Abstract
Phosphotriesters are one class of highly toxic synthetic compounds known as organophosphates. Wide spread usage of organophosphates as insecticides as well as nerve agents has lead to numerous efforts to identify enzymes capable of detoxifying them. A wide array of enzymes has been found to have phosphotriesterase activity including phosphotriesterase (PTE), methyl parathion hydrolase (MPH), organophosphorus acid anhydrolase (OPAA), diisopropylfluorophosphatase (DFP), and paraoxonase 1 (PON1). These enzymes differ widely in protein sequence and three-dimensional structure, as well as in catalytic mechanism, but they also share several common features. All of the enzymes identified as phosphotriesterases are metal-dependent hydrolases that contain a hydrophobic active site with three discrete binding pockets to accommodate the substrate ester groups. Activation of the substrate phosphorus center is achieved by a direct interaction between the phosphoryl oxygen and a divalent metal in the active site. The mechanistic details of the hydrolytic reaction differ among the various enzymes with both direct attack of a hydroxide as well as covalent catalysis being found.
1. Introduction
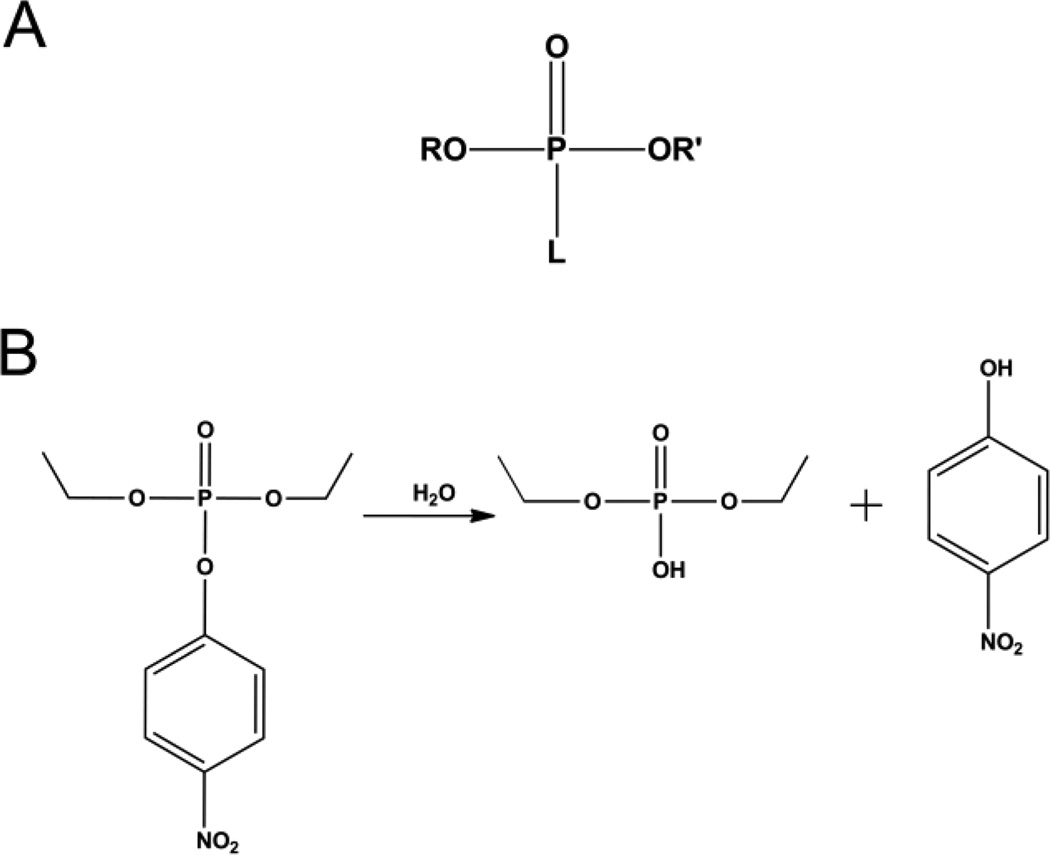
Phosphotriesters are included in the larger group of organophosphates which were first developed during the late 1940s and early 1950s. These highly toxic compounds have found widespread use as insecticides for the protection of agricultural crops. As insecticides, their mode of action is the inhibition of the enzyme acetylcholine esterase causing over-stimulation of the nervous system, ultimately resulting in paralysis and finally death [1, 2]. The general chemical structure of the organophosphates is a phosphate center with three ester linkages (Fig. 1A). Two of the ester linked groups are relatively stable, while the third ester linked group is fairly labile. It is the cleavage of the labile bond in the active site of acetylcholine esterase that leads to the irreversible inhibition. While the positive impact of these compounds on food production and security is hard to over estimate, the toxic effects are not limited to insects. Human exposure to organophosphates is a major hazard with thousands of cases of poisoning reported each year worldwide [2]. While serious efforts have been made to limit the toxicity of the pesticides available, some of the most toxic organophosphates have been developed as nerve agents for military use [3]. Concerns over the health and environmental impact of insecticide usage as well as the continued threat posed by nerve agents has fueled the search for enzymes that can hydrolyze and neutralize organophosphate compounds (Fig. 1B).
Figure 1.
A) General structure of an organophosphate where R and R' are alkyl groups and L can be a phenol, thiol, or fluoride group. B) Hydrolysis reaction of paraoxon catalyzed by phosphotriesterase.
