Abstract
Objective
Blood-sucking arthropods salivary glands (SGs) contain a remarkable diversity of antihemostatics. The aim of this study was to identify the unique salivary anticoagulant of the sand fly Lutzomyia longipalpis, which remained elusive for decades.
Methods and Results
Several L. longipalpis salivary proteins were expressed in HEK293 cells and screened for inhibition of blood coagulation. A novel 32.4-kDa molecule, named Lufaxin, was identified as a slow, tight, non-competitive, and reversible inhibitor of Factor Xa (FXa). Notably, Lufaxin primary sequence does not share similarity to any physiological or salivary inhibitors of coagulation reported to date. Lufaxin is specific for FXa and does not interact with FX, DEGR- FXa, or 15 other enzymes. In addition, Lufaxin blocks prothrombinase and increases both PT and aPTT. Surface plasmon resonance experiments revealed that FXa binds Lufaxin with a KD ~3 nM, and isothermal titration calorimetry determined a stoichiometry of 1:1. Lufaxin also prevents PAR2 activation by FXa in the MDA-MB-231 cell line and abrogates edema formation triggered by injection of FXa in the paw of mice. Moreover, Lufaxin prevents FeCl3-induced carotid artery thrombus formation and prolongs aPTT ex vivo, implying that it works as an anticoagulant in vivo. Finally, SG of sandflies was found to inhibit FXa and to interact with the enzyme.
Conclusion
Lufaxin belongs to a novel family of slow-tight FXa inhibitors, which display antithrombotic and antiinflamatory activities. It is a useful tool to understand FXa structural features and its role in pro-hemostatic and pro-inflammatory events.
Keywords: hematophagy, thrombosis, leishmaniasis, vector biology, microcirculation
Salivary glands (SGs) of blood-sucking arthropods display a notable variety of negative modulators of vascular biology.1 Several molecules have been studied in detail, including anticoagulants,2 platelet aggregation inhibitors,3 and vasodilators.4 These activities are aimed to block the host response to an injury (i.e., bite), therefore contributing to successful blood feeding.1 In some cases, saliva also contributes to transmission of infections agents to the vertebrate host, a phenomenon most notably demonstrated for leishmaniasis5—a vector-borne disease transmitted by the bite of the sand flies Lutzomyia longipalpis sp. and Phlebotomus papatasi sp, among others. Saliva of these phlebotomines expresses a number of pharmacologically active components which modulate hemostasis and the immune response. For instance, saliva of sand flies contains an apyrase that inhibits platelet aggregation,6 a potent vasodilator (maxadilan) that promotes an increase in blood flow,7 and at least one immunomodulator (e.g., SP15) that skews the host immune response toward Th1 polarization.8 However, several proteins coded by their corresponding SG transcripts remains without a defined function.9–11 Likewise, some biological functions described in the salivary gland have not been associated with a specific protein. For example, the anticoagulant of L. longipalpis or P. papatasi remained elusive for decades.
A remarkable diversity of anticoagulants targeting FVIIa/tissue factor(TF), FIX(a), FXa, thrombin, and of the contact pathway have been reported in other blood-sucking animals, including mosquitoes, ticks, bugs, leeches and bats, but not sand flies.2 Among several coagulation factors, FXa is a particularly attractive target because it plays a central role in the coagulation cascade, where both extrinsic and intrinsic pathways converge, leading to prothrombinase assembly with subsequent thrombin generation and fibrin formation.12 FXa also activates protease-activated receptor (PAR) 1 or PAR2 in different cell types, which enables this enzyme to promote inflammation and immune modulation in the absence of fibrin formation.13–16
The importance of FXa in the coagulation cascade is illustrated by the tight regulation of its activity by three physiological inhibitors: tissue factor pathway inhibitor (TFPI), antithrombin and protein Z. TFPI is a multidomain Kunitz-type inhibitor that binds to the active site of FXa by the second Kunitz domain, and this complex blocks FVII/TF.17 Antithrombin is a serpin that binds heparin and regulates proteolytic activity of FXa by binding to the active site, trapping the enzymes in an inactive complex.18,19 Protein Z serves as a cofactor for the inhibition of FXa by protein Z-dependent protease inhibitor.20 Notably, only 5 distinct salivary inhibitors targeting FXa have been molecularly cloned and expressed from blood-sucking animals, including Kunitz-type from ticks or black flies,21,22 Ascaris-type inhibitor from Ancylostoma sp,23 antistasin-like from leeches,24 serpins from mosquitoes,25 and Salp family inhibitors from ticks.26 These anticoagulants distinctly affect FXa function through a variety of mechanisms.2 This description underscores how evolutionary pressure has recruited different genes coding for families of proteins which exert anticoagulation through blockade of FXa.
To identify the anticoagulant of L. longipalpis, a number of salivary cDNAs were cloned in VR2001-TOPO vector, which enables protein expression in mammalian cells HEK293.27 Through screening of activities, it was discovered that one recombinant protein from the SG of L. longipalpis possesses potent and specific anticoagulant activity toward FXa. This inhibitor was named Lufaxin (Lutzomyia longipalpis Factor Xa inhibitor) and was found to tightly bind to FXa, attenuate inflammation triggered by the enzyme in vitro and in vivo, and to prevent arterial thrombosis in a mouse model.
Methods
Reagents
FXa, FX, and FIXa were from Enzyme Research Laboratories (South Bend, IN). Dansyl-Glu-Gly-Arg(DEGR)-FXa, FXIa, FVa, prothrombin and thrombin were from Hematologic Technologies (Essex Junction, VT). Fibrillar collagen (from equine tendons) was from Chronolog (Haverton, PA), and U46619 was purchased from Cayman Chemicals (Ann Arbor, MI). APTT (STA-PTT Automate) and PT (Neoplastine CI Plus) reagents were from Diagnostica Stago (Asnieres, France). S2222 (N-benzoyl-L-isoleucyl-L-glutamyl-glycyl-L-arginine-p-nitroaniline hydrochloride) and S2238 (H-d-phenylalanyl-l-pipecolyl-l-arginine-p-nitroaniline dihydrochloride) were obtained from Diapharma (West Chester, OH). Phosphatidylcholine and phosphatidylserine were from Sigma Co. (Saint Louis, MO). Anti-phospho-ERK 1/2 and polyclonal anti-ERK 1/2 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Secondary antibodies conjugated with biotin and peroxidase-conjugated streptavidin were obtained from Zymed (Invitrogen, San Diego, CA).
Sand Flies and Preparation of SG Homogenate (SGH)
L. longipalpis (Jacobina strain), P. papatasi, and P.duboscqi were reared at the Laboratory of Malaria and Vector Research (NIAID/NIH) using as larval food a mixture of fermented rabbit feces and rabbit food. SGH homogenates were obtained as reported in detail in the Supplemental data.
Lufaxin Properties
cDNA for mature Lufaxin (gi41397464; clone LJL143) codes for a protein of predicted molecular mass of 32495.78 da (278 amino acids [aa]) with an estimated pI 8.27. Extinction coefficient at 280 nm is 36180 (all disulfide bonds); A280 nm/cm0.1% (1 mg/ml), 1.0975. Potential N-, or O-glycosylation site in Lufaxin were predicted with NetNGlyc 1.0 or NetOGlyc 3.0 servers, respectively.
Cloning of L. longipalpis cDNAs in His-Tagged TOPO Vector
VR2001-TOPO is a topoisomerase adaptation of VR1020 plasmid (Vical, Inc., San Diego, CA) described in a previous report.27 cDNA of Lufaxin (and other candidates) were amplified by PCR using a specific forward primer deduced from the amino-terminus region and a specific reverse primer containing an ATGATGATGATGATGATG motif between the stop codon and the carboxy-terminus region to introduce a 6xHis tag. The expected amplified sequences were predicted to code for proteins starting after the natural cleavage site and containing a 6xHis tag at the C-terminus region.
Production and purification of recombinant proteins
VR2001-TOPO plasmids coding for Lufaxin and other salivary proteins containing a 3’ histidine tag were used for protein expression in HEK-293 F cells at the Protein Expression Laboratory at NCI-Frederick (Frederick, Maryland), and reported elsewhere.28 The supernatant was collected after 72 hours and concentrated from 500 ml to 300 ml using a Stirred Ultrafiltration Cell unit (Millipore) with a 30 kDa ultrafiltration membrane (Millipore). Purification of Lufaxin was achieved using a HiTrap Chelating HP columns (GE Healthcare) using a gradient of imidazole followed by a chromatography in a gel-filtration column, as described in detail in the Supplemental data.
Polyclonal Antibodies against Lufaxin
Antibodies were produced as described in detail in the Supplemental data.
PAGE and Western Blotting
The samples were treated with 4× NuPAGE LDS sample buffer and analyzed in NuPAGE 4–12% gels with MES running buffer.
Deglycosylation of Lufaxin
This was performed using the Enzymatic DeGlycoMx Kit from QA-Bio (Palm Desert, CA) which contains a mixture of PNGase F, Sialidase, β-Galactosidase, Glucosaminidase and O-Glycosidase. The assay was performed following the manufacturer instructions, and described in the Supplemental data.
Platelet Aggregation Assays
Platelet-rich plasma was obtained by plateletpheresis from medication-free platelet donors at the DTM/NIH blood bank. Aggregation was performed as described previously.28
Contraction of Rat Aorta
Contraction of rat aortic ring preparations by U46619 was measured isometrically and recorded with transducers from Harvard Apparatus Inc. (Holliston, MA) as reported,29 and described in detail in the Supplemental data. Briefly, aortic rings were suspended in a 0.5-mL bath kept at 36°C and were pre-constricted by 100 nM U-46619 before addition of Lufaxin to give final concentrations of 1 µM, or salivary gland homogenates of Rhodnius prolixus (0.04 of one pair of glands/ml, with an approximate final concentration of nitrophorins of 2 µM - positive control).
Recalcification Time
Clotting activity was measured by the recalcification time of human plasma using a Thermomax microplate reader (Molecular Devices, Menlo Park, CA) with a kinetic module, as described in the Supplemental data.
aPTT and PT Assays
The effect of Lufaxin on coagulation tests aPTT and PT was evaluated on an Amelung KC4A coagulometer (Labcon, Heppenheim, Germany) as reported in the Supplemental data.
