Abstract
Variability is the law of life,… and no two individuals react alike and behave alike under the abnormal conditions which we know as disease. Sir William Osler, The Principles and Practice of Medicine (1)
Introduction
Neurohormonal activation is an important driver of heart failure progression, and all pharmacologic interventions that improve heart failure survival inhibit this systemic response to myocardial injury. Adrenergic stimulation of β1 receptors in the kidney results in the release of plasma renin, the conversion of peptide precursors to angiotensin II (a2), and ultimately the production of aldosterone. Beta blockers, angiotensin converting enzyme (ACE) inhibitors, angiotensin receptor blockers (ARBs), and aldosterone receptor antagonists, all act by inhibiting the activity of critical protein of this core pathway: the beta 1 receptor, ACE, the a2 receptor and aldosterone synthase. Interestingly, there is significant genomic heterogeneity and functional polymorphisms at every step which impact mediator levels and influence therapeutic effectiveness (Figure 1). Investigation of the pharmacogenetic interactions of the ACE D/I polymorphism and heart failure therapy demonstrate the power of genomics to target therapeutics. This review will explore how genetic variation in genes involved in neurohormonal activation influence both heart failure outcomes and the impact of pharmacotherapy.
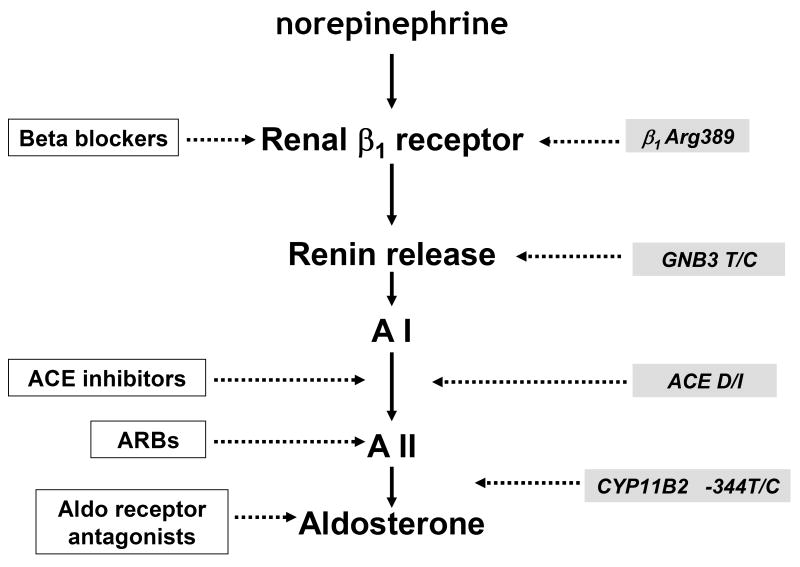
Figure 1. RAAS pathway and site of action of drug therapies and functional polymorphisms.
Major pharmacologic therapies which improve survival in heart failure in white boxes: beta blockers, ACE inhibitors, ARBs (angiotensin receptor blockers) and Aldo (aldosterone) receptor antagonists all act on RAAS. In grey boxes are major genetic polymorphisms which influence outcomes and the impact of therapy including β1Arg389, G protein β 3 subunit (GNB3 T haplotype linked to low plasma renin), ACE D/I (deletion/Insertion) and aldosterone synthase (CYP11B2) promoter
ACE D/I Polymorphism and Heart Failure Outcomes
The ACE deletion/insertion biallelic polymorphism of intron 16 is the most extensively studied cardiovascular polymorphism, and has been the subject of hundreds of investigations since its initial discovery (2, 3). While the clinical implications of this polymorphism have been controversial, the physiologic association of the ACE D/I polymorphism with enzymatic activity has been consistent. The D allele has been linked in nearly every clinical study to increased activity of the ACE enzyme and higher levels of the product of ACE activity, the peptide mediator angiotensin 2 (a2) (4). The cellular mechanism remains to be elucidated but this linkage of genotype with ACE activity is consistent across multiple different clinical paradigms from hypertension to myocardial infarction (5, 6, 7) and demonstrates a D allele “dose effect” for a2 levels. Subjects with the DD genotype have the highest levels of a2, heterozygotes are intermediate and, those homozygous for I allele have the lowest levels. For disease states such as heart failure where angiotensin II facilitates disease progression, the potential implications of this genetically “ordered” ACE activity are readily apparent.
Given the role of renin-angiotensin activation in heart failure, it is hypothesized that the ACE D allele functions as a genetic modifier, accelerates disease progression and worsens survival. Indeed, this has been demonstrated in three separate clinical investigations. The first was a population of 193 subjects with idiopathic dilated cardiomyopathy and demonstrated poorer survival for subjects homozygous for the D allele (8). Most recently the adverse impact of the ACE D allele was demonstrated in 978 subjects post myocardial infarction (9). The impact in this cohort was primarily in subjects with lower left ventricular ejections fraction (LVEF) or higher brain naturetic peptide (BNP) levels. Forty five percent of the subjects in the post myocardial infarction study were on ACE inhibitor therapy, compared to only 25% of subjects in the earlier study of idiopathic dilated cardiomyopathy. Neither study addressed the potential pharmacogenetic interactions of the ACE D/I polymorphism with the medical therapy of heart failure.