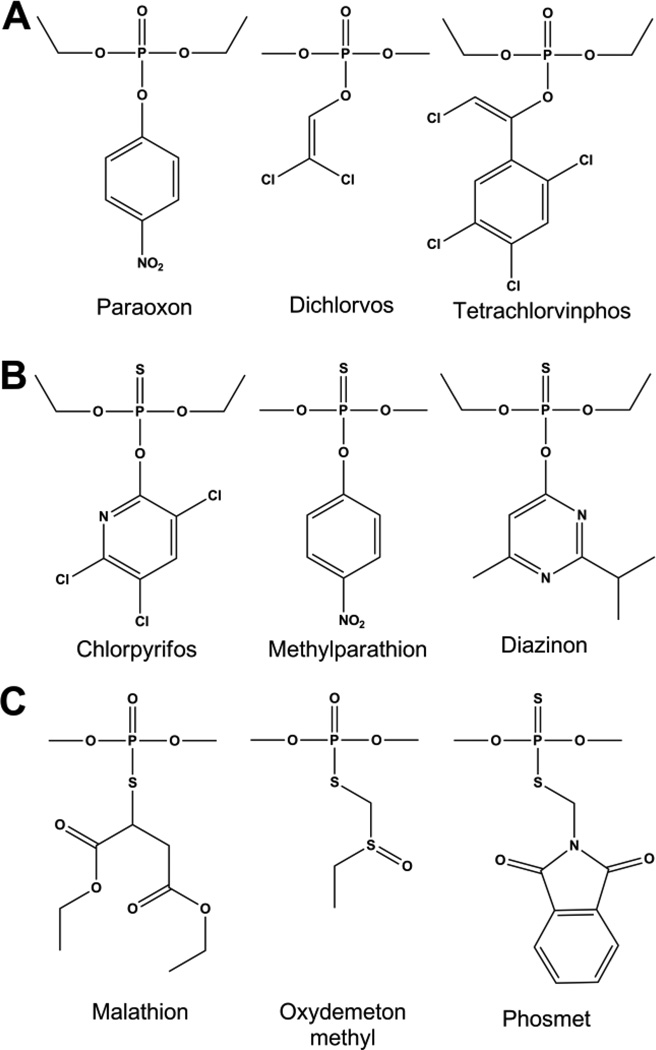
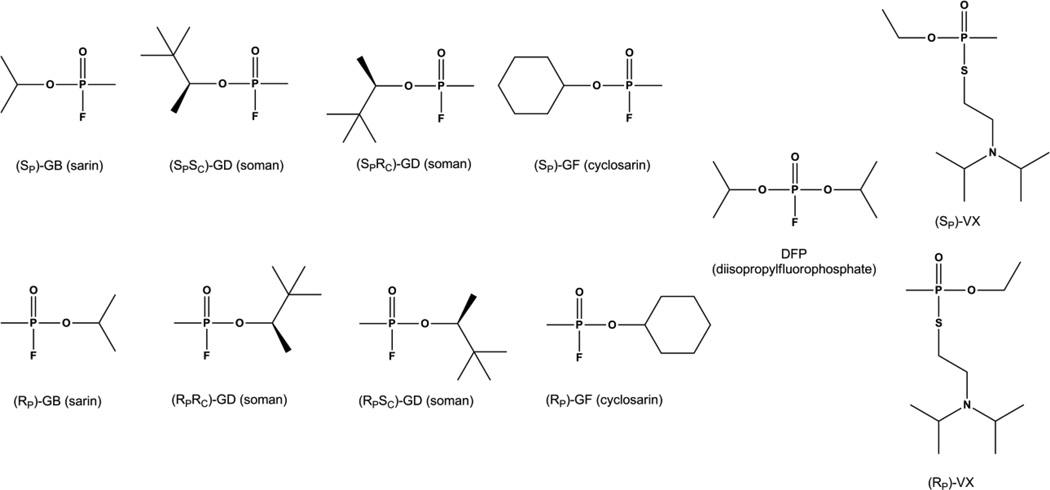
The organophosphate insecticides are primarily phosphotriesters, thiophosphotriesters, or phosphorothiolesters. Phosphotriesters such as paraoxon, contain a phosphate center with three O-linked groups (Fig. 2A). Thiophosphotriesters, such as methyl parathion, have the phosphoryl oxygen replaced by sulfur (Fig. 2B), and in phosphorothiolesters, such as malathion, one or more of the ester oxygens is replaced by a sulfur (Fig. 2C). A number of insecticides, such as phosmet, contain both a thiophosphate as well as a thiol-ester linkage. The labile group in organophosphate insecticides is most commonly a phenol or a thiol which can be straight chained, branched or aromatic. Insecticides typically have at least two identical ester groups which are small simple alkyl chains giving rise to an achiral phosphorus center. By contrast the nerve agents are all chiral phosphonate compounds with a direct P-CH3 linkage (Fig 3). The two best known nerve agents are the G-type, which utilize a fluoride group as the labile bond, and the V-type agents which use a branched thiol.
Figure 2.
Chemical structures of organophosphate insecticides. Common examples are shown for A) Phosphotriesters, B) Thiophosphates, C) Phosphorothiolates.
Figure 3.
Chemical structures for the individual enantiomers of chiral organophosphate nerve agents GB (sarin), GD (soman), GF (cyclosarin) and VX as well as the achiral analog DFP. Chiral phosphorus centers are drawn as Fischer projections. The Additional stereo-center in GD is drawn in standard stereochemical convention.
The organophosphates are an enormous class of compounds with widely varied chemical structures. Despite this structural variation a number of enzymes have been discovered that are capable of hydrolyzing the majority of these compounds. Phosphotriesterases have been identified in numerous bacteria as well as from squid and mammals [4–12]. The ability to hydrolyze these synthetic compounds is thought to have developed from a promiscuous activity due to physical similarities between the phosphoryl center and transition state of the enzyme's natural reaction [13]. In the case of the bacterial enzymes, the activity is thought to have evolved to take advantage of the organophosphate insecticides in the environment [5,7]. For many of these enzymes the phosphotriesterase activity appears to be derived from a lactonase function [13–16]. However, activity has been described for a group of prolidases capable of efficient hydrolysis of fluoride containing organophosphates [9,17]. The crystal structures of a number of the phosphotriesterases have been solved revealing that this activity can be achieved with numerous protein folds including the TIM-barrel, β-lactamase, "pita-bread," and β-propeller fold [18–23]. While the enzymes capable of hydrolyzing organophosphates seem nearly as varied as the class of compounds itself, there are some significant similarities. All of the phosphotriesterases are metal-dependent hydrolases. In all of the enzymes there is a requirement for a divalent metal which directly ligates to the substrate to facilitate catalysis [8,19,24,25]. While the protein folds are quite varied, the substrate binding sites are generally hydrophobic and can be divided into three binding pockets where the leaving group and the two other substituents interact to position the phosphorus center for catalysis [19,22–23, 26–29]. The mechanistic details of the reaction differs according to the protein fold and active site structure. The TIM-barrel enzymes and the β-lactamase enzymes are thought to share a catalytic mechanism [19,30]. The "pita bread" enzymes have a similar mechanism but are proposed to activate the substrates differently [22], while the β-propeller enzymes are postulated to utilize a covalent catalysis mechanism rather than direct attack by water [29,31].