Prothrombinase Assembly
Activation of prothrombin by human FXa was performed in TBS-Ca2+ (20 mM Tris-HCl, 150 mM NaCl, 5 mM CaCl2, 0.3% BSA, pH 7.5), using a discontinuous assay as reported before30 and described in the Supplemental data.
Kinetics Studies
This was performed as described31 using chromogenic substrate (S2222) hydrolysis specific for FXa and described in detail in the Supplemental data.
Determination of the active site of Lufaxin
In a PCR tube, Lufaxin (1.9 µM) was incubated with human FXa (2.7 µM) for 3 or 24 hours at 37°C, with and without 5 mM Ca2+. LDS loading buffer and DTT were added to the tubes, warmed for 10 min at 70°C, and the mixture was loaded in a 4–12% NU-PAGE gel (MES buffer) and Coomassie Blue stained. See Blue standard was used as molecular mass marker (Invitrogen).
Surface Plasmon Resonance (SPR)
All SPR experiments were carried out in a T100 instrument (Biacore Inc., Uppsala, Sweden) following the manufacturer's instructions. For immobilization using an amine coupling kit (Biacore), CM5 chips were activated with 1-ethyl-3-(dimethylaminopropyl) carbodiimide, and N-hydroxysuccinimide before injection of Lufaxin (6.5 µg/mL) or FXa (30 ug/ml) in acetate buffer, pH 5.5. Remaining activated groups were blocked with 1 M ethanolamine, pH 8.5, resulting in a final immobilization of 604.5 RU (for Lufaxin) or 661.7 RU (for FXa). Kinetic experiments were carried out by injecting FXa for a contact time of 180 seconds at a flow rate of 30 µL/minute at 25°C and described in the Supplemental data.
Isothermal Titration Calorimetry (ITC)
Lufaxin binding to FXa was performed using a VP-ITC microcalorimeter (Microcal, Northampton, MA) at 30°C. Prior to the run, the proteins were dialyzed against 20 mM Tris-HCl, 0.15 M NaCl, pH 7.4 for binding experiments. Titration experiments were performed by making successive injections of 5 µL each of 20 µM FXa into the 2-mL sample cell containing 2 µM Lufaxin until near-saturation was achieved as described in the Supplemental data.
Cell Culture
MDA-MB-231 breast cancer cells were maintained in ISCOVES medium (Invitrogen, CA) supplemented with 10% FBS in culture flasks in a 5% CO2-air mixture at 37°C. Subconfluent cultures were washed twice with PBS, and cells were detached with Hank’s solution containing 10 mM HEPES and 0.2 mM EDTA. Cells were seeded at 5 × 105 cells/well in 6-well tissue culture plates for signaling assays.
PAR2 Signaling Assay
MDA-MB-231 cells were serum starved for 90 minutes and stimulated with 10 nM FXa (in ISCOVES medium, no FBS) for 10, 15, 30, and 60 minutes. Lufaxin (50 nM) was added 1 hour prior to stimulation with FXa. To avoid PAR1 activation by thrombin, assays were performed in the presence of 10 nM hirudin as described.32 Detection of p-ERK was carried out as described in the Supplemental data.
Paw Edema in Mice
Female C57BL/6 mice, 6–8 weeks old, were purchased from the Jackson Laboratory (Bar Harbor, ME) and maintained in the NIAID Animal Care Facility and experiments were performed as reported.33 Briefly, the posterior footpad thickness of each mouse was recorded using a caliper, prior to each injection (Mitutoyo America Corp., Aurora, IL). Subsequently, PBS, FXa alone, or FXa incubated wiht Lufaxin were injected intradermally in the paw using a 0.3-mL syringe. As an index of edema formation, paw thickness was then measured at 15, 30, 45, and 60 minutes as described in the Supplemental data
FeCl3-Induced Artery Thrombosis
Thrombus formation was induced by applying a piece of filter paper (1 × 2 mm) saturated with 7.5% FeCl3 solution on the adventitial surface of the artery for 3 minutes. After exposure, the filter paper was removed, and the vessel was washed with sterile normal saline. Carotid blood flow was continuously monitored for 60 minutes or until complete occlusion (0 flow for at least 10 seconds) occurred as described in detail in the Supplemental data.
Tail Bleeding Assay
Bleeding was estimated by the tail transection method as described in the Supplemental data.
Protease inhibition assays
This was performed as described in the Supplemental data.
Statistical analysis
Results are expressed as means ± SE. Statistical differences among the groups were analyzed by t-test, or analysis of variance (ANOVA) using Bonferroni as a multiple comparison post-test. Significance was set at p≤0.05 (Graph-Pad Prisma software, La Jolla, CA).
Results
Identification of Lufaxin as a Novel Specific FXa Inhibitor
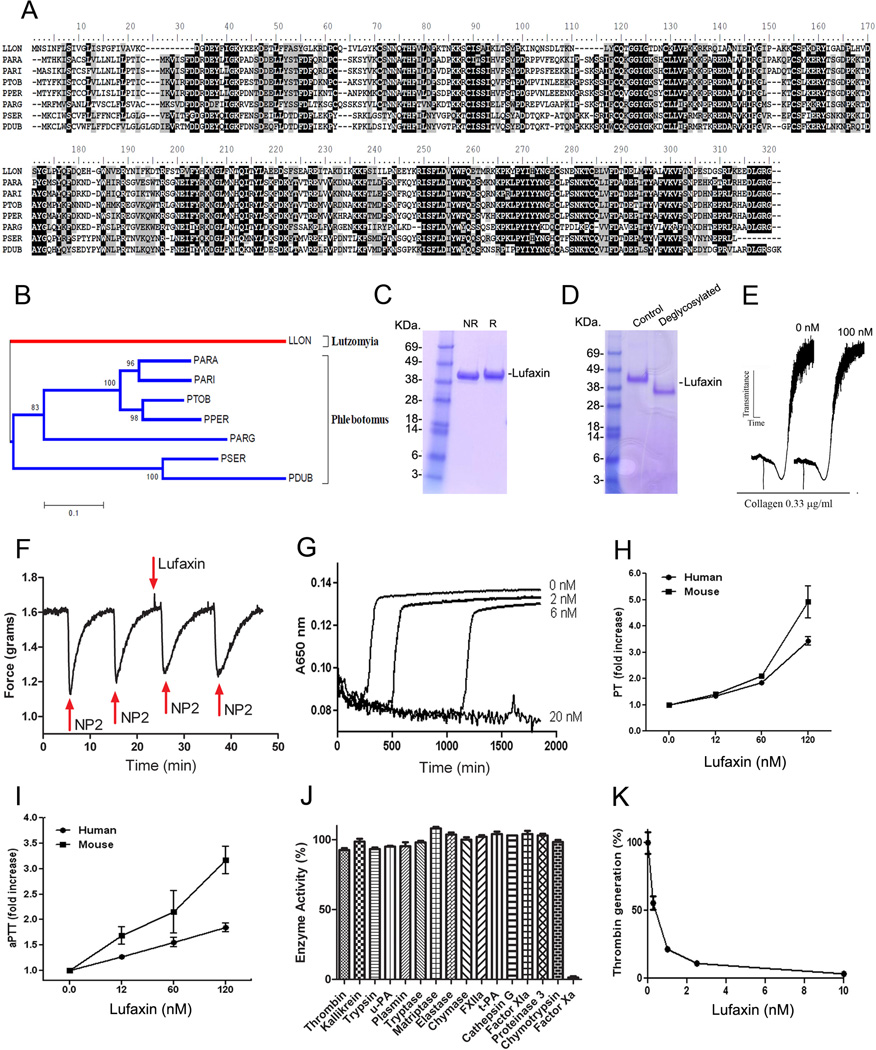
Lufaxin (gi 41397464) from L. longipalpis is a 32.4-kDa protein of 278 aa, 8 cysteines, and pI 8.27. It belongs to a family of highly conserved salivary proteins present in other Phlebotominae sp. including P. arabiensis, P. ariasi, P. perniciosus, P. tobbi, P. sergenti, P. argentipes, and P. dubosqi (Figure 1A). The function of this family of protein has remained elusive so far.9–11 Phylogenetic analysis revealed that these molecules are closely related suggesting that Lufaxin family of anticoagulant evolved from a common ancestor (Figure 1B). It is also evident that Lufaxin members found in the Phlebotominae clade are separated from the L. longipalpis homolog (Figure 1B), consistent with the separation of Old World and New World sand flies over 60 million years ago after the split of Gondwanaland.
Figure 1.
Lufaxin is a novel, specific FXa inhibitor from Phlebotominae sp. A, Clustal alignment of Lufaxin from L. longipalpis (Llon, gi41397464) and orthologues from P. arabiensis (Para, gi242564823), P. ariasi (Pari, gi61807168), P. tobbi (Ptob, gi299829388), P. perniciosus (Pper, gi76446599), P. argentipes (Parg, gi74486551), P. sergenti (Pser, gi299829440), and P. dubosqi (Pdub, gi112496879). B, Phylogeny for the sequences described in A. Analysis was performed using neighbor-joining analysis using Bootstrap support (10,000 replicates). The numbers indicate the 20% amino acid divergence in the sequences. C, Lufaxin was expressed in HEK293 cells and purified with a Ni2+ column followed by a gel-filtration chromatography. Samples were loaded in a NU-PAGE gel under non-reducing (NR) or reducing (R) conditions. Gels were stained with Coomassie Blue. On the left, molecular mass markers are indicated. D, Lufaxin is glycosylated. Lufaxin incubated with a combination of glycosidases migrates as a lower molecular mass protein in a NUPAGE gel. E, Lufaxin does not affect platelet aggregation. PRP was stimulated with collagen (0.66 µg/mL) with or without Lufaxin (100 nM). Aggregation response was monitored by turbidimetry using a Lumi-Aggregometer. F, Lufaxin does not produce vasodilation. Lufaxin (100 nM) was added to rat aorta strips at the time indicated (30 minutes) by the arrow. No vasodilation was detectable. As a control, 2 µM NP2 (NO-carrying protein) was delivered to the chamber (arrows) at different time points, which was followed by vasodilation. G, Indicated concentrations of Lufaxin were tested by the recalcification time assay. Citrated human plasma was mixed with recombinant proteins and reactions were initiated by the addition of CaCl2. Clotting formation was monitoring at 650 nm. H, Lufaxin prolongs PT. Lufaxin was added to plasma, followed by addition of PT reagent and Ca2+. Clotting was estimated using a coagulometer. PT time; human plasma, 16.67±0.20 s; mice plasma, 15.79±0.40 s. I, Lufaxin prolongs aPPT. Lufaxin was added to plasma, followed by addition of aPTT reagent and Ca2+. Clotting was estimated using a coagulometer. aPPT time; human plasma, 42.92±2.08 s; mice plasma, 37.61±3.70 s. J, Lufaxin does not inhibit activity of several enzymes, except FXa. Catalytic activity was estimated by fluorogenic substrate hydrolysis. K, Lufaxin inhibits thrombin formation by prothrombinase. FXa was incubated with Lufaxin, followed by addition of FVa, PC/PS, and prothrombin in the presence of Ca2+. After 6 minutes, aliquots were taken to estimate thrombin generation and expressed as % of control (no inhibitor). Data is the average ± SE of triplicate determinations (n=3).