Pharmacogenetics of the ACE D/I polymorphism: GRACE
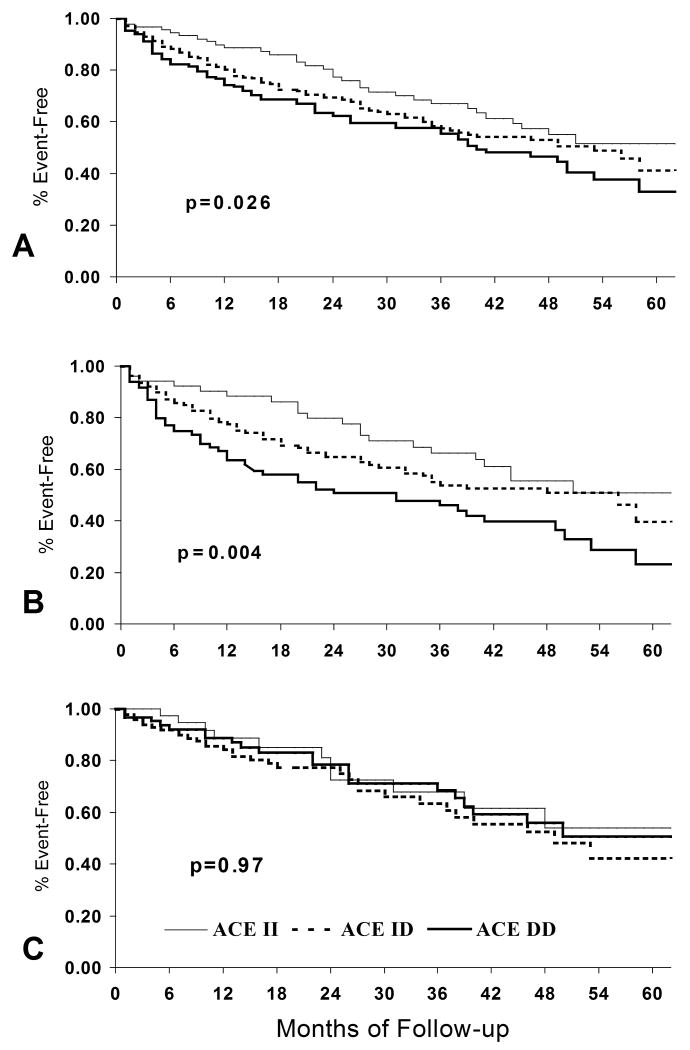
A similar overall impact of the ACE D allele on heart failure outcomes was demonstrated in a single center study at the University of Pittsburgh, the GRACE study of Genetic Risk Assessment of Cardiac Events. Four hundred seventy-nine with systolic dysfunction (mean LVEF 0.25 ± 0.08) with both ischemic and non-ischemic etiologies were followed for a median of 3 years until death or cardiac transplantation. The D allele was associated with poorer in transplant free survival, and demonstrated the same “gene ordered” effect shown earlier for ACE activity: II homozygotes demonstrated the best survival; DD homozygotes the poorest, with heterozygotes displayed the predicted “intermediate” survival between the homozygotes. The adverse impact of the D allele on survival for this cohort was first reported on an analysis of the first 328 subjects (10), and remained evident during analysis for the entire cohort (11) (Figure 2A).
Figure 2.
Transplant-free survival By ACE D/I genotype, GRACE study, University of Pittsburgh (reference 11). A. Overall cohort, ACE D allele associated with poorer event free survival, n=479, p=0.026 B: Subset with no beta blocker therapy, n=277, p=0.004 C. Subset treated with beta blocker therapy, n=202, p=0.97
β-blocker Therapy
Genetic background must interact with environment, and for the heart failure patient a critical aspect of the local “environment” is the pharmacologic milieu. The effect of genetic modulation of neurohormonal activation on heart failure outcomes may be altered by pharmacological inhibition, and this was examined in GRACE. Ninety-five percent of subjects in GRACE were on ACE inhibitors or ARBs, while only 42% were on beta-blockers reflecting the evolution of care at the time of the initiation of the study. For beta-blockers, analysis by treatment subset demonstrated the effects of ACE D on outcomes to be primarily in subjects not treated with β-blockers (10,11) (Figure 2B) and that impact of the polymorphism is eliminated by β-blocker therapy (Figure 2C). For subjects not on beta-blockers, the 2–year event-free survival for DD subjects was only 51%, versus 80% for the II homozygotes. In contrast, for subjects on beta-blockers, no effect of the D–allele on outcomes was apparent. The elimination of the effect of the D allele by beta-blocker therapy likely reflects their role as inhibitors of renin release. An important stimulus to the renin angiotensin activity in heart failure is sympathetic activation, and beta blockers markedly reduce angiotensin 2 levels (12).
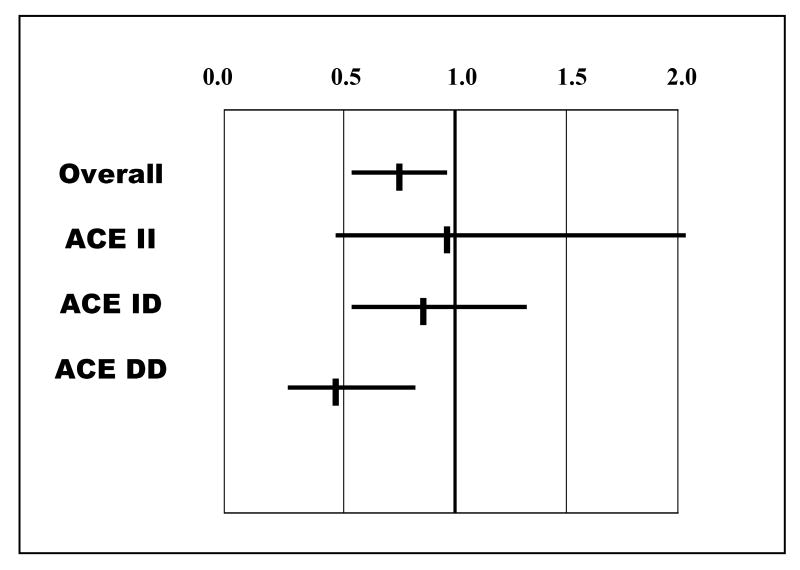
The potential utility of this pharmacogenetic interaction is evident when examining the impact of β-blocker therapy within ACE D/I genetic subsets (Figure 3). While beta-blockers overall improved survival in the GRACE cohort, much of that benefit is in the DD homozygotes genetically predicted to have the highest levels of ACE activity. The impact of therapy is diminished in the heterozygotes, and in this small cohort is no longer evident for subjects who are II homozygotes. These results suggests the ACE D/I polymorphism delineates genetic subsets of heart failure patients in whom the therapeutic impact of beta-blockade on heart failure survival is distinctly different. However, a recent study (13) (15) of the effect of beta-blockers for 199 subjects with chronic heart failure demonstrated similar improvements in LVEF in all three genetic subsets, and suggests the ACE D/I polymorphism does not predict this particular endpoint of beta-blocker response.