2. TIM-barrel Fold
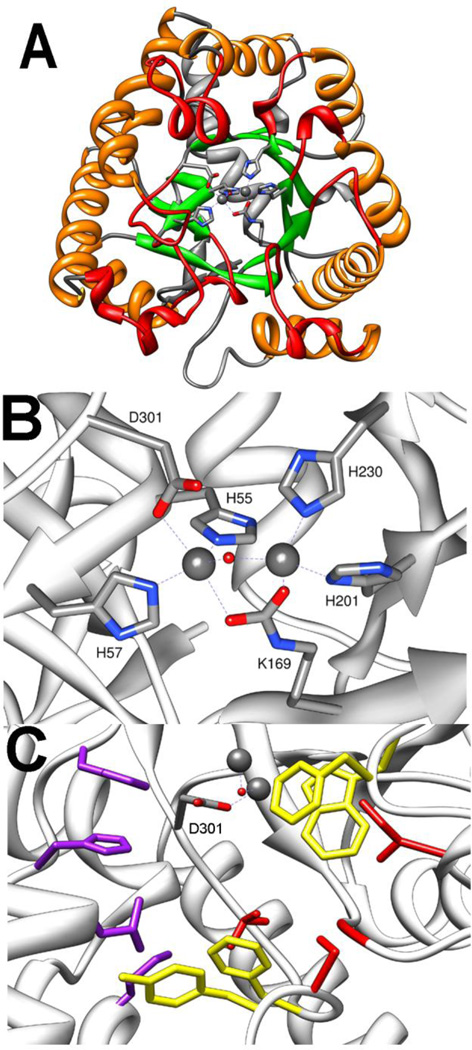
The amidohydrolase superfamily enzyme phosphotriesterase (PTE) is the most extensively studied enzyme capable of hydrolyzing organophosphates. PTE was first identified from the bacterium Pseudomonas diminuta and Flavobacterium sp. in parathion contaminated soil [32,33]. Two other highly similar enzymes have now been identified in the soil bacterium Agrobacterium radiobacter (OpdA) and Chryseobacterium balustinum [10,34]. All three unique variants of PTE have greater than 90% sequence identity and similar activity profiles [35]. Numerous crystal structures of PTE and OpdA have been solved, and the overall fold has been determined to be a distorted (β/α)8 or TIM-barrel (Fig. 4A) [18,35–39]. This protein fold contains a core barrel of 8 parallel β-strands surrounded by 8 α-helices with the ends of the barrel being formed by the loops connecting one β-strand to the subsequent α-helix. As with all members of the amidohydrolase superfamily, the active site of PTE is located at the C-terminal end of the TIM-barrel [18]. The active site contains a binuclear metal center ligated to residues from the C-terminal ends of the core β-strands with the substrate binding site being made of the loops that form the C-terminal end of the barrel. The more buried α-metal is ligated in a trigonal bipyramidal geometry to the protein by two histidines, H55 and H57, from β-strand 1 and an aspartate (D301) from β-strand 8 (Fig. 4B) [40,41]. The more solvent exposed β-metal is ligated by two histidines, one from β-stand 5 (H201) and one from β-strand 6 (H230). The two metals are bridged by a hydroxide and a carboxylated lysine (K169) from β-strand 4. The native metal found in PTE is Zn2+, but PTE is fully active with Cd2+, Mn2+, or Ni2+ [24].
Figure 4.
Crystal structure of PTE (pdb. 1dpm). A) TIM-Barrel fold of PTE shown with core α-helices in orange, core β-strands shown in green, N-terminal loops shown in grey, and C-terminal loops shown in red. Metals in active site are shown as spheres, and the metal ligating side chains shown as sticks. B) Metal binding site of PTE. C) Substrate binding pockets of PTE. Leaving group pocket residues W131, F132, F306, and Y309 are shown in yellow. Large group pocket residues H254, H257, L271, M317 are shown in purple. Small group pocket residues G60, I106, L303, and S308 are shown in red.
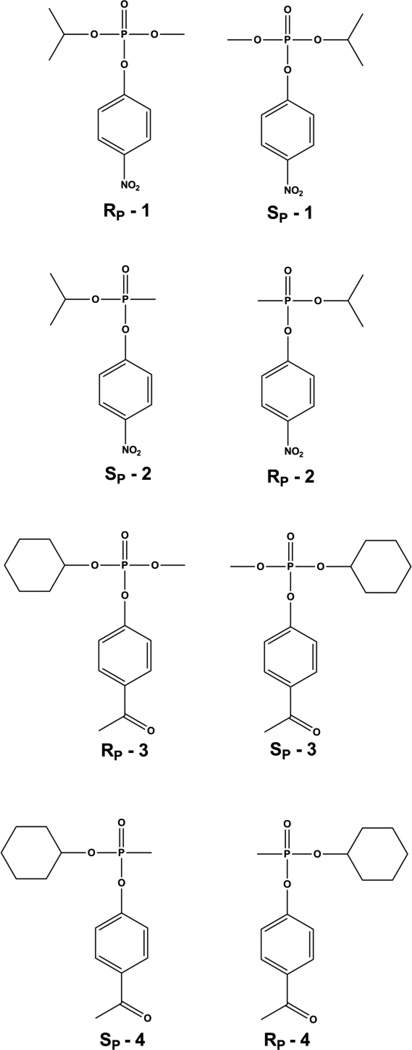
The best substrate for PTE is paraoxon. The catalytic efficiency with this substrate approaches the limits of diffusion, but the substrate specificity of PTE is extraordinarily large [35,42–48]. PTE is specific for triesters with very little diesterase activity [49]. However, PTE will readily hydrolyze phosphonate and phosphinate compounds in addition to the phosphate esters [43]. PTE is able to hydrolyze many insecticides including phosphotriesters, thiophosphotriesters, and phosphorothioesters [35,45,46,50–52]. PTE is most efficient at hydrolyzing substrates with electron withdrawing phenolic leaving groups [53], but PTE will also cleave thiol linkages and halide bonds [44,53]. This remarkably broad specificity is thought to be due to the non-specific nature of the substrate binding site. The three ester groups of the substrate interact with three hydrophobic pockets on the surface of the enzyme [36]. The leaving group pocket is made up by residues W131, F132, F306, and Y309 (Fig. 4C). Kinetic studies with mutant forms of PTE have verified that these residues largely dictate the enzyme specificity for the leaving group [26]. The two other ester groups interact with the large and small pockets. The large pocket is made up of residues H254, H257, L271, and M317, while the small pocket is made up of G60, I106, L303 and S308. The large and small pockets have been shown to determine the specificity for the side ester groups on the substrate including the stereochemical preference for chiral phosphorus centers [26]. The wild-type enzyme prefers the SP enantiomers for most phosphate substrates with the stereochemical selectivity increasing with the difference in size between the side groups [54]. For the compounds 1 and 3 PTE prefers the SP enantiomer over the RP enantiomer by 130- and 78,000-fold, respectively (Fig. 5) [43,54]. The same pattern is seen with the phosphonate compounds, however, due to a reversal in priority of the groups the corresponding chirality is RP [43,55]. Wild type PTE prefers the RP enantiomer of compound 2 by 20:1 and the RP enantiomer of compound 4 by 760:1 (Fig. 5)[55,56]. It has been shown that the stereochemical selectivity of PTE can be enhanced, diminished, or reversed by modulation of the large and small pockets [57]. Shrinking the small pocket by the mutation G60A increases the enantiomeric selectivity for the RP enantiomer of compound 4 from ~760-fold to 23,000-fold [55]. Mutation of the large pocket residues in the variant H254Q/H257F reduced the selectivity to ~ 6:1. Simultaneous alteration to both the large and small pocket with the mutations H254G/H257W/L303T reverses the selectivity so that the SP enantiomer is preferred by 100-fold. The ability to manipulate the enantiomeric preference along with the high levels of selectivity that can be achieved has made PTE a valuable enzymatic tool for the isolation of enantiomerically pure compounds and selective degradation of enantiomers [55–61]. Similarly, the detailed structural information available for PTE has allowed for directed evolution to enhance the activity against the G-type and V-type nerve agents as well as numerous insecticides [35,43,44,48,50–52–55,56,60,62]. These approaches have yielded variants of PTE enhanced for the most toxic enantiomers of GB and GD by over 2 orders-of-magnitude and approximately 25-fold for VX [52,55,62]. Activity against the insecticides chlorpyrifos, methyl parathion, and malathion have been enhanced 755-, 25-, and 77-fold respectively [50–52].