In an attempt to identify the function of Lufaxin, its corresponding cDNA was cloned in a VR2001-TOPO27 expression vector and described in Methods, and used for transfection of HEK293-F cells. Supernatants containing recombinant Lufaxin (containing a 6xHIS-tag) were loaded in a Ni2+-column followed by gel-filtration chromatography. Purified Lufaxin was analyzed by NU-PAGE and found to migrate as a single band of ~38 kDa under denaturing or reducing conditions (Figure 1C). The N-terminal for Lufaxin obtained by Edman degradation identified the sequence DGDEYFIGKYKED, which is in agreement with the N terminus predicted for the mature protein, according to the cDNA. To verify whether the higher molecular mass of Lufaxin was due to glycosylation, the inhibitor was incubated with a combination of glycosidases, or D.W. (control) followed by NU-PAGE. Figure 1D shows that the migration pattern of Lufaxin changes to lower molecular mass after incubation with the mixture of enzymes. This indicates that recombinant Lufaxin is glycosylated, explaining the difference between the actual and predicted molecular masses. This experimental finding is also in agreement with the prediction of one N-linked glycosylation (calculated value 0.6638; threshold at 0.5) in the sequence NKTC (aa 239) of mature Lufaxin. On the other hand, Lufaxin sequence does not display potential O-linked glycosylation sites.
Lufaxin was tested in screening assays in an attempt to identify its molecular target. Lufaxin (100 nM) did not inhibit platelet aggregation by collagen (Figure 1E) or promote vasodilation (Figure 1F). When tested in the recalcification time assay, Lufaxin dose-dependently prolonged clotting time, suggesting that it is an anticoagulant (Figure 1G). To determine whether Lufaxin blocks coagulation factors involved in the intrinsic, extrinsic, or common pathway, PT and aPTT were performed. Figure 1H shows that Lufaxin dose-dependently prolongs PT in both human and mouse plasma. Likewise, Lufaxin prolonged aPTT in a dose-dependent manner in either species and at a similar concentration (Figure 1I). These results indicated that Lufaxin interferes with a common pathway of the coagulation cascade. Concentrations of Lufaxin to block PT and aPTT were higher than observed for inhibition of the recalcification time likely due to the stronger activation of the coagulation (i.e. FXa production) by the PT or aPTT reagents when compared to CaCl2 only. In fact, control clotting time for PT and aPTT were 16.67±0.20 sec and 42.92±2.08 sec respectively, while it took 200–250 sec for the plasma to clot in the recalcification time assay. Next, specificity of Lufaxin was tested by incubating a molar excess of the inhibitor with FXa or other enzymes involved in coagulation/inflammation (e.g., thrombin, FXIa, FXIIa, kallikrein, chymase, tryptase, matryptase, elastase, cathepsin G, proteinase 3), fibrinolysis (e.g., plasmin, uPA,t-PA) or digestive processes (e.g., trypsin and chymotrypsin). Incubation was followed by addition of small fluorogenic substrates specific for each enzyme. Figure 1J demonstrates that only FXa was inhibitable by Lufaxin, which thus characterizes it as a specific inhibitor for FXa.
FXa plays a critical role in prothrombinase assembly, through conversion of prothrombin to thrombin in the presence of membrane phospholipids, FVa, and calcium.12 To ascertain whether Lufaxin blocks prothrombinase, inhibitor and enzyme were incubated followed by addition of other components of the complex. Lufaxin incubated with FXa dose-dependently blocks thrombin formation, indicating that the inhibitor not only prevents small chromogenic substrate hydrolysis by FXa but also interferes with the productive assembly of the prothrombinase (Figure 1K).
Lufaxin is Present in the SG of L. longipalpis
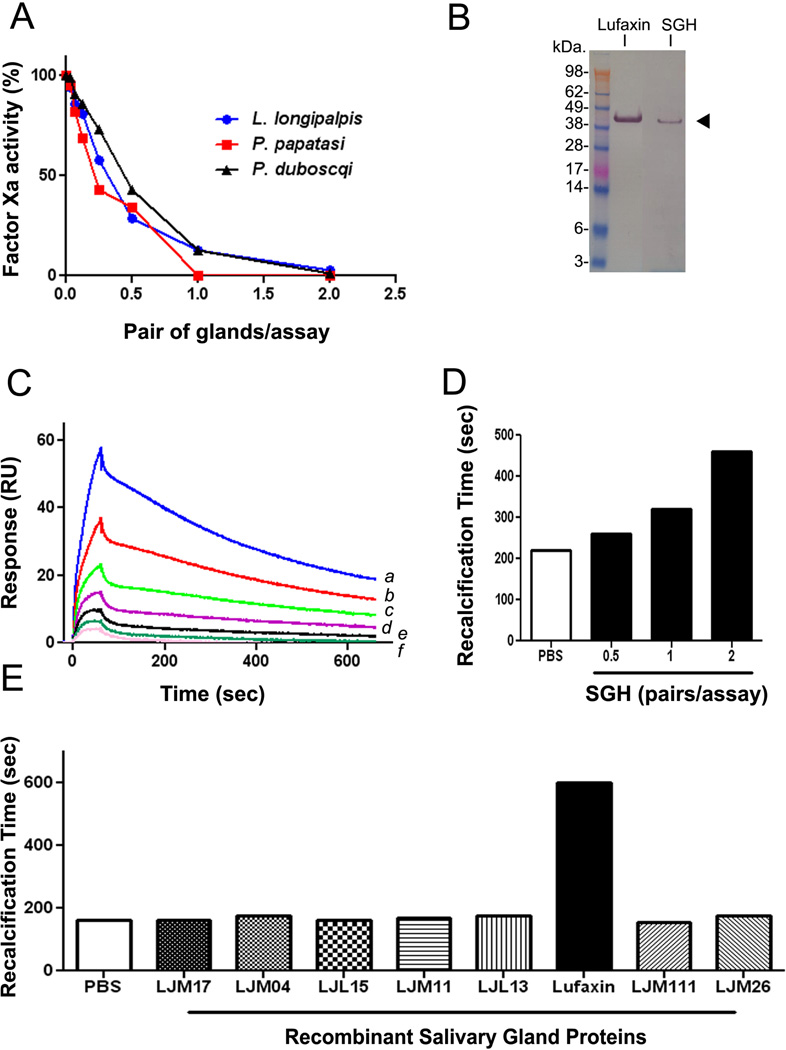
To verify whether an FXa inhibitor was expressed in the SG of L. longipalpis and other Phlebotominae, fresh SGHs were obtained and tested for inhibition of FXa catalytic activity using S2222. Figure 2A shows that SGH from L. longipalpis, P. papatasi, or P. duboscqi cause dose-dependent inhibition of FXa catalytic activity, with an IC50 ~ 0.5 pairs/assay. Experiments performed side by side with Lufaxin (0–60 nM) or SGH (0–2 pairs/assay), which were incubated with FXa (0.5 nM) followed by addition of S2222 (250 µM) indicate that the concentration of Lufaxin in 1 pair of gland is approximately15–30 nM (Figure 1S, Supplemental data). In addition, 2B demonstrates that both recombinant Lufaxin and a protein with similar migration properties from the SGH were recognized by anti-Lufaxin antibody. Next, SGH was tested for FXa-binding activity using SPR. FXa was immobilized in a CM5 chip, and L. longipalpis SGH was used as an analyte. Figure 2C shows that SGH produces an increase in resonance units in a dose-dependent manner, which is consistent with FXa-binding protein present in the SGH. This was congruent with the finding that L. longipalpis SGH also prolongs recalcification time (Figure 2D). Finally, a number of recombinant proteins from the salivary gland of L. longipalpis which were expressed in the same system as Lufaxin were tested side by side. Figure 2E demonstrates that Lufaxin is the only inhibitor that prolongs recalcification time, while 7 other recombinant proteins were without effect.
Figure 2.
Salivary glands (SGs) from Lutzomyia and Phlebotominae sp. exhibit anti-FXa activity. A, Salivary gland homogenate (SGH) from L. longipalpis, P. papatasi, and P. duboscqi were incubated with FXa (0.5 nM) in TBS-BSA buffer for 1 hour, followed by addition of S2222 (250 µM). Reactions were followed for 1 hour and inhibition of FXa activity estimated as percent of control (PBS only). B, Detection of Lufaxin in the SGH of L. longipalpis. Lufaxin or SGH were loaded in a NU-PAGE gel, followed by Western-blot using anti-Lufaxin polyclonal antibody. The arrowhead indicates the bands corresponding to recombinant Lufaxin and native Lufaxin (from the SGH). C, SPR experiments demonstrate that SGH of L. longipalpis binds to immobilized FXa. SGH (a, 2 pairs; b, 1 pair; c, 0.5 pair, d, 0.25 pair; e, 0.125 pair, and f, 0.06 pair in 100 µL of HBS-P) was used as an analyte to bind immobilized FXa. Association phase of 60 seconds was followed by 600-second dissociation. D, L. longipalpis SGH prolongs recalcification time. SGH (0–2 pairs/assay) was added to plasma and CaCl2 was added to start reactions (7.5 mM, final concentration). Clot formation was estimated by turbidimetry at 650 nm. Assay volume was 100 µL. E, Lufaxin and several recombinant proteins (50 nM each) from the SG of L. longipalpis were incubated with plasma and recalcification time estimated as in D. Only Lufaxin prolonged the clotting time. Experiments were performed at least three times and confirmed with different preparations of SGH or batches of recombinant Lufaxin.