Figure 3.
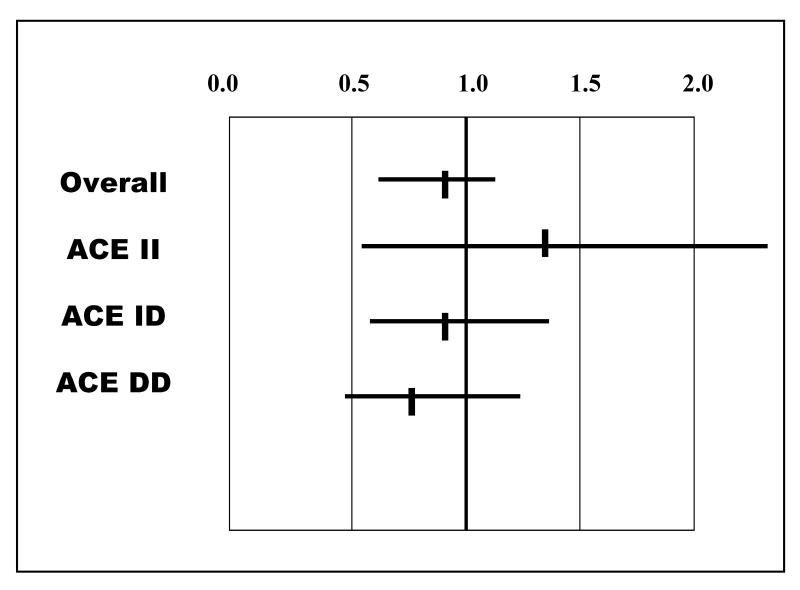
Relative Risk of event (death or transplantation) by beta blocker use, GRACE study: Overall cohort and by ACE D/I genotype, DD,DI and II (adapted from table 4, reference 11)
The relative merits of β1 selective antagonists (e.g. metoprolol) and non-selective β-blocker (e.g. carvedilol) are still debated, however if inhibition of renin release is the primary mechanism of this pharmacogenetic interaction, β1 selective and non-selective β-blocker should be equally effective. The role of β-receptor selectivity for the interaction between the angiotensin converting enzyme (ACE) insertion/deletion polymorphism and β-blocker therapy was investigated in a subset analysis from GRACE (14). Subjects were separated into no beta blocker, β1-selective and non-selective β-blocker treatment groups. The D allele adversely affected transplant-free survival for subjects not on β-blockers (p=0.004). Treatment with selective β1-blockers eliminated the impact of the D allele (p=0.51) in a manner similar to non-selective β1,2-blockers (p=0.80) (Figure 4). Treatment with β1-blockers was sufficient to eliminate the adverse impact of the ACE D allele, suggesting this pharmacogenetic interaction is mediated through the β1-receptor.
Figure 4.
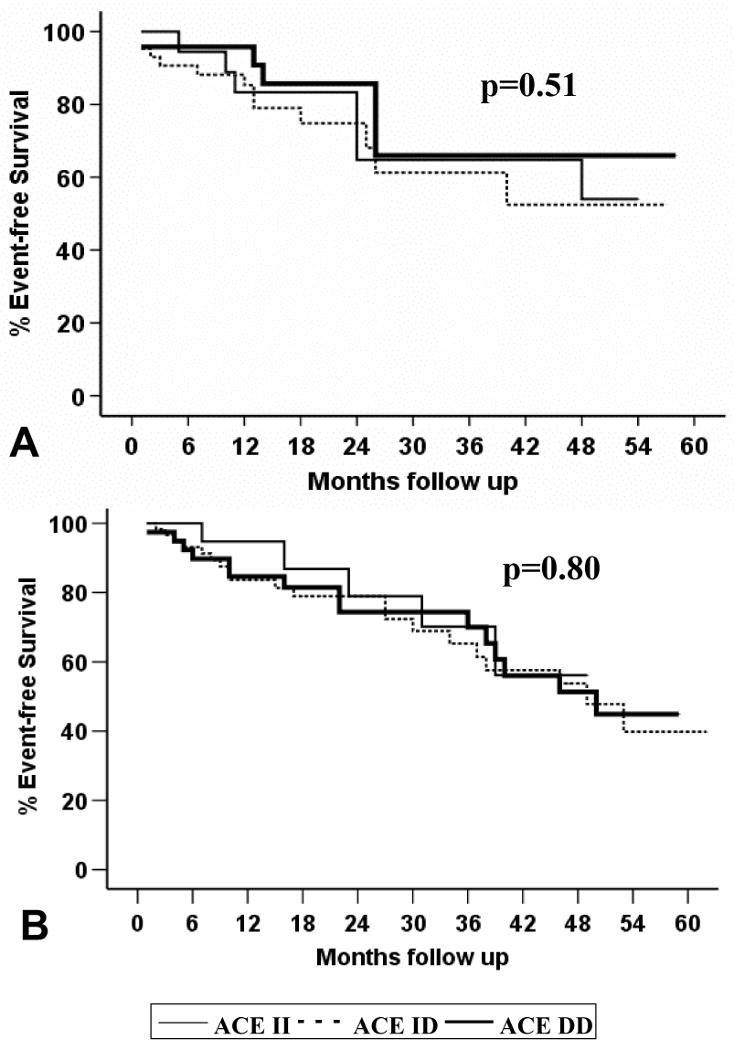
Transplant-free survival By ACE D/I genotype, GRACE study, University of Pittsburgh (reference 14). A. Event-free survival by ACE genotype, β1-selective beta blocker only (n=85), p=0.51. B. Event-free survival by ACE genotype, β1,2- non-selective beta blocker (n=117), p=0.80
ACE inhibitor Therapy
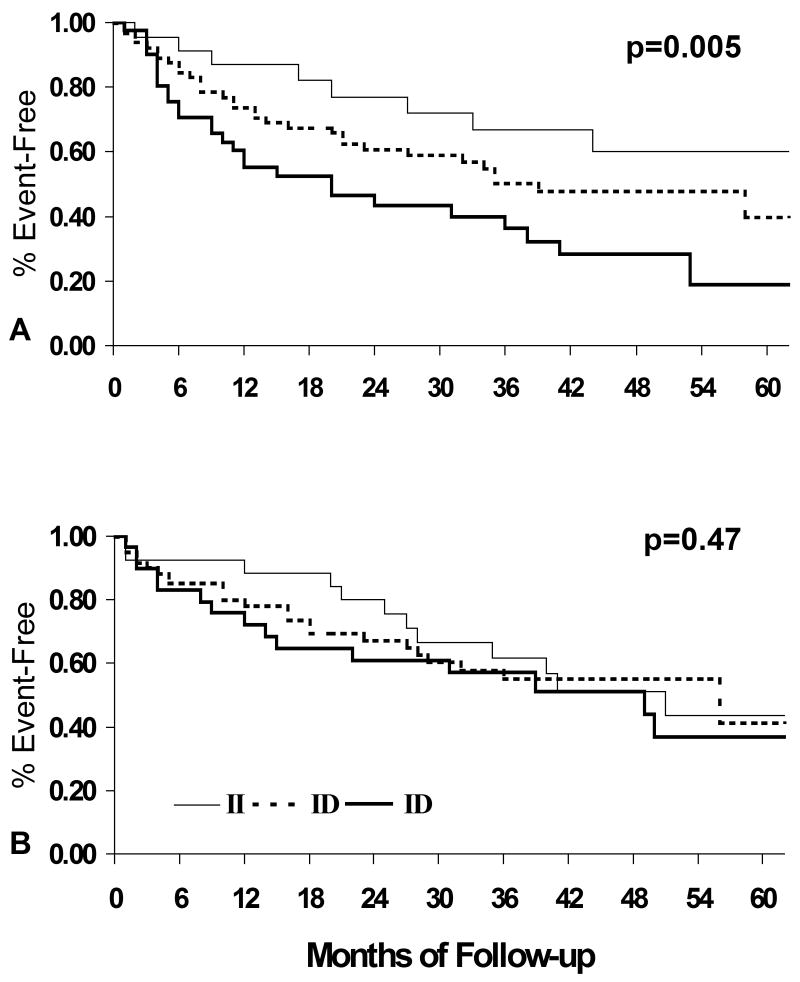
Based on ACE activity, it is also predicted that the ACE D/I polymorphisms should modulate the effect of ACE inhibitors. As ninety-five percent of the subjects in GRACE were either on an ACE inhibitor or an ARB, this pharmacogenetic interaction was investigated in this cohort using a dose analysis comparing the effect of higher dose ACE therapy (> 50% of target daily dose defined by national guidelines) on transplant free survival to low dose therapy (< or = 50%). This resulted in a roughly 3-fold difference in the mean dose for