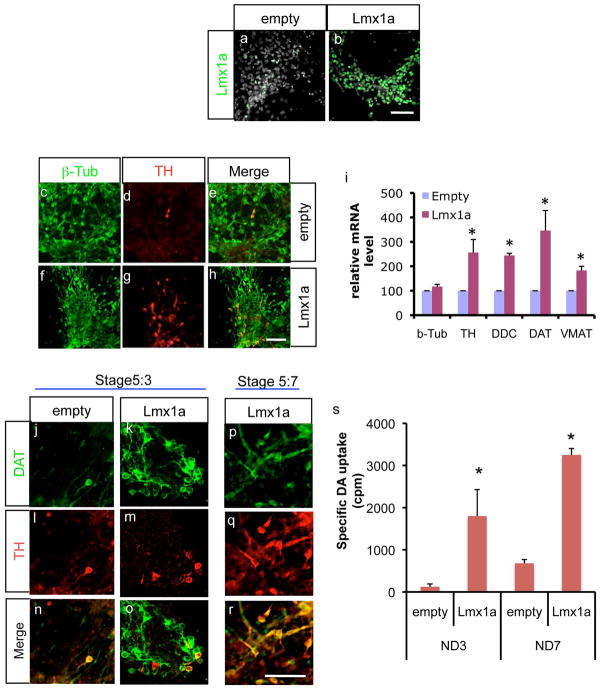
Previously we observed that Lmx1a controls mDA differentiation by regulating key regulators of the mDA differentiation pathway, such as Wnt1, Nurr1 and Pitx3 (Chung et al. 2009). Since Lmx1a continues to be expressed in the mature mDA neuronal domain (Ono et al. 2007), we speculated that Lmx1a also participates in the direct regulation of mature mDA neuronal markers. To address this, we infected differentiating J1 ESC with either empty or Lmx1a-expressing retrovirus at the late NP stage and analyzed the outcomes at the ND stage day 3 (ND3) (Fig. 1a–b). As expected from its role in mDA differentiation, there was a significant increase in TH+ neuronal cell numbers in Lmx1a-expressing cells, compared to empty vector cells (Fig. 1c–h; 0.39±0.10 vs. 3.32±0.86 TH+/β-tubulin+ cells). We further tested for changes in expression of other mDA markers genes, e.g., tyrosine hydroxylase (TH), dopa decarboxylase (DDC), DAT and vesicular monoamine transporter (VMAT), and found that expression of all these genes was significantly increased, whereas there was not much difference in expression of the general neuronal marker β-tubulin gene (Fig. 1i). Interestingly, DAT’s expression was more pronounced compared to other mDA marker genes. Thus, we further analyzed DAT gene expression after retroviral Lmx1a expression. As shown in Fig. 1j–o, retroviral Lmx1a expression led to robust increase in DAT-expressing cell numbers (0.18±0.12 vs. 6.32±3.73 DAT+/β-tubulin+ cells). Notably, some of these DAT+ cells were not TH+ (Fig. 1o), suggesting that DAT induction was the direct result of precocious regulation by Lmx1a, rather than being the consequence of an increase in DA differentiation. In support of this, at day 7 of the ND stage, most DAT+ cells became TH+ (Fig. 1p–r). To test whether induction of DAT by Lmx1a has functional consequences, we assayed DA uptake. As shown in Fig. 1q, we found that DA uptake was significantly increased in Lmx1a-overexpressing cells at both the ND3 and ND 7 stages (Fig. 1s).
Figure 1.
The mDA marker gene, DAT is a direct downstream target of Lmx1a. a–b. Exogenous Lmx1a expression by retroviral infection. Scale bar=50μm. c–h. J1 ES cells were infected with either empty virus or Lmx1a-expression virus at the NP stage, and analyzed by immunofluorescence at ND3. Scale bar=50μm. i. Lmx1a significantly increases the expression of DA markers, TH, DDC, DAT and VMAT2, shown by real time PCR analysis at stage ND3. Relative mRNA levels are shown where the value of empty vector is set at 100 (n=4 from independent differentiation, mean±SEM, p<0.05). j–o. DAT immunocytochemistry of either empty virus or Lmx1a-expressing virus treated cells at ND3. Note that some DAT+ cells in Lmx1a-expressing cells are not yet TH+. p–r. Most DAT+ cells are also expressing TH by ND7. Scale bar=50μm. s. DAT uptake assay. Empty virus or Lmx1a virus-treated cells were subject to DAT uptake assay at ND stage day 7 (n=3 from independent differentiation, mean±SEM, p<0.05).