Figure 5.
Chemical structures of enantiomers of chiral phosphate and phosphonate nerve agent analogs for GB and GF. Chiral phosphorus centers are drawn as Fischer projections.
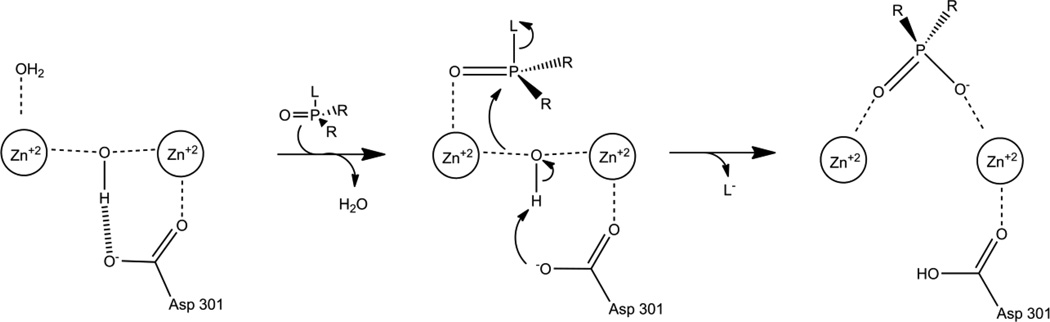
The proposed catalytic mechanism of PTE is shown in Figure 6 [30]. A combination of structural, labeling, kinetic, and physical methods have been employed to probe the various parts of the mechanism. The nature of the chemical reaction catalyzed by PTE was established utilizing 18O labeled water to demonstrate that the nucleophilic attack is directed at the phosphorus center rather than at the leaving group [63]. The same study demonstrated that there is an inversion of stereochemistry at the phosphorus center by analyzing the products of hydrolysis using O-ethyl O-(4-nitrophenyl) phenylphosphonothioate (ESP). ESP is a chiral substrate for PTE that yields a chiral hydrolysis product, and the inversion of stereochemistry indicates a direct attack by water on the substrate rather than a covalent enzyme bound intermediate.
Figure 6.
Proposed catalytic mechanism of PTE.
The presence of a hydroxide or water bridge between the metals is clearly seen in the X-ray structure of the PTE active site [36,40]. Probing the binuclear center of the Mn2+ containing enzyme with EPR demonstrated hyperfine splitting of the Mn2+ signal [64]. The coupling of the metal center was found to depend on pH with the protonation of the bridging hydroxyl resulting in a loss of coupling. Kinetic pH rate profiles of PTE demonstrate that a single group must be deprotonated for the reaction to take place with a catalytic pKa of ~6 which was found to match the pKa for protonation of the bridging hydroxide, thus confirming the bridging hydroxide as the attacking nucleophile [24,64].
Structural studies with Co2+ containing OpdA found an octahedral α-metal site with an additional water bound to the metal [38]. The α-bound water could potentially serve as the attacking nucleophile with the bridging hydroxyl acting as a general base. However, more recent work by the same group used magnetic circular dichroism to spectroscopically demonstrate that the α-metal is in a trigonal bipyramidal geometry in the resting state and during the course of the reaction with a slow substrate [64]. The direct probe of the metal site shows the ligation state of the α-metal unchanged between the resting state and the catalytically active state with substrate not inducing the switch to the octahedral geometry seen in the crystal structure. Analysis of the pH dependence of the reaction of Co2+ containing OpdA found that the pKa of the α-bound water was out of range for it to be deprotonated in the reaction, further confirming that the bridging hydroxide is the nucleophile [66].
The direct ligation of the substrate to the metal center during the course of the reaction has been observed structurally and verified by both EPR spectroscopy using the Mn2+ enzyme as well as by analysis of the transition state using Brønsted analysis and kinetic isotope effects. EPR studies on the coupled metal system demonstrate a substantial shift in the coupling from a hyperfine splitting at g = 2.2 to a hyperfine splitting at g = 4.3 on the addition of substrate analogs [66]. The shift in splitting to g = 4.3 is likely due to a transition allowed by a reduction of symmetry between the metals by the ligation of substrate to the metal center, presumably at the β-metal. Brønsted analysis using a series of paraoxon analogs was conducted to probe the transition state of the mechanism [53]. It was found that leaving groups with pKa values >8 give a linear relationship indicating that the chemistry is fully rate limiting with these substrates. The βLg value of −2.2 indicates a late transition state. Comparing the effects of an oxo vs. thiol phosphates on the transition state with Zn2+, Cd2+, or Mn2+ containing enzyme further supported the direct ligation of the substrate to the metal. The βLg value is similar for the oxo (−2.2) and thiol (−2.0) series with Zn2+ at the active site, but when the metal is substituted for Cd2+ the values are −3.0 for oxo-substrates and −1.4 for thiol-substrates. The softer Cd2+ chelates the softer thiols more strongly and causes a shift to an earlier transition state for the thiol substrates. Similarly, for the harder metal, Mn2+, the transition state for the thiol comes later than that of oxo substrates as indicated by βLg values of −3.2 and −4.3 respectively. Similar results have been obtained by measurement of the primary and secondary 18O kinetic isotope effects on the hydrolysis of paraoxon and O,O-diethyl O-(4-carbamoylphenyl) phosphate [68]. As observed with the Brønsted analysis, the primary isotope effects indicate an associative mechanism with a pentavalent transition state coming late compared to chemical hydrolysis. For O,O-diethyl O-(4-carbamoylphenyl) phosphate the maximal primary 18O isotope effect is expected to be 1.039 which corresponds to complete bond cleavage in the transition state. The experimentally observed effect of 1.036 indicates that for this substrate the cleavage of the leaving group is nearly complete in the transition state. Alkaline hydrolysis gave a smaller primary isotope effect of 1.027 which indicates that the enzymatic reaction has a later transition state. Labeling the substrate at the phosphoryl oxygen gives secondary isotope effects which report on the bond order between the oxygen and phosphorus with a maximal value of 1.04 for a change of bond order from 2 to 1. The measured secondary effect for alkaline hydrolysis of 1.02 indicates a bond order of approximately 1.4 in the transition state. The secondary isotope effect for the enzymatic reaction is diminished to 1.0144 indicating a bond order of 1.53 probably due to the interaction with the metal [68].