the high dose group compared to low dose group. In contrast, the multi-center Assessment of Treatment with Lisinopril and Survival (ATLAS) trial (15) evaluated the impact of a 10 fold difference in the mean dose of ACE inhibitor. Analysis by ACE dose treatment subset suggests higher doses of ACE inhibitors limit the adverse impact of the ACE D allele (11), and this effect of high dose therapy was particularly evident in subjects not treated with beta-blockers (figure 5).
Figure 5.
Transplant-free survival By ACE D/I genotype, GRACE study, University of Pittsburgh (reference 11). A. Subset on low dose ACE inhibitors and no beta-blockers, ACE D allele associated with poorer event free survival, n=130, p=0.005 B. Subset with high dose ACE inhibitor therapy and no beta blockers, n=117, p=0.47
In overall event-free survival, the benefits of higher dose ACE inhibitors were modest with the relative risk of high dose therapy of 0.88. This 12% reduction was not significant statistically, but was similar to the trend in survival benefit seen in ATLAS. The relative risk reduction with higher dose ACE inhibitor therapy within ACE genotype class demonstrated a gene ordered effect similar to that seen with β-blockers, with the greatest impact within the DD subset though this failed to reach significance (figure 6).
Figure 6.
Relative Risk of event (death or transplantation) by ACE inhibitor dose use, GRACE study: Overall cohort and by ACE D/I genotype, DD,DI and II (adapted from table 4, reference 11)
The effect of high dose therapy in DD homozygotes is consistent with greater resistance to ACE inhibition in this subset. In subjects with heart failure, the blood pressure response to captopril demonstrates minimal effects in DD subjects, an intermediate response in heterozygotes, and the greatest impact in II patients (16). A similar interaction was demonstrated for chronic therapy with angiotensin receptor antagonists in hypertensive subjects (17). The studies suggest a diminished response to ACE inhibition or blockade with the ACE D allele. The higher ACE activity evident in ACE DD homozygotes is the most likely mechanism of this genetically determined drug “resistance”.