Lufaxin Is a Slow, Tight and non-competitive Inhibitor of FXa
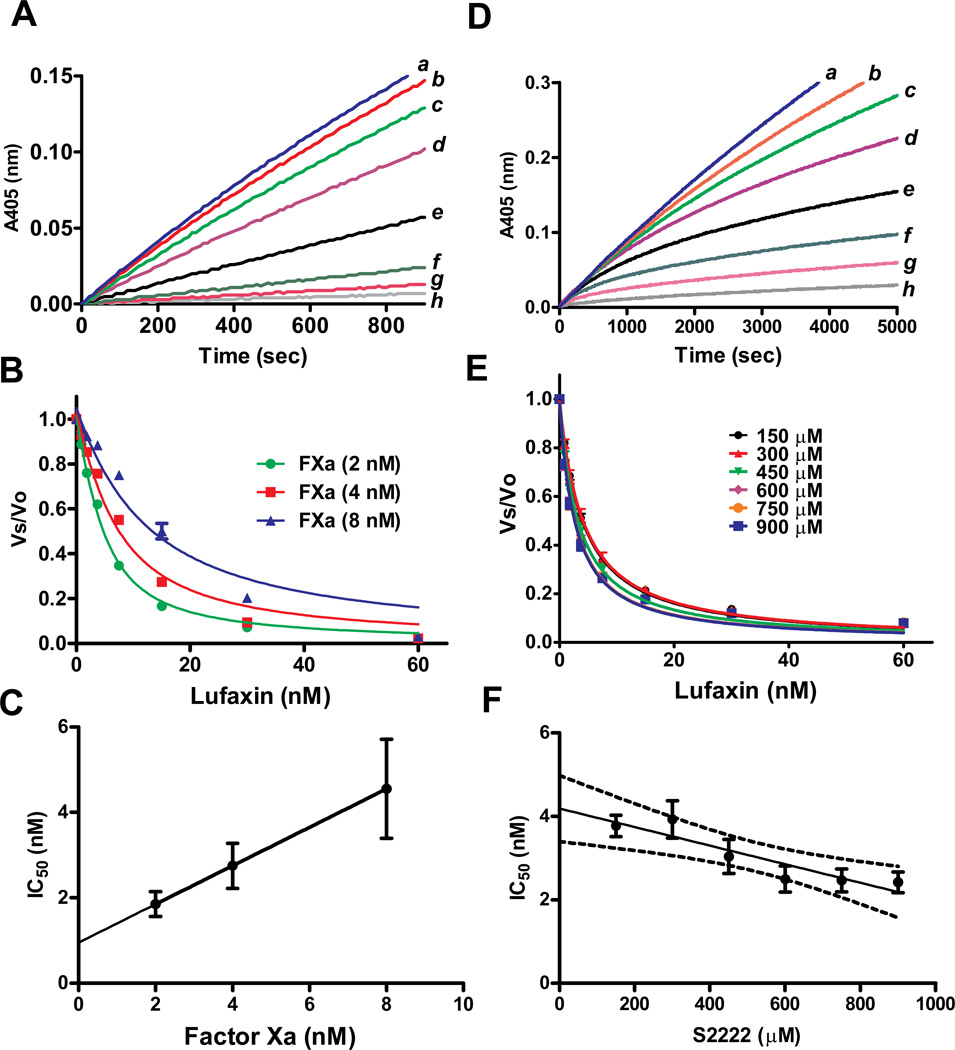
We examined whether inhibition by Lufaxin could be characterized as a tight inhibitor of FXa using S2222. For these experiments, Lufaxin (0–60 nM) and FXa (2, 4, and 8 nM) were incubated for 60 minutes followed by addition of S-2222 (250 µM) to start reactions. Figure 3A shows that inhibition was immediate, resulting in linear progress curves. The data were transformed as Vs/Vo and fitted with the Morrison equation for tight inhibitors,34,35 which allows calculation of the IC50 for each FXa concentration tested in the assay (Figure 3B). It has been demonstrated that an IC50 value obtained from this treatment of the data is similar to the concentration of total enzyme in the sample (i.e., within a factor of 10) when an inhibitor is of the tight-binding type. This is because a tight-binding inhibitor interacts with the enzyme in nearly stoichiometric fashion.34,35 In other words, the higher the concentration of enzyme present, the higher the concentration of inhibitor required to reach half-maximal saturation of the inhibitor binding sites. Thus, a plot of IC50 as a function of [E] (at a single, fixed substrate concentration) is expected to yield a straight line with a slope of 0.5, and y intercept equals apparent Ki (Ki*). The value Ki* is related to true Ki by factors involving substrate concentration and Km, depending on the mode of interaction between inhibitor and enzyme.34,35 Figure 3C reveals that inhibition of FXa by Lufaxin obeys typical graphic properties of a tight inhibitor, with a slope of 0.4504 and calculated Ki* of 0.95 ±0.07 nM.
Figure 3.
Lufaxin is a tight and slow inhibitor of FXa. A, Typical progress curves for FXa-mediated S2222 hydrolysis in the absence (curve a) and presence of Lufaxin (curves b, 0.93 nM; c, 1.8 nM; d, 3.75 nM; e, 7.5 nM; f, 15 nM; g, 30 nM, and h, 60 nM). Reactions started with addition of S2222 (250 µM) to a mixture containing Lufaxin incubated for 1 hour with FXa (2 nM). Substrate hydrolysis was followed for 2 hours at 37°C, at 405 nm. B, The experiment was performed as in A but FXa concentrations were 2, 4, and 8 nM. The ratio of Vs/Vo was plotted against Lufaxin concentration and data fitted with the Morrison equation to calculate the IC50 at each enzyme concentration. C, Plot of IC50 and FXa produces a straight line typical of tight inhibitors. D, Slow-type inhibition of FXa by Lufaxin. Typical progress curves for FXa-mediated S2222 (1 mM) hydrolysis in the absence (curve a) and presence of Lufaxin (curves b, 0.93 nM; c, 1.8 nM; d, 3.75 nM; e, 7.5 nM; f, 15 nM; g, 30 nM and h, 60 nM). Reactions started with addition of FXa (0.5 nM) to a mixture containing Lufaxin and S2222 (1 mM). Substrate hydrolysis was followed for 2 hours at 37°C and 405 nm. E, Experiments were performed as in D but S2222 concentrations were 150, 300, 450, 600, 750, 900 µM. The ratio Vs/Vo obtained between 45 – 60 min was plotted against S2222 concentration and data fitted with the Morrison equation to calculate the IC50 at each substrate concentration. F, Plot IC50 and S2222 concentrations. Value were fitted by linear regression. The pattern of curves is typical for non competitive inhibitors. Six experiments were performed, and each data point is the average of duplicate determinations.
To verify whether Lufaxin is a slow or fast inhibitor of FXa, the enzyme was added to reaction mixture containing inhibitor and S2222. Reactions were initiated with FXa. This protocol also allows the characterization of the inhibitor as competitive, uncompetitive, or noncompetitive. Figure 3D shows that the progress curves displayed a downward concavity, which is followed by a linear component after 40 min which indicates that the reaction reached a steady state. This kinetic pattern is typical of slow-binding inhibitor. The Vs/Vo values obtained between 45–60 min (Vmax mode) were fitted by the Morrison equation for each substrate concentration.34,35 Figure 3E shows that the curves obtained using this approach were almost superimposable and the calculated IC50 was determined from six repeated duplicates experiments with a less than 10% error.
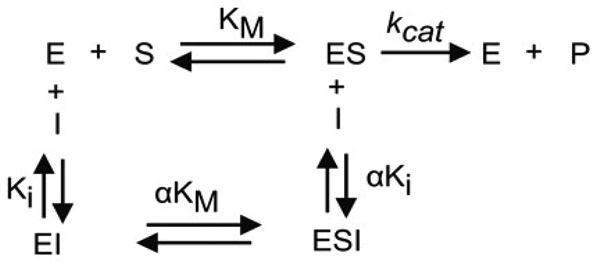
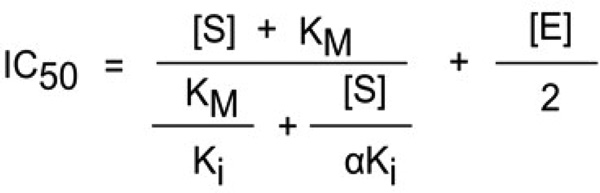
For tight inhibitors, a plot of IC50 vs substrate concentration produces and intercept yielding the true Ki.34,35 For a competitive inhibitor, the IC50 value will increase linearly with increasing substrate concentrations. For an uncompetitive inhibitor the IC50 will curve downward sharply. For the noncompetitive type, the relationship between IC50 values and substrate concentration varies depending on the dissociation constant of the inhibitor-enzyme-substrate complex (αKi) and the inhibitor-enzyme complex (Ki) (Scheme 1). When Ki equals αKi, the equation for a tight binding competitive inhibitor reduces to IC50 = Ki + [E]/2, indicating that substrate binding does not influence the binding of inhibitor to FXa. If αKi < Ki, the inhibitor-enzyme-substrate complex formation is favored over the inhibitor-enzyme complex, indicating that the substrate has a positive influence on inhibitor binding to the enzyme. If αKi > Ki, the substrate must have a negative influence on inhibitor binding to the enzyme.34,35 Accordingly, when αKi = Ki the curve determining IC50 as a function of substrate concentration is a straight line with zero slope, when αKi > Ki the curve has an upward concavity and positive slope, and when αKi < Ki the curve has downward concavity and negative slope. Scheme 1 predicts the general mechanisms which describe a non competitive inhibitor.