Crystal structures of PTE show hydrogen bonding between the bridging hydroxide and D301 supporting the role of D301 as the species that deprotonates the hydroxyl as the nucleophilic attack takes place [30,40]. D301 serves as a metal ligand making establishing other roles difficult, but the proton is proposed to be shuttled to the solvent through D233 [30]. Mutation of D233 to either alanine or asparagine substantially slows the reaction with paraoxon consistent with the loss of the proton shuttle. However, these mutations substantially increase the rate with the substrate analog diethyl, p-chlorophenylphosphate. This analog contains the relatively poor leaving group p-chlorophenol. While the disruption of the proton shuttle slows fast reactions, the trapped proton is apparently able to protonate the poor leaving group and speed the reaction.
The proposed dissociation of the hydroxide from the β-metal during the course of the reaction is supported by EPR and structural data. EPR experiments with substrate analogs bound to the active site show a decoupled mononuclear signal at g = 2 and g = 3.8 [67]. While a fraction of the enzymes maintains a coupled metal system, a portion of the enzyme sample undergoes a decoupling of the system indicating the dissociation of the hydroxide form the β-metal. This decoupling is dependent on the binding of the substrate analog and does not occur in the resting enzyme. The binding of the product diethylphosphate results in a complete loss of coupling in agreement with the proposed bidentate binding mode of the product. While EPR cannot rule out the existence of the hydroxide in the enzyme/product complex, the crystal structures of enzyme/product complex of both PTE and OpdA clearly show the presence of diethylphosphate (PTE and OpdA) or diethylthiophosphate (OpdA) bound in a bidentate manner [37–39].
The mechanism of paraoxon hydrolysis has been probed computationally by a number of groups employing different levels of theory and including more or less of the protein structure into the simulations. A high level density functional theory (DFT) simulation involving only the bridging hydroxide, metals and protein side chains that ligate the metals predicted a two transition state system with a high energy intermediate [69]. The first transition state is predicted to involve the binding of the substrate and a distortion of the phosphorus center to bring the phosphorus center into proximity of the hydroxide. The intermediate is predicted to be the pentavalent species. In the second transition state the hydroxide proton is transferred to D301, and the bond to the leaving group breaks. The overall energy of activation calculated by DFT simulations for the hydrolysis of paraoxon was between 10.5 and 13.3 kcal/mol which compares favorably to the 12.8 kcal/mol found experimentally [70].
Two studies have utilized quantum mechanical/molecular mechanical (QM/MM) simulations including the entire protein as well as active site waters and a water shell [70,71]. The first study predicted a single transition state, but reported a transition state plateau in the reaction coordinate similar in nature to the intermediate state of the DFT study [69,70]. In the predicted transition state the phosphorus is brought into proximity to the hydroxide while the bond to the leaving group is lengthened and the bonds between the hydroxyl and both metals are dissociated. These simulations predict a solvent water replacing the hydroxide as the α-metal ligand as catalysis takes place. The predicted enzyme/product complex has the new α-bound water molecule form an H-bond to the hydroxyl now attached to the phosphorus center. The simulations support some of the observed kinetic experiments [53,68], but there is no bidentate binding mode for the product observed in the crystal structure [37]. While this proposed mechanism was attractive because it avoids a strongly bound product, no crystal structures have been obtained with a monodentate product.
The later QM/MM study was published utilizing reoptimized parameters to describe the phosphorus atom and Zn2+ ions [71]. The result is a prediction of a series of discreet transition steps and intermediates that describe all steps of the reaction from substrate binding to hydrolysis and finally product release. The reoptimized parameters eliminated the need for the breaking of the bond between the hydroxide and the metals as the hydrolytic chemistry takes place, and the prediction is for the bidentate product as observed in the crystal structures. The most interesting prediction of this study was that the highest energy barrier along the reaction pathway was for the distortion of the phosphorus center which must occur to bring it into proximity to the hydroxide. This transition state was similar to the first transition state seen in the DFT simulations and may help to explain why the bond length between the β-metal and substrate analogs in crystal structures is longer than typical coordinate bonds [36,69]. The reoptimized QM/MM simulations also suggest a substantial barrier to the release of the diethylphosphate product. While qualitatively the QM/MM simulations seem to have identified many of the key characteristics of the PTE catalyzed hydrolysis of paraoxon, the validity of the predictions from QM/MM simulations remains somewhat suspect due to the significant disparity between the calculated energies of activation and the experimentally observed kinetics. The first QM/MM study calculated the minimum energy of activation to be 18.3 kcal/mol while the latter found the energy of activation to be 18.7 kcal/mol both of which are significantly higher than the 12.8 kcal/mol measured experimentally [70,71].
3. β-lactamase Fold
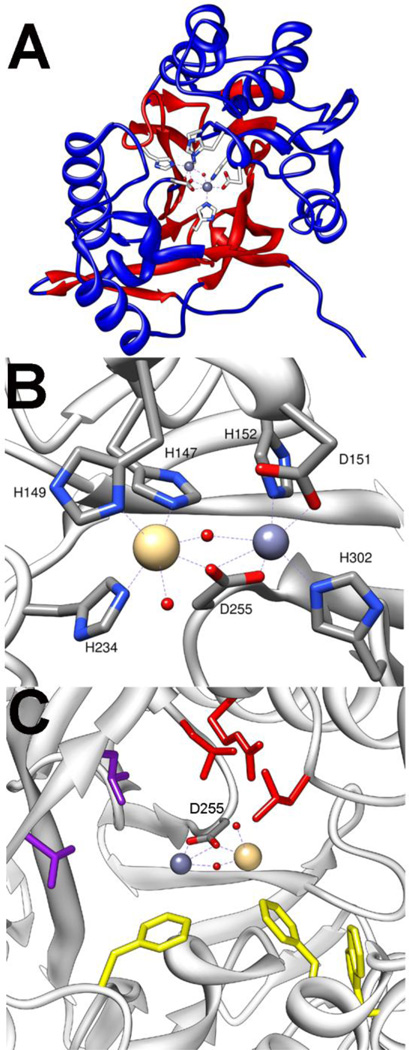
The enzyme methyl parathion hydrolase (MPH) has been identified in three bacterial species that are able to use the insecticide methyl parathion as their sole source of carbon and nitrogen [5–7]. These organisms were identified separately from organophosphate contaminated soils, but the sequenced genes show a remarkably high similarity. While the enzymes were initially suspected to be related to PTE, the sequences showed no relationship [5]. The structure of the methyl parathion hydrolase form Pseudomonas sp. WBC3 was solved, and it revealed that MPH has a β-lactamase fold rather than the TIM-barrel fold [19]. Each monomer in the dimeric enzyme interact as an αβ/βα sandwich, typical of the β-lactamase fold (Fig.7A). Despite the dissimilar sequence and protein fold, the active site of MPH shares common features with PTE. The active site is made up of three hydrophobic pockets. Molecular modeling studies have predicted the leaving group would interact with a pocket made up of resides F119, W179, and F196 (Fig. 7C) [19]. Mutation of any of these residues to alanine led to a substantial loss of activity. One of the additional pockets is lined by residues R72, L258, and L273 while the other is lined by L65 and L67.