RAAS polymorphisms and Blood Pressure
The ACE D/I genotype does appear to predict the time course of blood pressure response to ACE inhibitors but not other anti-hypertensives (18). Genetic variability in the ACE pathway is not limited to the ACE D/I polymorphisms, and additional RAAS polymorphisms also influence blood pressure response to ACE inhibitors and may ultimately affect clinical outcomes. In a study of over 1,400 hypertensive patients the impact of ACE inhibitor therapy on blood pressure was linked to polygenetic variation in both the angiotensinogen and angiotensin II receptor genes (19). In a second study in 450 heart failure patients, haplotypes of the angiotensinogen gene were associated with poorer outcomes (20). Increasingly, genetic outcome analysis is moving toward more complex polygenic analysis and an interaction between the ACD allele and the alpha adducin Gly460TRP polymorphisms has been reported to worsen cardiovascular in a Flemish population study of Environment, Genetics and Health outcomes (21).
The prevalence of “aldosterone escape” on ACE inhibitors is greatest in the ACE DD genotype (22). The impact of the aldosterone receptor antagonist spironolactone on left ventricular remodeling is also diminished in ACE DD subset compared to the ID and II subsets (23) in a manner which closely parallels the diminished clinical response to ACE inhibitors. The influence of the ACE D/I polymorphism on the effect of aldosterone receptor antagonists on survival remains to be determined.
Aldosterone synthase
The aldosterone synthase gene (CYP11B2) promoter polymorphism (C-344T) has been investigated extensively with the -344C allele consistently associated with higher aldosterone synthase activity, linked to hypertension (24) and greater left ventricular remodeling (25). The African American Heart Failure Trial (AHeFT) investigated the addition of a fixed combination of isosorbide dinitrate and hydralazine (I/H) to standard therapy of ACE inhibitors and beta blockers. The genetic sub -study of AHeFT, Genetic Risk Assessment of Heart Failure in African Americans or “GRAHF”, was initiated to determine the genomic basis for the apparent racial differences in therapeutic efficacy of I/H and ACE inhibitors. In GRAHF, the aldosterone synthase promoter polymorphism was evaluated and the -344C allele was associated with significantly poorer event-free survival during the course of follow up (26).
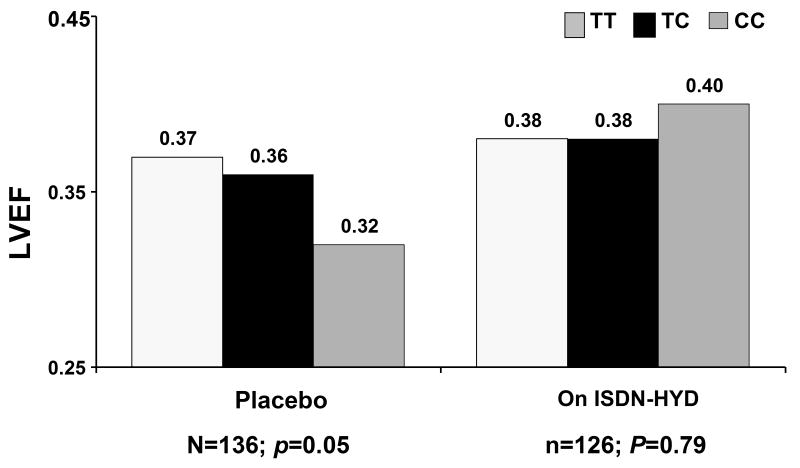
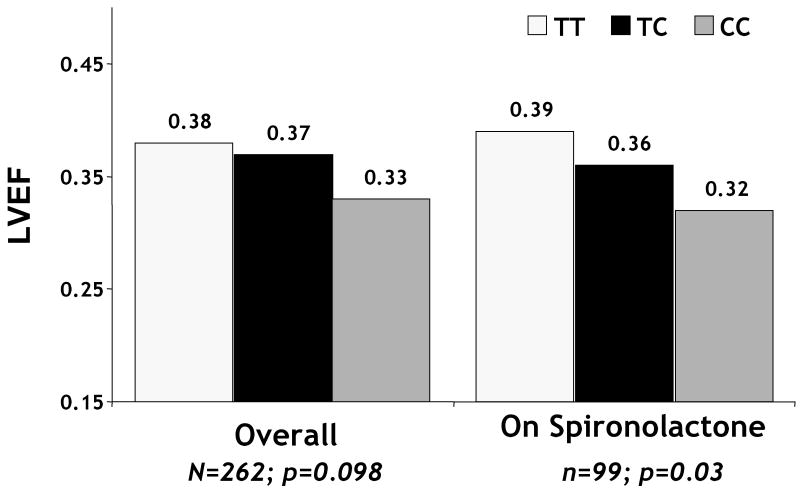
Consistent with previous studies, the -344C allele was also linked to LV remodeling; however its impact was diminished by treatment with I/H. In subjects randomized to placebo, an association of the -344C allele with lower LVEF at 6 months was apparent (p=0.05) which was not evident in patients treated with I/H (p=0.79) (Figure 7). The impact of therapy on remodeling for the - 344 CC genotype mirrors a study in a separate African Americans heart failure cohort which suggested improvements at one year in left ventricular dimension were greater among patients with the -344 CC genotype (27). A randomized study of the effect of the -344 promoter polymorphism in 500 hypertensive subjects suggest the impact of ACE inhibitors on blood pressure is diminished in those with the -344CC genotype (28). Surprisingly, subset analysis from AHeFT trial suggests treatment with the aldosterone receptor antagonist spironolactone did not diminish the adverse affect of the C allele on LV remodeling (figure 8) (26).