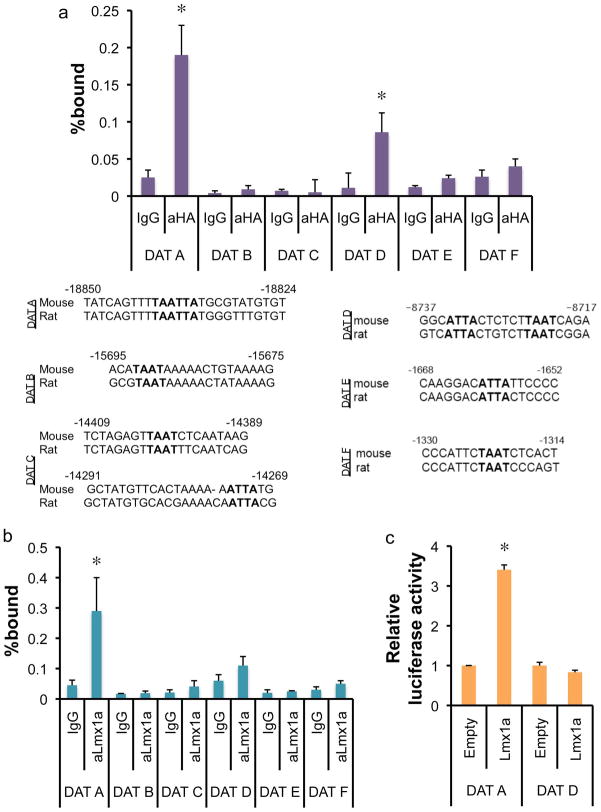
Since Lmx1a regulates critical regulators of mDA development (e.g., Wnt1, Nurr1, and Pitx3), upregulation of mDA marker genes can be indirectly caused by these upregulated factors. At the same time, it is still possible that DAT is subject to direct regulation by Lmx1a. Thus, we performed chromatin immunoprecipitation (ChIP) analysis to address whether Lmx1a directly regulates DAT gene expression. In vitro differentiated J1 cells were transduced with retroviral construct expressing HA-tagged Lmx1a at the NP stage, and were harvested at ND3 for ChIP analysis. Crosslinked chromatin complexes were immunoprecipitated using anti-HA antibody or control IgG. Crosslinks were reversed and purified ChIP DNAs were analyzed by real time PCR. To find potential Lmx1a binding site(s), we searched for ECR (evolutionary conserved region) in the entire DAT gene promoter. Six well-conserved homeodomain-binding sites were identified in these ECRs (Fig. 2a). Using primer sets encompassing these well-conserved homeodomain-binding sites, real time PCR analysis showed significant Lmx1a binding at the DAT A and D fragment. On the other hand, we could not detect any significant binding of Lmx1a to the other potential homeodomain-binding sites of the DAT promoter, indicating the specificity of Lmx1a binding (Fig. 2a). Next, we further analyzed the Lmx1a binding during normal mouse development. E13.5 VM was dissected and ChIP was performed using anti-Lmx1a antibody. Among the potential binding sites of Lmx1a, only DAT-A showed significant binding to Lmx1a (Fig. 2b). DAT-D showed only marginal and insignificant binding to Lmx1a in VM. To further confirm the functionality of this binding site, we did reporter analysis using reporter constructs containing the DAT-A or DAT-D sites. Lmx1a significantly activated reporter gene expression by the DAT-A reporter construct but not by the DAT-D reporter construct (Fig. 2c). Taken together, we conclude that Lmx1a plays an important role during specification of mature mDA phenotype, at least in part by directly regulating DAT gene expression.
Figure 2.
Lmx1a works through a well-conserved binding site in the DAT promoter région. a. ChIP-Real time PCR analysis on DAT promoter region. In vitro differentiated cells were transduced with HA-tagged Lmx1a-expressing retrovirus and fixed for ChIP at ND3. ChIP fragments were immunoprecipitated with normal rabbit IgG or anti-HA antibody and analyzed by qPCR. The average of three independent ChIP analyses (n=3, mean±SEM, p<0.05) are presented. b. E13.5 VMs were dissected and used for ChIP using Lmx1a antibody. Binding of Lmx1a to the DAT promoter was assayed by qPCR (n=3 from independet ChIP reactions, mean±SEM, p<0.05). c. Reporter construct containing multiple copies of DAT-A mediates transactivation by Lmx1a (n=3 from independent transfection reactions, mean±SEM, p<0.05). Reporter constructs were cotransfected with either empty vector or Lmx1a expression vector to human neuroblastoma SK-N-BE(2)C cells and Luciferase activaty was measured 24–48 hours after transfection and normalized by β-galactosidase activity.