Figure 7.
Crystal structure of MPH (pdb. 1p9e). A) αβ/βα structure of monomer shown with central β-strands in red and the sandwiching α-helices shown in blue. Metals at active site are shown as spheres and ligating residues shown as sticks. B) Metal site shown with the smaller Zn2+ in α-site and the larger Cd2+ in the β-site. C) Substrate binding pockets of MPH. Leaving group pocket residues are shown in yellow. Side pockets are shown in purple and red.
The binuclear metal sites of PTE and the MPH are very similar though there are differences in the ligation patterns [19]. The α-metal in MPH is ligated by two histidines (H152 and H302) and by aspartate-151 (Fig. 7B). As in PTE the binuclear center is bridged by a carboxylate ligand (D255) as well as a bridging hydroxide. However, unlike PTE, the carboxylate bridge ligates both the α- and β-metals with a single oxygen while the other carboxylate oxygen functions as an additional ligand to the α-metal. The presence of the additional ligand makes the geometry of the α-site octahedral rather than the trigonal bipyramidal seen in PTE. The β-metal of MPH has an octahedral geometry due to an additional histidine ligand (H147, H149, H234) along with the two bridging ligands.
No mechanistic work and only limited kinetic characterization has been done with MPH. However, the strong similarity between the active sites and metal centers, including the presence of the bridging hydroxide, it is believed that the catalytic mechanism is the same as the one for PTE (Fig 6). This notion is supported in the finding that MPH has extremely high activity for the insecticide methyl parathion (kcat/Km ~106 M−1s−1) and is the only enzyme aside from PTE to display this level of activity against compounds with a p-nitrophenol leaving group[19].
4. Pita Bread Fold
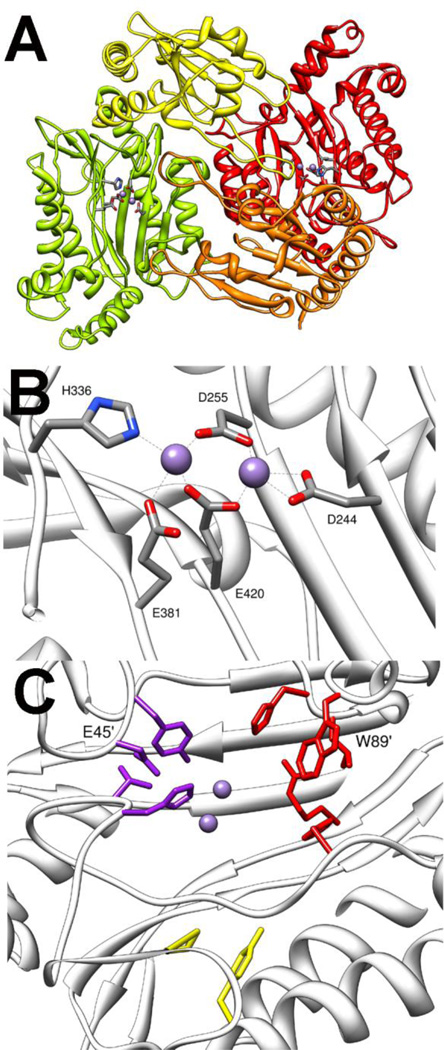
The next class of enzymes with phosphotriesterase activity is the bacterial prolidases. The best characterized enzyme is the organophosphate acid anhydrase (OPAA) from Alteromonas sp. [8]. The related enzyme aminopeptidase P (AMPP) from E. coli has been shown to have some phosphotriesterase activity [72]. The functional oligomeric state of the enzyme is a tetramer that is best described as a dimer of dimers [17,22]. Each subunit of the dimer contains a globular N-terminal domain and a C-terminal domain with a "pita bread" fold (Fig. 8A) [22]. The C-terminal domain contains the binuclear Mn2+ site in the central cleft of the fold. The more solvent exposed A-metal ligated to H336 and E381 (Fig. 8B) [22]. The B-metal is ligated in a bidentate fashion to D244, and the two metals are bridged by two additional carboxylate ligands, E420 and D255. Structures of OPAA have been solved as enzyme/product complexes and show the metals bridge an oxygen from the product. It is presumed that the metal center in the resting state will be bridged by a hydroxide. The structure of AMPP, which has an identical metal site was solved without product bound, and the bridging hydroxide is unambiguously seen [17].
Figure 8.
Crystal structure of OPAA (pdb. 3l7g). A) Dimeric structure of OPAA. One protomer is shown with N-terminal domain in yellow and C-terminal domain green. The second protomer is shown with N-terminal domain in orange and C-terminal domain in red. Mn2+ is shown as spheres, and metal ligating residues are shown as sticks. B) Expanded view of the OPAA metal center. C) Substrate binding site of OPAA. Small binding pocket residues are shown in purple. Large binding pocket residues are colored red. Leaving group pocket is colored yellow. The residues in large and small pockets from opposing subunit are labeled.
The native activity of OPAA and AMPP appears to be the degradation of proline containing peptides generated during the break down of collagen-like proteins [9,72]. OPAA has been demonstrated to be a prolidase with activity specific for dipeptides with proline as the second residue [9]. AMPP has been shown to cleave dipeptides, but has an order-of-magnitude better activity with tripeptides containing proline as the second residue [73,74]. Interestingly, the activity of OPAA is reported to be ~5-fold higher against fluoride containing phosphotriesters than it is against dipeptide substrates [9]. The best substrate reported is the nerve agent GD with a kcat of ~3000 s−1 [9]. OPAA also catalyzes the hydrolysis of nerve agent analogs containing a p-nitrophenol leaving group, though with rates nearly two orders-of-magnitude slower [75,76].
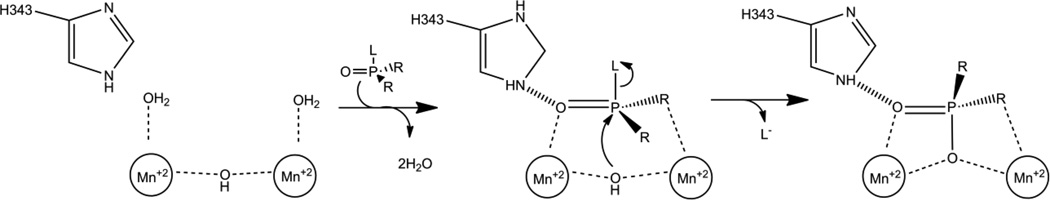
Like the other phosphotriesterases, the substrate binding site in OPAA is comprised of three pockets: the small pocket, the large pocket and the leaving group pocket (Fig 8C) [22]. The small pocket is lined by residues Y212, V342, H343 and capped by D45' from the N-terminal domain of the opposite subunit in the dimer. The large pocket is lined by residues L225, H226, H332, R418 and capped by W89' from the other subunit. The leaving group pocket is very small by comparison to other phosphotriesterase enzymes and is mainly made up of F292 and L366. In addition to being part of the small pocket, H343 forms a hydrogen bond with the free phosphoryl oxygen helping to position the substrate for catalysis. The stereoselectivity of OPAA is for the RP enantiomers of methyl phosphonates with selectivity of better than 1000-fold with analogs of GD [76]. To date, no systematic analysis of active site residues has been done, but the large preference for the fluoride leaving group is thought to be in part a consequence to the small leaving group site.