Figure 7.
LVEF at 6 months in AHeFT: Comparison by treatment group, placebo or ISDN-HYD (fixed combination of isosorbide dinitrate and hydralazine) of the impact of Aldosterone promoter genotype. The -344C allele associated with lower LVEF at six months for subjects on placebo (p=0.05) but not on ISDN-HYD (p=0.79) (reference 26)
Figure 8.
AHeFT: Impact of Aldosterone promoter genotype on LVEF at 6 months Overall and in subjects treated with aldosterone receptor antagonist (sprironolactone) The -344C allele associated with a trend toward lower LVEF at six months overall (p=0.098) which was more pronounced for subjects on spironolactone (p=0.03). (reference 26)
Endothelial Nitric Oxide Synthase (NOS3)
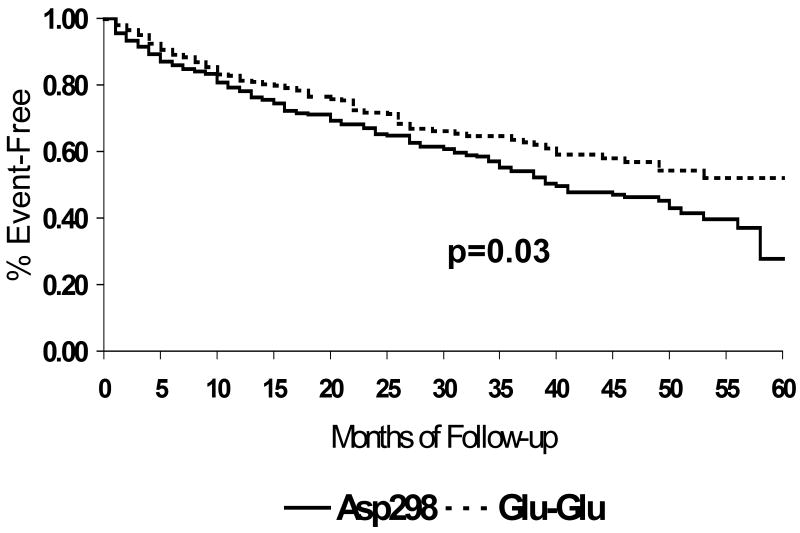
Nitric oxide (NO) plays an important protective role in heart failure (29, 30) and endothelial nitric oxide synthase (NOS3) is the predominant source of vascular NO. A common polymorphism (G894T) exists on exon 7 (codon 298: Glu298Asp) for which the wild type glutamate is replaced with aspartic acid (31). The functional role of this apparently charge neutral amino acid change remains controversial; however the Asp298 variant has a shorter half-life and therefore less NO activity in endothelial cell culture (32). The Asp298 variant has been linked to the risk of coronary disease, hypertension and stroke. For subjects with heart failure, functional assessment by metabolic stress testing demonstrates significantly higher VO2 max in subjects with the Glu298Glu phenotype compared to those homozygous or heterozygous for the Asp298 variant (33). This modulation of functional capacity by the Asp298Glu polymorphism parallels the impact on survival as poorer transplant-free survival was evident for subjects with the Asp298 variant compared to individuals homozygous for Glu298 (Figure 9) (33). While the Asp298 variant has been previously investigated as a risk factor for coronary disease, the impact in GRACE reflects a role as a modulator of heart failure, and was more apparent in the non-ischemic cohort.
Figure 9.
Transplant-free survival by endothelial nitric oxide synthase (NOS3) codon 298 polymorphism, GRACE study, University of Pittsburgh: Overall cohort (n=469): Asp298 variant (solid line, n=266), Glu298 homozygotes (dashed line, n=203). Event-free survival significantly poorer in subjects with the Asp298 variant, p=0.03 (reference 33)
Pharmacogenetic Interactions with NOS3 and ACE inhibitors
Clinical and animal studies demonstrate that NOS3 plays a central role in the therapeutic effects of ACE inhibitors. Murine knockout models have demonstrated that the post infarction benefit of ACE inhibitors on remodeling is dependent on NOS3 (34). In clinical studies of vascular reactivity, the effects of ACE inhibitors are diminished by pretreatment with NOS inhibitors. In GRACE, the examination of high dose versus low dose ACE inhibitor suggest a pharmacogenetic interaction with the NOS3 Glu298Asp polymorphism, as the impact of Asp298 was primarily in subjects on low dose ACE inhibitor, and was not evident in those receiving high dose therapy (35). Analysis of the impact of ACE inhibitor dose by genetic subset suggests the variable benefit of ACE inhibitors may be partially explained by variation at the NOS3 locus, however this will need additional investigation in larger cohorts.
Summary: Genetic Targeting of Heart Failure Therapy
For the ACE D/I polymorphism, linkage of the D allele to higher ACE activity results in a predictable impact for therapies inhibiting A-II production: ACE inhibitors and beta receptor antagonists. Both therapies appear to have their greatest impact on the 30% of the population who are homozygous for the D allele, genetically predicted to have the highest levels of neurohormonal activation. The adverse impact of the ACE D allele in heart failure has been demonstrated by several independent groups of investigators and is consistent with the role of renin-angiotensin activation in clinical progression. Genotyping of the ACE locus may identify large subsets patients who receive maximal benefit for aggressive neurohormonal blockade.