Scheme 1.
In our experiments with Lufaxin and FXa, the IC50 values were higher at or below the Km of the enzyme (0.3 mM) but essentially the same at higher substrate concentration, indicating a noncompetitive pattern and α < 1 (Figure 3F). Linear regression analysis indicates a negative slope (−.00222) with P=0.0104, leading us to accept the hypothesis of Lufaxin being a non-competitive inhibitor with an α < 1. Next, the IC50 values obtained for each substrate concentration of the six experiments were used to calculate the best α and Ki (as determined by the least sum of squares to the observed points) using the equation for non competitive tight binding inhibitors
This transformation yield α value of 0.28±0.049 and Ki of 7.9 ± 0.70 nM, which is the Ki in the limiting condition of substrate absence. The product αKi is thus equal to 2.21 nM, being the Ki observed when all the enzyme is in the ES complex form (see Discussion).
Ca2+ is not required for FXa-Lufaxin complex formation
Progress curves initiated by addition of FXa to a mixture containing Lufaxin and S2222 were superimposable when 0, 0.5 mM, or 5 mM Ca2+ was added to the reaction mixture (not shown, n=3), indicating that Ca2+ is not required for the reaction. Our results also did not detect interaction of Lufaxin with vesicles of PC/PS (10 µM), in the presence of Ca2+ (not shown). Inhibition of FXa by Lufaxin was also unaffected by PC/PS (20 µM) (not shown).
Lufaxin Displays High-Affinity and Stoichiometric Interaction with FXa
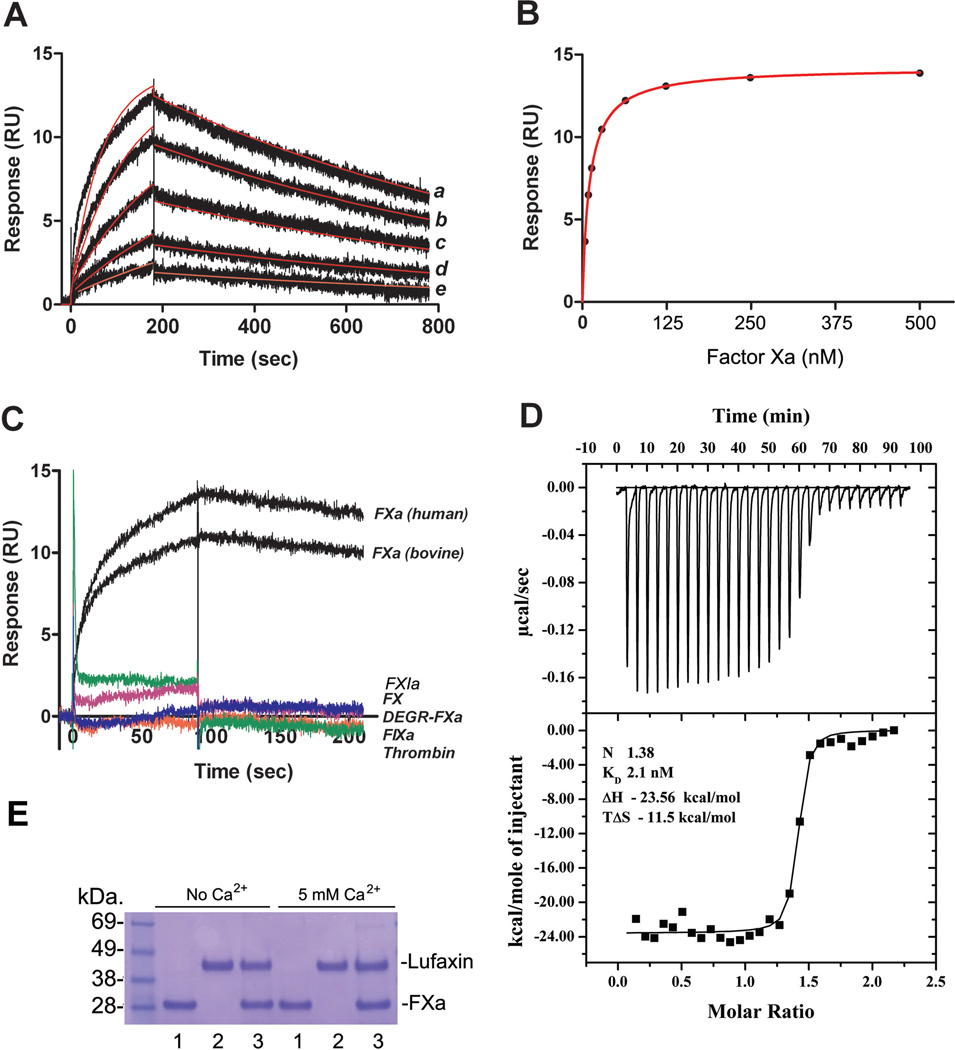
Kinetics of FXa interaction with Lufaxin was studied by SPR. Lufaxin was immobilized in a CM5 sensor chip, and FXa was used as analyte. Typical sensograms are shown in Figure 4A. Best fit was attained using a 1:1 Langmuir equation, with a calculated kon 2.296 ± 1.325 × 105 M−1s−1 and koff 7.203 ± 1.709 × 10−4s−1 and equilibrium constant (KD) 3.86 ± 2.7 nM (χ2 = 0.44 ± 0.1). Fitting was not significantly improved using other models (e.g., two-state reaction). When data points were fitted using steady-state kinetics, binding was saturable and yielded a KD of 9.69 ± 0.38nM (χ2 = 0.083 ± 0.03) (Figure 4B). Our results also show that Lufaxin interacts with bovine FXa but does not bind to catalytic site-blocked FXa (DEGR-FXa), thrombin, FXIa, FIXa, or zymogen FX (Figure 4C). Lufaxin binding to FXa is non-covalent, as it was readily dissociated by the acidic pH needed to regenerate the sensor chip.
Figure 4.
Kinetics and stoichiometry of Lufaxin interaction with FXa. A, SPR experiments demonstrate that FXa interacts with immobilized Lufaxin. Factor Xa at 62.5 nM (a), 31.25 nM (b), 15.6 nM (c), 7.8 nM (d), and 3.9 nM (e) were injected over immobilized Lufaxin for 180 seconds. Dissociation of the Lufaxin-FXa complex was monitored for 1200 seconds, and a global 1:1 binding model was used to calculate kinetic parameters. Representative sensograms are shown in black, and fitting of the data points using the Langmuir equation is depicted in red. B, Lufaxin-FXa interaction calculated by steady-state kinetics. C, Lufaxin binds to human and bovine FXa but does not interact with FX, DEGR-FXa, FIXa, FXIa, or thrombin. All analytes were tested at 100 nM and injected in a sensor chip containing immobilized Lufaxin. D, Solution binding of Lufaxin to FXa as measured by isothermal titration calorimetry. Upper panels, base line-adjusted heats per injection of FXa (20 µM) into Lufaxin (2.0 µM). Lower panels, molar enthalpies per injection for FXa interaction with Lufaxin. Filled squares, measured enthalpies; solid line, fit of experimental data to a single site binding model. Thermodynamic parameters: ΔH in kcal/mol, TΔS in kcal/mol, and KD are indicated in the inset. E, Lufaxin (1.9 µM) was incubated with FXa (2.7 µM) for 3 or 24 hours, with and without 5 mM Ca2+. The mixture was loaded in a NU-PAGE gel. The bands corresponding to FXa or Lufaxin are indicated. 1, FXa; 2, Lufaxin; 3, mixture of FXa and Lufaxin. No cleavage of Lufaxin is observed.
To determine the stoichiometry and enthalpy of the interaction between FXa and Lufaxin, ITC was performed. Results are shown in Figure 4D. Fitting of the observed enthalpies to a single-site binding model revealed a KD of 2.1 ± 0.62 nM for Lufaxin binding to FXa with a stoichiometry of binding (n = 1.38 ± 0.006), compatible with 1:1 enzyme/inhibitor complex formation (Figure 4D). Binding was exothermic, with a favorable enthalpy (ΔH) of −23.56 kcal/mol and unfavorable entropy (ΔS = –11.5 cal/mol K).
Lufaxin is Not Cleaved by FXa
Our results demonstrate that no change in the pattern of Lufaxin migration was observed after overnight incubation at 37°C with molar excess of FXa, with or without 5 mM Ca2+ (Figure 4E).