Despite AMPP having at least an order of magnitude less activity against organophosphates, the similar active sites and identical metal centers lead to the expectation that the chemical mechanism for organophosphate cleavage by OPAA and AMPP is the same [17,22,72,75]. The proposed chemical mechanism of OPAA shown in Figure 9 is similar to that of PTE but with some significant differences [22]. The most complete mechanistic study reported was carried out using AMPP with p-nitrophenol containing methyl phosphonates to specifically address the phosphotriesterase reaction [77]. As with PTE, mass spectroscopy analysis of the products from the reaction in 18O labeled water resulted in the label appearing on the phosphorus product confirming the nucleophilic attack comes at the phosphorus center. NMR analysis of the products from the hydrolysis of a chiral thiophosphonate demonstrate an inversion of stereochemistry at the phosphorus center indicating a direct attack by the bridging hydroxide rather than a covalent intermediate. The most significant difference in the proposed mechanisms for PTE and OPAA is in the initial binding of the substrate [22]. Structural evidence shows that the free phosphoryl oxygen binds to the more solvent exposed A-metal of OPAA, and the ester oxygen extending to the small pocket ligates to the B-metal. The bidentate ligation of the substrate is proposed to allow the hydroxide to attack the phosphorus center without dissociation from one of the metals. The leaving group is expelled, and the phosphoryl product is bound in a tridentate manner. This tridentate ligation of product is not observed in PTE, but has been observed in crystal structures of OPAA with an organophosphate analog and is analogous to the proposed ligation with the natural peptide substrates of AMPP [22,74]. The substrate profile of OPAA makes the mechanistic importance of the interaction between the ester oxygen and the B-metal somewhat uncertain. Studies with nerve agent analogs found the best analog substrate for OPAA is the RPSC phosphonate analog of GD [76]. This compound has a methyl group directly linked to the phosphorus center which precludes the interaction with the B-metal when the larger side group extends into the large pocket. However, experiments with methylphosphonate and O-methyl phosphate analogs of GB demonstrated that the selectivity varies dramatically when the ester oxygen is present [75,76]. The SP enantiomer of compound 1 (Fig. 5) can bind with the larger ester group extending into the large pocket while the ligation of the B-metal by the opposing ester oxygen is still possible. The enantiomers of compound 1 differ by an order of magnitude in kcat and have a selectivity of 23-fold [75]. By contrast, the phosphonate compound 2 (Fig. 5) lacking the ester oxygen on the small group gives only 2-fold selectivity for the RP enantiomer indicating that while not strictly required the ligation to the B-metal may indeed be important for catalysis [75].
Figure 9.
Proposed catalytic mechanism for OPAA cleavage of phosphotriesters. L is leaving group which can be fluoride, or p-nitrophenol. R is an ester or methyl group.
5. β-Propeller Fold
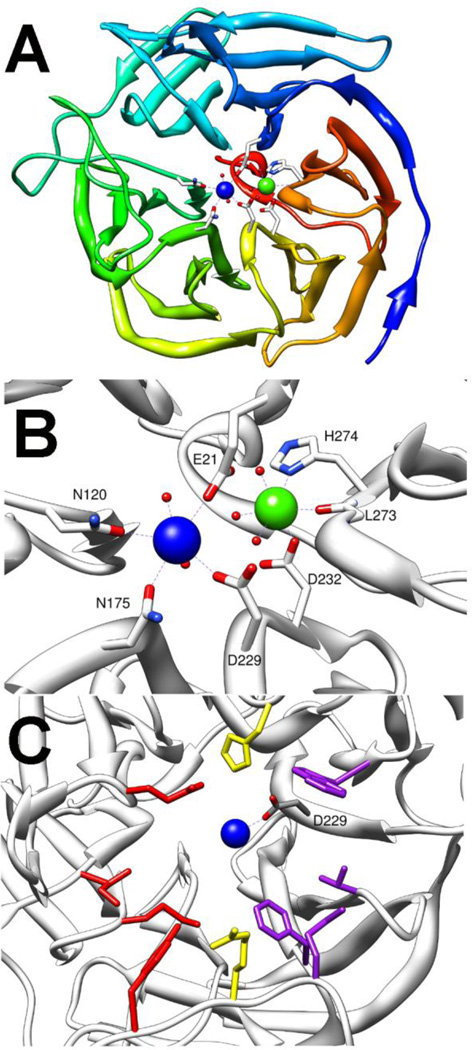
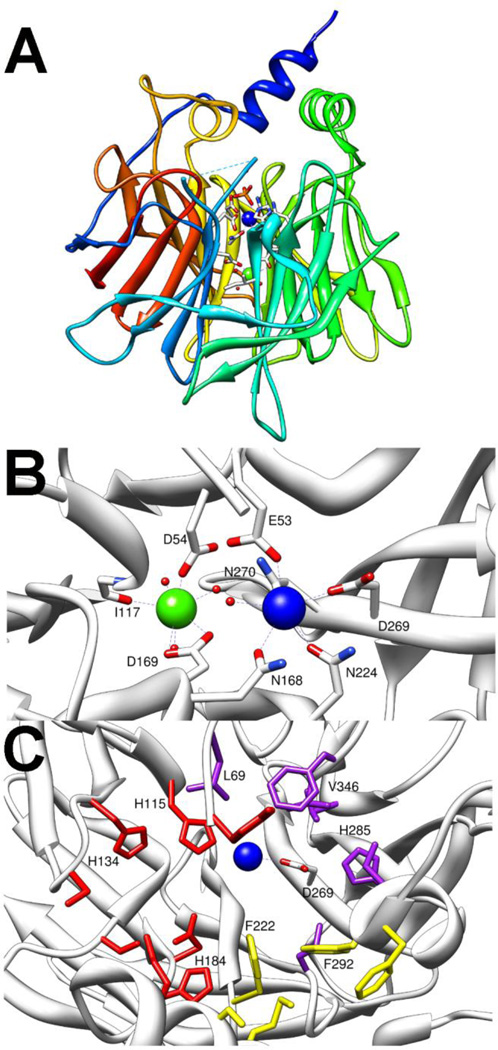
The mammalian serum paraoxonase (PON1) and squid diisopropylfluorophosphatase (DFPase) make up the final class of enzymes known to function as phosphotriesterases [11,12]. The X- ray structure of DFPase was solved to 0.85 Å with the wild-type enzyme, but the structure of PON1 could only be solved using an evolved variant with kinetic constants similar to the wild-type enzyme [20,78,79]. DFPase and PON1 are calcium containing hydrolases with a 6-bladed β-propeller fold (Fig. 10A and 11A) [20,23]. Each propeller is made of 4 pleated β-sheets, while the two calcium ions are located in a central water filled core. The more deeply buried calcium is commonly referred to as the structural calcium and is required for maintaining the fold of the enzyme, but is not proposed to play any role in catalysis [25,80]. It is ligated to the protein by a backbone carbonyl group and by two amino acid side chains, D232 and H274 in the case of DFPase, or D169 and D54 in PON1 (Fig. 10B and 11B) [21,23]. Additionally, the structural calcium is ligated to three ordered waters and functions to anchor an extended structural water network in the core of the protein fold. The more solvent exposed calcium, known as the catalytic calcium, serves to facilitate the hydrolytic reaction. In DFPase the catalytic calcium is ligated by D229, E21, N120 and N175 (Fig. 10B) [21]. Two waters from the central core ligate the catalytic calcium from below the active site while a third water was found ligated to the metal in the substrate binding site. Similarly, the catalytic calcium of PON1 is bound by D269 and E53, but PON1 has three asparagine ligands (N168, N224, and N270) to the catalytic calcium with the third asparagine ligand replacing one of the ordered waters from the core of the enzyme (Fig. 11B) [23]. The structure of PON1 was solved with a phosphate ligated to the calcium suggesting that, like the other phosphotriesterases, the mechanism proceeds via a direct interaction between the catalytic metal and the substrate [23]. Despite relatively little sequence homology, the overall fold of DFPase and PON1 are quite similar. The major deviation between the structures of the two enzymes is the presence of two additional α-helixes in PON1 that are thought to anchor it to high density lipoprotein (HDL) complex in vivo [23]. These additional helices sit above and cap the active site resulting in a much more enclosed active site in PON1 compared to that of DFPase (Fig. 11A and C).