Predictably, genetic influences on heart failure therapy are polygenic. In addition to ACE, the therapeutic impact of beta-receptor antagonists is influenced by variation at the beta 1 receptor locus. The effectiveness of ACE inhibitor therapy may be influenced by variation of both ACE and NOS3. Pharmacogenetic investigations to date have not integrated more than one genetic locus into a clinical outcomes model, and larger cohorts will be required for adequate statistical power to address polygenic “background” effects. Analysis of genetic background has become a potential clinical tool for predicting heart failure outcomes and targeting therapeutic intervention. Prospective validation of the predictive impact of genetic variants will be required prior to the routine implementation of genetically individualized treatments.
Footnotes
This article was adapted from “Pharmacogenomics for Neurohormaonal Intervention in Heart failure” by Dennis M. McNamara, Heart Failure Clinics, volume 1, issue 1.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Osler W. The principles and practice of medicine. D. Appleton and Co.; New York: 1892. pp. 623–640. [Google Scholar]
- 2.Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–6. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rigat B, Hubert C, Corvol P, Soubrier F. PCR detection of the insertion/deletion polymorphism of the human angiotensin converting enzyme gene (DCP) (dipeptidyl carboxypeptidase 1) Nucleic Acids Res. 1992;20:1433. doi: 10.1093/nar/20.6.1433-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tiret L, Rigat B, Visvikis S, Breda SC, Corvol P, Cambien F, et al. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am J Hum Genet. 1992;51:197–205. [PMC free article] [PubMed] [Google Scholar]
- 5.Danser AH, Derkx FH, Hense HW, Jeunemaitre X, Riegger GA, Schunkert H. Angiotensinogen (M235T) and angiotensin-converting enzyme (I/D) polymorphisms in association with plasma renin and prorenin levels. J Hypertens. 1998;16:1879–1883. doi: 10.1097/00004872-199816121-00005. [DOI] [PubMed] [Google Scholar]
- 6.Cambien F, Poirier O, Lecerf L, Evans A, Cambou JP, Arveiler D, et al. Deletion polymorphism in the gene for angiotensin-converting enzyme is a potent risk factor for myocardial infarction. Nature. 1992;359:641–644. doi: 10.1038/359641a0. [DOI] [PubMed] [Google Scholar]
- 7.Ihnken R, Verho K, Gross M, Marz W. Deletion polymorphism of the angiotensin I-converting enzyme gene is associated with increased plasma angiotensin-converting enzyme activity but not with increased risk for myocardial infarction and coronary artery disease. Ann of Intern Med. 1996;125(1):19–25. doi: 10.7326/0003-4819-125-1-199607010-00004. [DOI] [PubMed] [Google Scholar]
- 8.Andersson B, Sylven C. The DD genotype of the angiotensin-converting enzyme gene is associated with increased mortality in idiopathic heart failure. J Am Coll Cardiol. 1996;28:162–7. doi: 10.1016/0735-1097(96)00098-8. [DOI] [PubMed] [Google Scholar]
- 9.Palmer BR, Pilbrow AP, Yandle TG, et al. Angiotensin-converting enzyme gene polymorphism interacts with left ventricular ejection fraction and brain naturetic peptide levels to predict mortality after myocardial infarction. J Am Coll Cardiol. 2003;41:729–36. doi: 10.1016/s0735-1097(02)02927-3. [DOI] [PubMed] [Google Scholar]
- 10.McNamara DM, Holubkov R, Janosko K, et al. Pharmacogenetic Interactions Between (β-Blocker Therapy and the Angiotensin-Converting Enzyme Deletion Polymorphism in Patients with Congestive Heart Failure. Circulation. 2001;103:1644–8. doi: 10.1161/01.cir.103.12.1644. [DOI] [PubMed] [Google Scholar]
- 11.McNamara DM, Holubkov R, Postava L, et al. Phamacogenetic interactions between ACE inhibitor therapy and the angiotensin-converting enzyme deletion polymorphism in patients with congestive heart failure. J Amer Coll Cardiol. 2004;44:2019–26. doi: 10.1016/j.jacc.2004.08.048. [DOI] [PubMed] [Google Scholar]
- 12.Campbell D, Aggrarwal A, Esler M, et al. Beta blockers, angiotenisn II, and ACE inhibitors in patients with heart failure. Lancet. 2001;358:1609–10. doi: 10.1016/S0140-6736(01)06660-0. [DOI] [PubMed] [Google Scholar]
- 13.de Groote P, Helbecque N, Lamblin N, et al. Beta-adrenergic receptor blockade and the angiotensin-converting enzyme deletion polymorphism in patients with chronic heart failure. Eur J Heart Fail. 2004;6:17–21. doi: 10.1016/j.ejheart.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Ishizawar D, Teuteberg JJ, Cadaret LM, Mathier MA, McNamara DM. The β1-adrenergic receptor mediates the pharmacogenetic interaction of the ACE D allele and beta-blockers. Clinical Translational Science. 2008;1(2):151–154. doi: 10.1111/j.1752-8062.2008.00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Packer M, Poole-Wilson PA, Armstrong PW, et al. Comparative effects of low and high doses of the angiotensin-converting enzyme inhibitor, lisinopril, on morbidity and mortality in chronic heart failure. Circulation. 1999;100:2312–8. doi: 10.1161/01.cir.100.23.2312. [DOI] [PubMed] [Google Scholar]