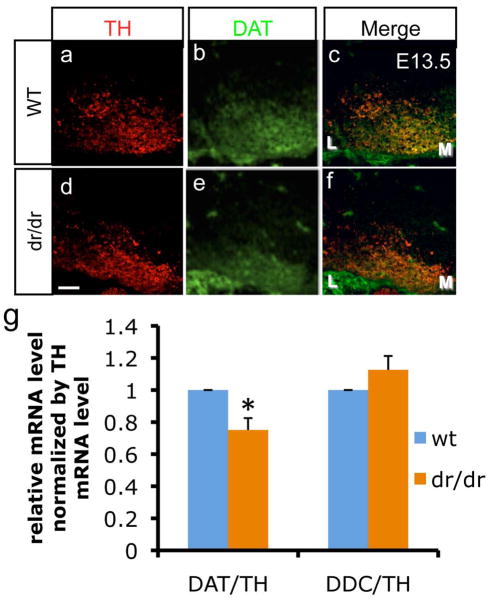
To further test the in vivo influence of Lmx1a on DAT gene regulation during mouse embryonic development, we analyzed DAT expression in the midbrain of littermate wild type (wt) and dr/dr embryos by immunocytochemistry. In agreement with previous studies (Ono et al. 2007), the number of TH+ neurons was decreased in dr/dr mice at E13.5 (Fig. 3a–f). Our analysis revealed that there is a decrease in DAT expression levels in mDA neurons of dr/dr mice, apart from the previously shown decrease in mDA neuronal numbers (Fig. 3a–f). This result was confirmed by real time PCR analysis from dissected E13.5 VM thus showing that Lmx1a mutation leads to greater decrease in DAT mRNA levels compared to TH or DDC mRNA levels (Fig. 3g).
Figure 3.
Expression of DAT is reduced by Lmx1a mutation. a–f. immunohistochemistry analysis of VM in E13.5 littermates’ wt and mutant embryos using anti-DAT and anti-TH. M denotes medial VM and L denotes lateral VM. Scale bar represents 50μm. g. Real time PCR analysis on dissected VM. To avoid variablility caused by difference in dissection, the results are normalized using the value of TH (n=3 from independent VM dissections, mean±SEM, p<0.05).
DAT is a membrane protein that regulates DA neurotransmission by reuptake of released dopamine from the extracellular space, and its abnormal function/expression has been implicated in multiple neurological and psychiatric diseases such as Parkinson’s disease (PD), psychostimulant abuse, and attention deficit hyperactivity disorder (ADHD) (Bannon 2005). In addition, some DAT gene polymorphisms are associated with these brain disorders (Bai & Burton 2009, Bannon et al. 2001, Michelhaugh et al. 2001, Greenwood & Kelsoe 2003). The DAT gene shows very specific expression pattern in DA neurons in the CNS and has been shown to be regulated by multiple complex regulatory elements present in the 5′ and 3′ flanking sequences as well as intronic sequences (Bannon et al. 2001). Thus, furthering our knowledge of the molecular mechanisms underlying the regulation of DAT in normal and diseased brains will likely lead to better understanding of disease mechanisms as well as the development of novel therapeutics.
Later regulators of mDA differentiation, such as Nurr1 and Pitx3 have been shown to regulate tissue specific expression of DAT during embryonic development or in adult brain (Hwang et al. 2009, Jacobs et al. 2009, Kadkhodaei et al. 2009). In addition, Nurr1 and Pitx3 collaborate with each other to regulate mDA specific marker genes expression including DAT (Martinat et al. 2006, Jacobs et al. 2009). Other than that, our understanding of the molecular mechanisms and regulators underlying cell type specific expression of the DAT gene are elusive. Here, we showed that the early transcription factor Lmx1a, in addition to its well-known role in mDA NP specification, directly regulates expression of the DAT gene using both ES cell in vitro differentiation and Lmx1a mutant embryos. Induction of DAT expression by Lmx1a was accompanied by functional consequences as evidenced by a significant increase in specific DA uptake by Lmx1a expression.
As we have shown previously, Lmx1a also regulates the expression of later regulators, Nurr1 and Pitx3 (Chung et al. 2009). Such multi-layer regulation of key DA marker gene expression by both direct regulation of Lmx1a and indirect regulation through other mDA regulators could serve to ensure proper expression of these important mDA phenotype genes.
Acknowledgments
This study was supported by NIH grants (NS070577, MH087903, and MH048866) and International Collaboration Grants from Korea Research Institute of Bioscience and Biotechnology (KRIBB) and CHA University, Korea.
Abbreviations
- mDA
midbrain dopamine
- NP
neuronal precursor
- ESC
embryonic stem cell
- DAT
dopamine transporter
- EB
embryoid body
- ND
neuronal differentiation
- ChIP
chromatin immunoprecipitation
- TH
tyrosine hydroxylase
- DDC
dopa decarboxylase
- VMAT
vesicular monoamine transporter
- ECR
evolutionally conserved region
- PD
Parkinson’s disease
- ADHD
attention deficit hyperactivity disorder
Footnotes
We declare that there is no conflict of interest.