Lufaxin Displays Antiinflammatory Activity
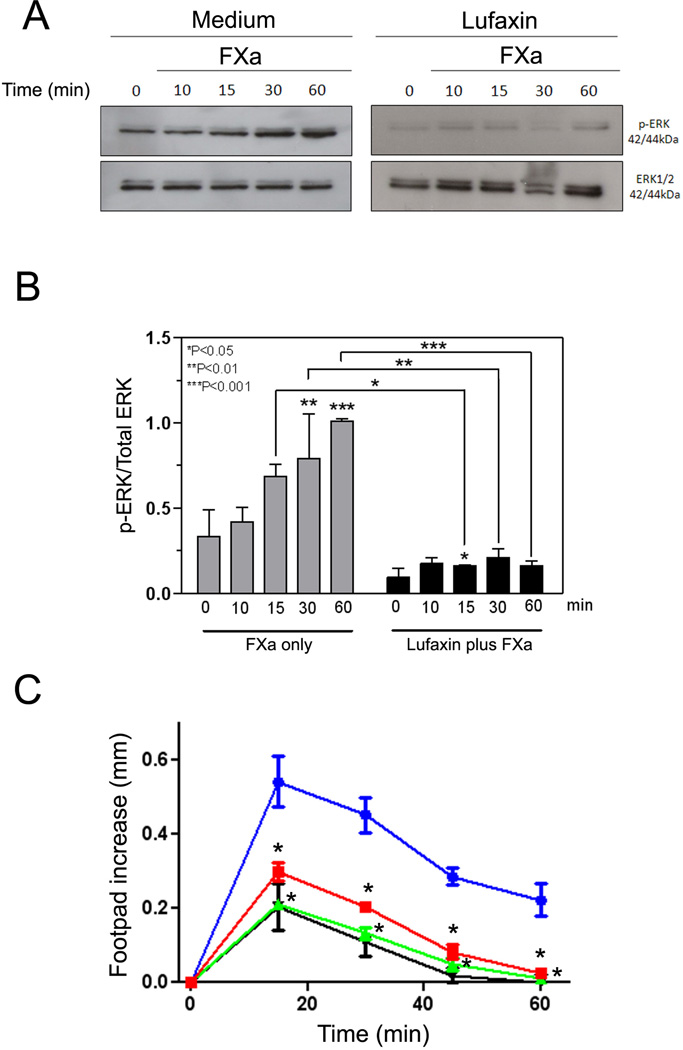
While FXa is a critical component of the coagulation cascade, it also induces cell activation through binding to PARs. This response induces cell signaling, which is accompanied by expression of adhesion molecules, procoagulant tissue factor, and cytokine production.13–16 To verify whether Lufaxin blocks FXa-induced PAR2 activation in vitro, MDA-MB-231 cells—which express high levels of PAR236,37 were incubated for 1 hr with Lufaxin, and stimulated with FXa for 10, 15, 30 or 60 min. Read-out for PAR2 activation was phosphorylation of extracellular signal-regulated kinase (pErk 1/2), which migrates as a p42–44 protein in the gel. All experiments were performed in the presence of hirudin to block (ectopic) thrombin activation of PAR1.32 As a control for loading, non-phosphorylated Erk was also employed in western-blots and probed with anti-Erk antibodies. Figure 5A demonstrates that FXa induces a robust response of the cells in a time-dependent manner that reaches a maximum at 60 minutes. In contrast, Lufaxin incubated with the cells for 1 hour, followed by addition of FXa, completely blocked pErk phosphorylation. Figure 5B depicts the quantification of the signal for pErk:Erk ratio by band densitometry for the experiments presented in Figure 5A.
Figure 5.
Lufaxin displays antiinflammatory activity. A, Lufaxin blocks PAR2 activation induced by FXa in the MDA-MB-231 cell line. Medium (left panel) or Lufaxin (50 nM, right panel) was added to the cells 60 min, followed by addition of FXa (10 nM) for 10 min, 15 min, 30 min, and 60 min. FXa was not added to the cells at time zero. Then, cell lysates were obtained and used for detection of p-ERK by western blot. As a control for protein loading, ERK detection was also performed. Detection for both panels were performed side by side using the same film and time of exposure. B, band densitometry for the gel presented in A. The ratio of pERK/Erk is reported. C, Lufaxin blocks the inflammatory effects of FXa. Posterior paw edema was induced by intradermal injection of 30 µL of FXa (10 µg, 7.3 µM) in the presence of PBS (circles) or FXa previously incubated with 3.2 µM (squares) or 9.8 µM (up triangles) of Lufaxin. Edema caused by 30 µL of PBS only is shown by down triangles. Edema formation (increase in paw thickness in mm) was estimated with a caliper before injection of FXa or after 15, 30, 45, and 60 minutes FXa. Four posterior paws were used for each data point. *, P<0.05 (for both concentrations of Lufaxin).
In vivo, sub-plantar injection of FXa produces a time- and dose-dependent edema in the paws of mice that resembles the effects observed after administration of carrageenan. This effect has been demonstrated to be mediated by PAR2 activation.38 In an attempt to determine whether Lufaxin also inhibits the inflammatory activity of FXa in vivo, the enzyme was injected either alone in the mouse paw or in the presence of two concentrations of Lufaxin. Figure 5C shows that FXa induces a time-dependent increase in paw edema that reached a maximum at 15 minutes and returned to basal level at 60 minutes, as previously reported.38 When a mixture of FXa (7.3 µM) and sub-saturating concentrations of Lufaxin (2.5 µM) were injected in the paw, edema was partially reduced; however, when FXa (7.3 µM) was injected in the presence of molar excess of Lufaxin (9.3 µM), complete blockade of edema formation was attained.
Lufaxin Is Antithrombotic in Vivo
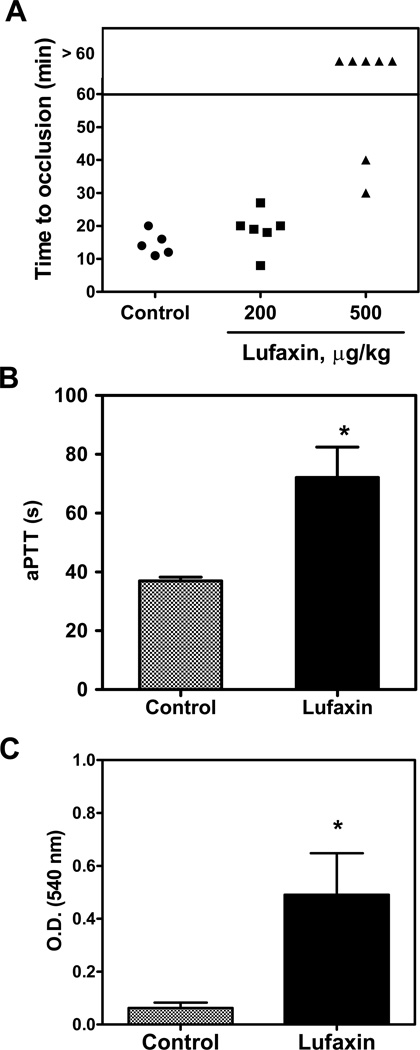
To test whether Lufaxin displays antithrombotic activity, a mouse model of thrombosis was employed in which FeCl3 was used to induce carotid artery injury.39 Thrombus formation was estimated using a Doppler flow probe that allows monitoring of carotid blood flow for 60 minutes or until complete occlusion takes place. Times to occlusion were not significantly different between control and mice treated with 200 µg/kg Lufaxin (16.11 ± 1.47 versus 17.0 ± 1.63 minutes); however, mice treated with 500 µg/kg Lufaxin were resistant to arterial occlusion. In these cases, occlusion did not take place before 60 minutes for most animals (Figure 6A). Injection of Lufaxin (500 µg/kg) in mice also prolongs aPTT ex vivo (Figure 6B). Next, the effects of Lufaxin in bleeding were estimated using the tail transection method. Figure 6C shows that Lufaxin (500 µg/kg) produces significant bleeding, as would be expected for an inhibitor targeting FXa.
Figure 6.
Lufaxin exhibits antithrombotic in vivo. A, A paper filter imbibed with 7.5% FeCl3 was applied to carotid artery, and blood flow was monitored with a perivascular flow probe for 60 minutes or until stable occlusion took place. Fifteen minutes before injury, Lufaxin was injected into the caudal vein of the mice. Each symbol represents one individual. B, aPTT was estimated ex vivo by addition of aPTT reagent to plasma collected from mice injected with Lufaxin (500 µg/Kg). Clot was determined using a coagulometer (n=6). C, Bleeding was estimated by the tail transection model after intravenous infection of Lufaxin (n=6). *, p<0.05.
Discussion
Sand flies are bloodsucking arthropods that rely on the composition of their saliva to obtain a blood meal. A number of pharmacologically active components have been identified in these secretions, including inhibitors of platelet aggregation,3,6,9 vasodilators,1,4,7 and immunomodulators.5,8 Notably identification of the major anticoagulant in sand fly saliva remained elusive for decades. Furthermore, analysis of the SG transcriptomes from several Phlebotominae has not revealed a candidate (e.g., Kunitz-type proteins) with the potential to block FXa.9–11 In an attempt to identify the major anticoagulant of L. longipalpis, a number of molecules found in the SG were selected for transient expression in mammalian cells (HEK293 cells). Recombinant proteins obtained using this approach were initially tested for inhibition of platelet aggregation or clotting, or for vasodilation activity. As a result of this screening, we discovered that one candidate, herein named Lufaxin, prolonged recalcification time and both PT and aPTT, suggesting inhibition of the common pathway. A screening for inhibition of a panel of enzymes identified only FXa, out of 16 enzymes, as the target for Lufaxin. Inhibition of FXa was also observed with the salivary glands of L. longipalpis, P. papatasi, and P. duboscqi. In addition, components from the SGH of L. longipalpis dose-dependently interact with FXa immobilized in a CM5 sensor chip (Figure 2). Moreover, a band corresponding to Lufaxin was identified by Western blot using homogenates implying that Lufaxin is expressed in the salivary glands of sand flies.
Notably, the primary sequence of Lufaxin is novel and it does not display similarity to the sequences or molecular domains of other proteins deposited in the databases. Therefore, it does not belong to any known family of physiological or salivary coagulation inhibitors such as Kunitz-type proteins (e.g., TFPI, TAP, simulikunin, ixolaris), Ascaris-type inhibitors (e.g., NAP5, NPA6, AceAP1), antistasin-like inhibitors from leeches (e.g., antistasin, ghilanten, therostasin), serpins (e.g., AT, mosquitoes serpin), or members of the Salp family (e.g., Salp14, Salp9).2 Remarkably, each of these inhibitors displays a particular mode of FXa inhibition, either mechanistically or through unique structural features, or both. For example, in TAP, the three N-terminal residues of TAP make multiple contacts with the catalytic site of FXa noncanonically,40 while in NAP5 or antistasin, both inhibitors canonically interact with FXa active site through the reactive-site loop that possesses a P1 Arg.41,42 In this respect, FXa has specificity for Ile-Glu-Gly-Arg-X, and many protein inhibitors of serine proteases inhibit their target enzyme by presenting a substrate- or “bait”-like region that resembles the enzyme`s natural substrate which display Arg or Lys as P1 position.43,44 In Lufaxin, a Lys residue is present adjacent to the 5th cysteine; several other positively charged residues that form a consensus C(K/L)LVFKK(R/K)(K/E) sequence between the 5th and 6th cysteines have also been identified (Figure 1). However, our attempt to identify the P1-P1′ reactive site of Lufaxin was unsuccessful, as no cleavage of the inhibitor was observed after 3- and 24-hour incubation with a molar excess of FXa, with or without Ca2+ (Figure 4). This result is not entirely surprising, because cleavage at the active-site cleft (P1-P1′) of other FXa inhibitors (e.g. TFPI) occurs only very slowly or not at all.17 Therefore, it remains to be identified how Lufaxin, a molecule whose sequence has no precedents, interacts with FXa at the molecular level.