- 16.O'Toole L, Stewart M, Padfield P, Channer K. Effect of the insertion/deletion polymorphism of the angiotensin-converting enzyme gene on the response to angiotensin-converting inhibitors in patients with heart failure. J Cardiovasc Pharmacol. 1998;32:988–94. doi: 10.1097/00005344-199812000-00017. [DOI] [PubMed] [Google Scholar]
- 17.Kurland L, Melhus H, Karlsson J, et al. Angiotensin-converting enzyme gene polymorphism predicts blood pressure response to angiotensin II receptor type 1 antagonist treatment in hypertensive patients. J Hypertens. 2001;19:1783–87. doi: 10.1097/00004872-200110000-00012. [DOI] [PubMed] [Google Scholar]
- 18.Bhatnagar V, O'Connor DT, Schork NJ, et al. Angiotensin-converting enzyme gene polymorphism predicts the time-course of blood pressure response to angiotensin converting enzyme inhibition in the AASK trial. J Hypertens. 2007;25:2082–92. doi: 10.1097/HJH.0b013e3282b9720e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su X, Lee L, Li X, et al. Association between angiotensinogen, angiotensin II receptor genes, and blood pressure response to an angiotensin-converting enzyme inhibitor. Circ J Am Heart Assoc. 2007;115:725–32. doi: 10.1161/CIRCULATIONAHA.106.642058. [DOI] [PubMed] [Google Scholar]
- 20.Pilbrow AP, Palmer BR, Frampton CM, et al. Angiotensinogen M235T and T174M gene polymorphisms in combination doubles the risk of mortality in heart failure. J Am Heart Assoc Hyper. 2007;49:322–27. doi: 10.1161/01.HYP.0000253061.30170.68. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Zagato L, Kuznetsova T, et al. Angiotensin-converting enzyme I/D and α-adducin Gly460Trp polymorphisms. Hyper. 2007;49:1291–97. doi: 10.1161/HYPERTENSIONAHA.106.085498. [DOI] [PubMed] [Google Scholar]
- 22.Cicoira M, Zanolla L, Rossi A, et al. Failure of Aldosterone suppression despite angiotensin-converting enzyme (ACE) inhibitor administration in chronic heart failure associated with the ACE DD genotype. J Am Coll Cardiol. 2001;37:1808–12. doi: 10.1016/s0735-1097(01)01237-2. [DOI] [PubMed] [Google Scholar]
- 23.Ciocoira M, Rossi A, Bonapace S, et al. Effects of ACE gene insertion/deletion polymorphism on response to sprironolactone in patients with chronic heart failure. Am J Med. 2004;116:657–661. doi: 10.1016/j.amjmed.2003.12.033. [DOI] [PubMed] [Google Scholar]
- 24.Materson BJ. Variability in response to antihypertensive drugs. Am J Med. 2007 Apr;120(4 Suppl 1):S10–20. doi: 10.1016/j.amjmed.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Tiago AD, Badenhorst D, Skudicky D, et al. An aldosterone synthase gene variant is associated with improvement in left ventricular ejection fraction in dilated cardiomyopathy. Cardiovasc Res. 2002 Jun;54(3):584–9. doi: 10.1016/s0008-6363(02)00281-x. [DOI] [PubMed] [Google Scholar]
- 26.McNamara DM, Tam SW, Sabolinski ML, et al. Aldosterone synthase promoter polymorphism predicts outcome in African Americans with heart failure. J Am Coll Cardiol. 2006;48:1277–82. doi: 10.1016/j.jacc.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 27.Biolo A, Chao T, Duhaney TAS, et al. Usefulness of the aldosterone synthase gene polymorphism C-344-T to predict cardiac remodeling in African-Americans versus non-African-Americans with chronic systolic heart failure. Am J Cardiol. 2007;100:285–90. doi: 10.1016/j.amjcard.2007.02.097. [DOI] [PubMed] [Google Scholar]
- 28.Yu HM, Lin SG, Liu GZ, et al. Associations between CYP11B2 gene polymorphisms and the response to angiotensin-converting enzyme inhibitors. Clin Pharmacol Ther. 2006;79:589–9. doi: 10.1016/j.clpt.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Kelly RA, Balligand JL, Smith TW. Nitric oxide and cardiac function. Circ Res. 1996;79:363–80. doi: 10.1161/01.res.79.3.363. [DOI] [PubMed] [Google Scholar]
- 30.Drexler H. Nitric oxide synthases in the failing human heart: A double-edged sword? Circulation. 1999;99:2972–5. doi: 10.1161/01.cir.99.23.2972. [DOI] [PubMed] [Google Scholar]
- 31.Philip I, Plantefeve G, Vuillaumier-Barrot S, et al. G894T polymorphism in the endothelial nitric oxide synthase gene is associated with an enhanced vascular responsiveness to phenylephrine. Circulation. 1999;99:3096–8. doi: 10.1161/01.cir.99.24.3096. [DOI] [PubMed] [Google Scholar]
- 32.Tesauro M, Thompson WC, Rogliani P, et al. Intracellular processing of endothelial nitric oxide synthase isoforms associated with differences in severity of cardiopulmonary diseases: Cleavage of proteins with aspartate vs. glutamate at position 298. Proc Natl Acad Sci USA. 2000;97:2832–5. doi: 10.1073/pnas.97.6.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McNamara DM, Holubkov R, Postava L, et al. The Asp298 variant of endothelial nitric oxide synthase: Effect on Survival for patients with congestive heart failure. Circulation. 2003;107:1598–602. doi: 10.1161/01.CIR.0000060540.93836.AA. [DOI] [PubMed] [Google Scholar]
- 34.Yang XP, Liu YH, Sheseley EG, et al. Endothelial nitric oxide gene knockout mice cardiac phenotypes and the effect of angiotensin-converting enzyme inhibitor on myocardial ischemia/reperfusion injury. Hypertension. 1999;34:24–30. doi: 10.1161/01.hyp.34.1.24. [DOI] [PubMed] [Google Scholar]
- 35.Bedi M, Murali S, MacGowan G, et al. High dose ACE inhibitors reduces the impact of the NOS3 Asp298 variant on heart failure survival. Circulation. 2003;1085:IV–444. abstract. [Google Scholar]