References
- Andersson E, Tryggvason U, Deng Q, Friling S, Alekseenko Z, Robert B, Perlmann T, Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Ang SL. Transcriptional control of midbrain dopaminergic neuron development. Development. 2006;133:3499–3506. doi: 10.1242/dev.02501. [DOI] [PubMed] [Google Scholar]
- Bai Q, Burton EA. Cis-acting elements responsible for dopaminergic neuron-specific expression of zebrafish slc6a3 (dopamine transporter) in vivo are located remote from the transcriptional start site. Neuroscience. 2009;164:1138–1151. doi: 10.1016/j.neuroscience.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Bannon MJ. The dopamine transporter: role in neurotoxicity and human disease. Toxicol Appl Pharmacol. 2005;204:355–360. doi: 10.1016/j.taap.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Bannon MJ, Michelhaugh SK, Wang J, Sacchetti P. The human dopamine transporter gene: gene organization, transcriptional regulation, and potential involvement in neuropsychiatric disorders. Eur Neuropsychopharmacol. 2001;11:449–455. doi: 10.1016/s0924-977x(01)00122-5. [DOI] [PubMed] [Google Scholar]
- Chung S, Leung A, Han BS, et al. Wnt1-lmx1a forms a novel autoregulatory loop and controls midbrain dopaminergic differentiation synergistically with the SHH-FoxA2 pathway. Cell Stem Cell. 2009;5:646–658. doi: 10.1016/j.stem.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Sonntag KC, Andersson T, Bjorklund LM, Park JJ, Kim DW, Kang UJ, Isacson O, Kim KS. Genetic engineering of mouse embryonic stem cells by Nurr1 enhances differentiation and maturation into dopaminergic neurons. Eur J Neurosci. 2002;16:1829–1838. doi: 10.1046/j.1460-9568.2002.02255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nat Protoc. 2008;3:1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- Greenwood TA, Kelsoe JR. Promoter and intronic variants affect the transcriptional regulation of the human dopamine transporter gene. Genomics. 2003;82:511–520. doi: 10.1016/s0888-7543(03)00142-3. [DOI] [PubMed] [Google Scholar]
- Hwang DY, Hong S, Jeong JW, Choi S, Kim H, Kim J, Kim KS. Vesicular monoamine transporter 2 and dopamine transporter are molecular targets of Pitx3 in the ventral midbrain dopamine neurons. J Neurochem. 2009;111:1202–1212. doi: 10.1111/j.1471-4159.2009.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FM, van Erp S, van der Linden AJ, von Oerthel L, Burbach JP, Smidt MP. Pitx3 potentiates Nurr1 in dopamine neuron terminal differentiation through release of SMRT-mediated repression. Development. 2009;136:531–540. doi: 10.1242/dev.029769. [DOI] [PubMed] [Google Scholar]
- Kadkhodaei B, Ito T, Joodmardi E, et al. Nurr1 is required for maintenance of maturing and adult midbrain dopamine neurons. J Neurosci. 2009;29:15923–15932. doi: 10.1523/JNEUROSCI.3910-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Metzakopian E, Mavromatakis YE, et al. Foxa1 and Foxa2 function both upstream of and cooperatively with Lmx1a and Lmx1b in a feedforward loop promoting mesodiencephalic dopaminergic neuron development. Dev Biol. 2009;333:386–396. doi: 10.1016/j.ydbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Martinat C, Bacci JJ, Leete T, et al. Cooperative transcription activation by Nurr1 and Pitx3 induces embryonic stem cell maturation to the midbrain dopamine neuron phenotype. Proc Natl Acad Sci U S A. 2006;103:2874–2879. doi: 10.1073/pnas.0511153103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelhaugh SK, Fiskerstrand C, Lovejoy E, Bannon MJ, Quinn JP. The dopamine transporter gene (SLC6A3) variable number of tandem repeats domain enhances transcription in dopamine neurons. J Neurochem. 2001;79:1033–1038. doi: 10.1046/j.1471-4159.2001.00647.x. [DOI] [PubMed] [Google Scholar]
- Millonig JH, Millen KJ, Hatten ME. The mouse Dreher gene Lmx1a controls formation of the roof plate in the vertebrate CNS. Nature. 2000;403:764–769. doi: 10.1038/35001573. [DOI] [PubMed] [Google Scholar]
- Ono Y, Nakatani T, Sakamoto Y, et al. Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development. 2007;134:3213–3225. doi: 10.1242/dev.02879. [DOI] [PubMed] [Google Scholar]
- Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci U S A. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash N, Brodski C, Naserke T, et al. A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development. 2006;133:89–98. doi: 10.1242/dev.02181. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Burbach JP. How to make a mesodiencephalic dopaminergic neuron. Nat Rev Neurosci. 2007;8:21–32. doi: 10.1038/nrn2039. [DOI] [PubMed] [Google Scholar]