Kinetic studies using chromogenic substrate S2222 revealed that Lufaxin is a slow, tight, reversible, Ca2+-independent and non competitive inhibitor of FXa, which blocks the enzyme with a KD~8.0 nM (Figure 3F). Consistent with these results, SPR experiments revealed the KD of Lufaxin interaction to FXa as ~3.5 nM. This value is also in good agreement with the KD (~2 nM) calculated by ITC experiments which is based on thermodynamic parameters.45 ITC experiments also determined that one molecule of Lufaxin binds to one molecule of FXa, validating Lufaxin as a tight inhibitor. Of note, Lufaxin-FXa complex formation is considered fast (Kon ~2.2 × 105M−1.s−1) according to SPR results. Therefore, the slow-inhibition of FXa by Lufaxin observed with small substrate may in part be due to the use of the low molecular mass of S2222 instead of the natural macromolecular substrate prothrombin. Conceivably, this complex slowly converts to a stable one involving the catalytic site, which explains why slow-binding kinetics was observed in the experiments using S2222. It also suggests that exosites may be involved in the interaction of Lufaxin with FXa. Also, inhibition of catalytic activity of FXa assessed with S2222 was not complete at equimolar concentration of Lufaxin/FXa (Figure 3C), despite a 1:1 interaction confirmed by ITC. It is concluded that S2222 retains some degree of accessibility to the active site of FXa even when Lufaxin is bound to the enzyme. This is congruent with the finding that Lufaxin is a non competitive type inhibitor of FXa with respect to S2222. This means that the inhibitor is not likely to have an overlapping binding site with that of the chromogenic substrate. Furthermore, the smaller αKi compared with Ki indicated that the conformational changes in FXa upon S2222 binding have positive influence on Lufaxin interaction with the enzyme. It is also important to recognize that SPR experiments did not detect binding of Lufaxin to DEGR-FXa, which in many respects mimics the enzyme-substrate transition state complex (ES). This result suggests that occupancy of the catalytic site of FXa by DEGR may not entirely reproduce the interactions observed with S2222; of note, DEGR is an irreversible glu-gly-arg-chloromethyl ketone containing a dansyl group which is not present in the cleavable chromogenic substrate. Conceivably, the kinetics of FXa inhibition by Lufaxin in the presence of S2222 and described by Scheme 1 may not fully apply when DEGR-FXa is used as [S]. Additional kinetics and structural studies with catalytic sited-mutated FXa (e.g. FXaS195A) among other residues will be required to identify which specific amino acids contribute to FXa/Lufaxin complex formation and how DEGR interferes with this interaction.
The fact that Lufaxin inhibits FXa has relevant implications to the feeding behavior of the sand fly. FXa is central enzyme in coagulation and plays a critical role in prothrombinase complex assembly leading to amplification of the coagulation cascade.12 FXa is also a direct inflammatory molecule that acts in PARs, producing a number of effects such as mitogenic activity in smooth muscle, inflammatory responses in endothelial cells—including cytokine induction and nitric oxide-mediated hypotension—and hyperalgesia through activation of afferent sensory neurons.13–16 In other words, PAR expressed around the vessel wall may be part of a first response that helps link tissue injury to recruitment of platelets, leukocytes and, by promoting transudation, immune effector proteins to examine the locale for damage and infection.13–16 Accordingly, PAR2 knockout mice have impaired leukocyte rolling,46 antagonists of PAR2 prevent experimental colitis,47 and cell-penetrating peptides antagonizing PAR2 is associated with less inflammation in vivo.48 Notably, we demonstrated that Lufaxin is very efficient in blocking PAR2 activation by FXa in vitro using the breast cancer cell line MDAMB-231.36,37 Actually, baseline levels of pERK phosphorylation were also diminished in the presence of Lufaxin, likely due to blockade of PAR2 activation by ectopic FXa.32 Because PAR activation in different cell types occurs through a similar mechanism that involves cleavage of the receptor N-terminus,13–16 our in vitro results suggest that Lufaxin prevents activation of endothelial cells, leukocytes, and smooth muscles. This contention was supported by the finding that Lufaxin completely inhibits edema formation provoked by FXa in vivo. These results are relevant because administration of FXa to the dermis is associated with mast cell degranulation, a major component of acute inflammatory response.38,49
Injections of Lufaxin in the tails of mice 15 minutes before carotid injury with FeCl3 were accompanied by resistance of artery occlusion and prolongation of the aPTT ex vivo. This implies that attenuation of thrombin generation in the prothrombinase by Lufaxin results in inhibition of the coagulation cascade in vivo on one hand, and attenuation of thrombin (PAR1)-dependent activation of platelets and endothelial cells, on the other. Presumably, Lufaxin anticoagulant and antiinflammatory properties decreases the pro-hemostatic tonus at the interface of vector-host interactions. In fact, sand flies mouth parts work as “scissors”, which produce tissue damage, creating microhemorrhages from which blood is taken.1 Conceivably, intense exposure of tissue factor leads to strong activation of the coagulation cascade at sites of bites. Accordingly, blocking FXa is relevant to prevent clot formation under the pro-hemostatic environment generated by the feeding behavior of sand flies. Identification of Lufaxin as a novel FXa inhibitor is also relevant experimentally in order to study the participation of FXa in ischemic events, tumor growth or metastasis.13–16,50 Due to its unique sequence, Lufaxin may also be regarded as a useful tool to understand FXa structure and function.
Supplementary Material
Scheme 2.
Acknowledgments
We thank NIAID intramural editor Brenda Rae Marshall (NIAID) for editorial assistance, Van My-Pham (NIH), and Jorgeane Freire e Souza (UFRJ) for technical assistance.
Sources of Funding
This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health (USA). R.Q.M. received a grant from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro Carlos Chagas Filho (FAPERJ), Fundação do Câncer and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). M.K. received support from grant number P502/12/2409 of the Grant Agency of the Czech Republic, from a Jan Evangelista Purkyne fellowship of the National Academy of Sciences of the Czech Republic and from a Marie Curie Reintegration grant of the EU-FP7 (PIRG07-GA-2010-268177).
Because I.M.B.F., J.G.V., and J.M.C.R. are government employees and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
Footnotes
Disclosures
The authors declare no competing financial interests.
References
- 1.Ribeiro JM, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- 2.Koh CY, Kini RM. Molecular diversity of anticoagulants from haematophagous animals. Thromb Haemost. 2009;102(3):437–453. doi: 10.1160/TH09-04-0221. [DOI] [PubMed] [Google Scholar]
- 3.Francischetti IM. Platelet aggregation inhibitors from hematophagous animals. Toxicon. 2010;56(7):1130–1144. doi: 10.1016/j.toxicon.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takac P, Tsujimoto H, Champagne DE. Hypotensive proteins from hematophagous animals. In: Kini RM, Clemetson KJ, Markland FS, McLane MA, Morita T, editors. Toxins and Hemostasis From bench to bedside. Vol. 2010. Springer: New York; pp. 673–696. [Google Scholar]
- 5.Titus RG, Bishop JV, Mejia JS. The immunomodulatory factors of arthropod saliva and the potential for these factors to serve as vaccine targets to prevent pathogen transmission. Parasite Immunol. 2006;28(4):131–141. doi: 10.1111/j.1365-3024.2006.00807.x. [DOI] [PubMed] [Google Scholar]
- 6.Valenzuela JG, Belkaid Y, Rowton E, Ribeiro JM. The salivary apyrase of the blood-sucking sand fly Phlebotomus papatasi belongs to the novel Cimex family of apyrases. J Exp Biol. 2001;204(Pt 2):229–237. doi: 10.1242/jeb.204.2.229. [DOI] [PubMed] [Google Scholar]
- 7.Lerner EA, Ribeiro JM, Nelson RJ, Lerner MR. Isolation of maxadilan, a potent vasodilatory peptide from the salivary glands of the sand fly Lutzomyia longipalpis. J Biol Chem. 1991;266(17):11234–11236. [PubMed] [Google Scholar]
- 8.Valenzuela JG, Belkaid Y, Garfield MK, et al. Toward a defined anti-Leishmania vaccine targeting vector antigens: characterization of a protective salivary protein. J Exp Med. 2001;194(3):331–342. doi: 10.1084/jem.194.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charlab R, Valenzuela JG, Rowton ED, Ribeiro JM. Toward an understanding of the biochemical and pharmacological complexity of the saliva of a hematophagous sand fly Lutzomyia longipalpis. Proc Natl Acad Sci U S A. 1999;96(26):15155–15160. doi: 10.1073/pnas.96.26.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hostomska J, Volfova V, Mu J, et al. Analysis of salivary transcripts and antigens of the sand fly Phlebotomus arabicus. BMC Genomics. 2009;10:282. doi: 10.1186/1471-2164-10-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson JM, Oliveira F, Kamhawi S, et al. Comparative salivary gland transcriptomics of sandfly vectors of visceral leishmaniasis. BMC Genomics. 2006;7:52. doi: 10.1186/1471-2164-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishnaswamy S. Exosite-driven substrate specificity and function in coagulation. J Thromb Haemost. 2005;3(1):54–67. doi: 10.1111/j.1538-7836.2004.01021.x. [DOI] [PubMed] [Google Scholar]
- 13.Rothmeier AS, Ruf W. Protease-activated receptor 2 signaling in inflammation. Semin Immunopathol. 2012;34(1):133–149. doi: 10.1007/s00281-011-0289-1. [DOI] [PubMed] [Google Scholar]
- 14.Rattenholl A, Steinhoff M. Proteinase-activated receptor-2 in the skin: receptor expression, activation and function during health and disease. Drug News Perspect. 2008;21(7):369–381. doi: 10.1358/dnp.2008.21.7.1255294. [DOI] [PubMed] [Google Scholar]
- 15.Cottrell GS, Amadesi S, Schmidlin F, Bunnett N. Protease-activated receptor 2: activation, signalling and function. Biochem Soc Trans. 2003;31(Pt 6):1191–1197. doi: 10.1042/bst0311191. [DOI] [PubMed] [Google Scholar]
- 16.Coughlin SR. Protease-activated receptors in hemostasis, thrombosis and vascular biology. J Thromb Haemost. 2005;3(8):1800–1814. doi: 10.1111/j.1538-7836.2005.01377.x. [DOI] [PubMed] [Google Scholar]
- 17.Broze GJ, Jr, Girard TJ. Tissue factor pathway inhibitor: structure-function. Front Biosci. 2012;17:262–280. doi: 10.2741/3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olson ST, Gettins PG. Regulation of proteases by protein inhibitors of the serpin superfamily. Prog Mol Biol Transl Sci. 2011;99:185–240. doi: 10.1016/B978-0-12-385504-6.00005-1. [DOI] [PubMed] [Google Scholar]
- 19.Rezaie AR. Heparin chain-length dependence of factor Xa inhibition by antithrombin in plasma. Thromb Res. 2007;119(4):481–488. doi: 10.1016/j.thromres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Broze GJ., Jr Protein Z-dependent regulation of coagulation. Thromb Haemost. 2001;86(1):8–13. [PubMed] [Google Scholar]
- 21.Waxman L, Smith DE, Arcuri KE, Vlasuk GP. Tick anticoagulant peptide (TAP) is a novel inhibitor of blood coagulation factor Xa. Science. 1990;248(4955):593–596. doi: 10.1126/science.2333510. [DOI] [PubMed] [Google Scholar]
- 22.Tsujimoto H, Kotsyfakis M, Francischetti IM, Eum JH, Strand MR, Champagne DE. Simukunin from the Salivary Glands of the Black Fly Simulium vittatum Inhibits Enzymes That Regulate Clotting and Inflammatory Responses. PLoS One. 2012;7(2):e29964. doi: 10.1371/journal.pone.0029964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cappello M, Vlasuk GP, Bergum PW, Huang S, Hotez PJ. Ancylostoma caninum anticoagulant peptide: a hookworm-derived inhibitor of human coagulation factor Xa. Proc Natl Acad Sci U S A. 1995;92(13):6152–6156. doi: 10.1073/pnas.92.13.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunwiddie C, Thornberry NA, Bull HG, et al. Antistasin, a leech-derived inhibitor of factor Xa. Kinetic analysis of enzyme inhibition and identification of the reactive site. J Biol Chem. 1989;264(28):16694–16699. [PubMed] [Google Scholar]
- 25.Stark KR, James AA. Isolation and characterization of the gene encoding a novel factor Xa-directed anticoagulant from the yellow fever mosquito, Aedes aegypti. J Biol Chem. 1998;273(33):20802–20809. doi: 10.1074/jbc.273.33.20802. [DOI] [PubMed] [Google Scholar]
- 26.Narasimhan S, Koski RA, Beaulieu B, et al. A novel family of anticoagulants from the saliva of Ixodes scapularis. Insect Mol Biol. 2002;11(6):641–650. doi: 10.1046/j.1365-2583.2002.00375.x. [DOI] [PubMed] [Google Scholar]
- 27.Oliveira F, Kamhawi S, Seitz AE, et al. From transcriptome to immunome: identification of DTH inducing proteins from a Phlebotomus ariasi salivary gland cDNA library. Vaccine. 2006;24(3):374–390. doi: 10.1016/j.vaccine.2005.07.085. [DOI] [PubMed] [Google Scholar]
- 28.Ma D, Assumpcao TC, Li Y, Andersen JF, Ribeiro J, Francischetti IM. Triplatin, a platelet aggregation inhibitor from the salivary gland of the triatomine vector of Chagas disease, binds to TXA(2) but does not interact with glycoprotein PVI. Thromb Haemost. 2012;107(1):111–123. doi: 10.1160/TH11-10-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Assumpcao TC, Alvarenga PH, Ribeiro JM, Andersen JF, Francischetti IM. Dipetalodipin, a novel multifunctional salivary lipocalin that inhibits platelet aggregation, vasoconstriction, and angiogenesis through unique binding specificity for TXA2, PGF2alpha, and 15(S)-HETE. J Biol Chem. 2010;285(50):39001–39012. doi: 10.1074/jbc.M110.152835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monteiro RQ, Rezaie AR, Ribeiro JM, Francischetti IM. Ixolaris: a factor Xa heparin-binding exosite inhibitor. Biochem J. 2005;387(Pt 3):871–877. doi: 10.1042/BJ20041738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francischetti IM, Valenzuela JG, Ribeiro JM. Anophelin: kinetics and mechanism of thrombin inhibition. Biochemistry. 1999;38(50):16678–16685. doi: 10.1021/bi991231p. [DOI] [PubMed] [Google Scholar]
- 32.Camerer E, Huang W, Coughlin SR. Tissue factor- and factor X-dependent activation of protease-activated receptor 2 by factor VIIa. Proc Natl Acad Sci U S A. 2000;97(10):5255–5260. doi: 10.1073/pnas.97.10.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kotsyfakis M, Sa-Nunes A, Francischetti IM, Mather TN, Andersen JF, Ribeiro JM. Antiinflammatory and immunosuppressive activity of sialostatin L, a salivary cystatin from the tick Ixodes scapularis. J Biol Chem. 2006;281(36):26298–26307. doi: 10.1074/jbc.M513010200. [DOI] [PubMed] [Google Scholar]
- 34.Williams JW, Morrison JF. The kinetics of reversible tight-binding inhibition. Methods Enzymol. 1979;63:437–467. doi: 10.1016/0076-6879(79)63019-7. [DOI] [PubMed] [Google Scholar]
- 35.Copeland RA. Tight binding inhibitors. In: Copeland RA, editor. Enzymes: a practical introduction to strucuture, mechanism and data analysis. 2nd Edition. New York: Wiley-VCH Inc; 2000. pp. 305–317. [Google Scholar]
- 36.Morris DR, Ding Y, Ricks TK, Gullapalli A, Wolfe BL, Trejo J. Protease-activated receptor-2 is essential for factor VIIa and Xa-induced signaling, migration, and invasion of breast cancer cells. Cancer Res. 2006;66(1):307–314. doi: 10.1158/0008-5472.CAN-05-1735. [DOI] [PubMed] [Google Scholar]
- 37.Ryden L, Grabau D, Schaffner F, Jonsson PE, Ruf W, Belting M. Evidence for tissue factor phosphorylation and its correlation with protease-activated receptor expression and the prognosis of primary breast cancer. Int J Cancer. 2010;126(10):2330–2340. doi: 10.1002/ijc.24921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cirino G, Cicala C, Bucci M, et al. Factor Xa as an interface between coagulation and inflammation. Molecular mimicry of factor Xa association with effector cell protease receptor-1 induces acute inflammation in vivo. J Clin Invest. 1997;99(10):2446–2451. doi: 10.1172/JCI119428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Owens AP, 3rd, Lu Y, Whinna HC, Gachet C, Fay WP, Mackman N. Towards a standardization of the murine ferric chloride-induced carotid arterial thrombosis model. J Thromb Haemost. 2011;9(9):1862–1863. doi: 10.1111/j.1538-7836.2011.04287.x. [DOI] [PubMed] [Google Scholar]
- 40.Wei A, Alexander RS, Duke J, Ross H, Rosenfeld SA, Chang CH. Unexpected binding mode of tick anticoagulant peptide complexed to bovine factor Xa. J Mol Biol. 1998;283(1):147–154. doi: 10.1006/jmbi.1998.2069. [DOI] [PubMed] [Google Scholar]
- 41.Rios-Steiner JL, Murakami MT, Tulinsky A, Arni RK. Active and exo-site inhibition of human factor Xa: structure of des-Gla factor Xa inhibited by NAP5, a potent nematode anticoagulant protein from Ancylostoma caninum. J Mol Biol. 2007;371(3):774–786. doi: 10.1016/j.jmb.2007.05.042. [DOI] [PubMed] [Google Scholar]
- 42.Lapatto R, Krengel U, Schreuder HA, et al. X-ray structure of antistasin at 1.9 A resolution and its modelled complex with blood coagulation factor Xa. EMBO J. 1997;16(17):5151–5161. doi: 10.1093/emboj/16.17.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otlewski J, Jelen F, Zakrzewska M, Oleksy A. The many faces of protease-protein inhibitor interaction. EMBO J. 2005;24(7):1303–1310. doi: 10.1038/sj.emboj.7600611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zogg T, Brandstetter H. Complex assemblies of factors IX and X regulate the initiation, maintenance, and shutdown of blood coagulation. Prog Mol Biol Transl Sci. 2011;99:51–103. doi: 10.1016/B978-0-12-385504-6.00002-6. [DOI] [PubMed] [Google Scholar]
- 45.Leavitt S, Freire E. Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr Opin Struct Biol. 2001;11(5):560–566. doi: 10.1016/s0959-440x(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 46.Lindner JR, Kahn ML, Coughlin SR, et al. Delayed onset of inflammation in protease-activated receptor-2-deficient mice. J Immunol. 2000;165(11):6504–6510. doi: 10.4049/jimmunol.165.11.6504. [DOI] [PubMed] [Google Scholar]
- 47.Lohman RJ, Cotterell AJ, Suen J, et al. Antagonism of protease-activated receptor 2 protects against experimental colitis. J Pharmacol Exp Ther. 2012;340(2):256–265. doi: 10.1124/jpet.111.187062. [DOI] [PubMed] [Google Scholar]
- 48.Sevigny LM, Zhang P, Bohm A, et al. Interdicting protease-activated receptor-2-driven inflammation with cell-penetrating pepducins. Proc Natl Acad Sci U S A. 2011;108(20):8491–8496. doi: 10.1073/pnas.1017091108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moormann C, Artuc M, Pohl E, et al. Functional characterization and expression analysis of the proteinase-activated receptor-2 in human cutaneous mast cells. J Invest Dermatol. 2006;126(4):746–755. doi: 10.1038/sj.jid.5700169. [DOI] [PubMed] [Google Scholar]
- 50.Borensztajn K, Spek CA. Blood coagulation factor Xa as an emerging drug target. Expert Opin Ther Targets. 2011;15(3):341–349. doi: 10.1517/14728222.2011.553608. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.