Figure 10.
Crystal structure of DFPase (pdb 2gvv). Catalytic calcium is shown in blue while structural calcium is shown in green. A) Top down view of DFPase showing the 6 bladed β-propeller structure. B) Metal center ligation of DFPase. C) Substrate binding site in DFPase. Side pocket residues Y144, M90, I72 and E37 are shown in red. In purple are W244, T195, F173, and M148 from the second side pocket. The central cleft residues R146 and H287 are shown in yellow. Catalytic aspartate is labeled.
Figure 11.
Crystal structure of PON1 (pdb. 1v04). A) Side view of β-propeller fold with HDL anchoring helices extending above. Catalytic calcium shown in blue, structural calcium shown in green, and ligating residues shown as sticks. B) Metal centers and ligating residues of PON1. Catalytic calcium is blue structural calcium is green. C) Substrate binding pockets of PON1. Large group pocket residues are colored red. Small group pocket is colored purple, and leaving group pocket is colored yellow. Catalytic aspartate (D269) and residues known to be important for the phosphotriesterase reaction are labeled.
The native activities of DFPase and PON1 remain unclear. DFPase is found in the central nervous system of the squid where its role is not understood due to its strong preference for the hydrolysis of P-F or P-CN bonds which are not found in natural compounds [11,81,82]. PON1 is thought to serve in a protective role against lipid oxidation as part of the HDL particles of mammalian blood [83]. The native function is proposed to be a lactonase which serves to hydrolyze long chain fatty acid lactones which are generated by oxidation of lipids [16,83].
The substrate specificity of both PON1 and DFPase have been examined. DFPase will readily hydrolyze the fluoride containing diisopropylfluorophosphate as well as the nerve agents GB, GD and GF, in addition to the cyanide containing nerve agent tabun [81]. DFPase was initially only found to hydrolyze P-F and P-CN bonds and to be inert toward P-O or P-S bonds. However some more recent reports have found an extremely limited ability to hydrolyze P-O bonds in a GD analog with a coumarin leaving group [82]. PON1 has a much broader substrate specificity being able to hydrolyze γ- and δ-lactones, various aryl-esters as well as a wide variety of organophosphates [16]. Among the organophosphates hydrolyzed by PON1 are the nerve agents GB, GD, GF, tabun, VX and the insecticide paraoxon and the oxo forms of diazinon and chlorpyrifos [84–89].
In DFPase the active site is described as a hydrophobic cleft with the catalytic calcium centrally located at the bottom of the cleft [21,28,31]. The cleft is lined to one side of the calcium by W244, T195, F173 and M148 (Fig. 10C)[29]. The other side of the cleft is lined by residues Y144, M90, I72, and E37. Residues R146 and H287 appear opposite each other and fill in the central gap in the cleft. Site directed mutants of DFPase with chiral nerve agents have been used to confirm the importance of these residues in defining the substrate specificity of DFPase [28,90]. The wild-type DFPase prefers the RP enantiomer of GF over the SP enantiomer by a ratio of 50:1 [90]. In order to bind for catalysis, the compounds must position their ester groups extending toward opposite sides of the active site cleft. The variant E37A/Y144A/R146A/T195M which was predicted to shrink the T195 side of the cleft, where the large side substituent of the RP enantiomer must bind, and simultaneously expand the Y144 side of the cleft, where the larger group on the SP enantiomer binds, was found to reverse the stereoselectivity of DFPase with the mutant preferring the SP enantiomer by a ratio of 5:1 [90]. The mutant E37D/Y144A/R146A/T195M, which failed to fully expand the Y144 side of the cleft, was shown have little selectivity for GF mainly due to a decrease in activity against the RP enantiomer without an increase for the SP enantiomer.
The presence of the capping α-helices in PON1 makes the active site significantly more restricted [23]. The binding site can be divided into three regions. The large side pocket is an extended cavity lined by residues Y71, H115, H134, S137, S166, D183, H184, and K192 (Fig. 11C)[23,85,91]. There is a central pocket which is lined by residues S193, M196, F222, F292, and F293. Finally, there is a small side pocket which is lined by residues L69, L240, H285, I291, I332, V346 and F347. Residues 192,193, and 196 are contributed by α-helix 2 while residues 292 and 293 are from α-helix 3, neither of which is present in DFPase. One of the more unique facets of the PON1 enzyme is its ability to catalyze multiple types of reactions [16]. Kinetic analysis of mutant enzymes has revealed that the active site residues do not contribute equally to the different types of reactions. It has been shown that H115 and H134 are absolutely required for lactonase activity, but H115W is more active as a phosphotriesterase than the wild-type enzyme [23,92,93]. Similarly, mutations to reposition F292 significantly diminished phosphotriesterase and lactonase activity but have very little effect on esterase activity [23]. The active site residues L69, H115, and H184 from the large side pocket, F222 and F292 from the central pocket and residues, H285 and V346 from the small pocket appear most important for phosphotriesterase activity [23,85,91]. Directed evolution of the active site of PON1 has yielded improved activity against the nerve agents GD, GF and VX [85,86,88]. With GF, the activity against the most toxic enantiomer was improved more than 2-orders of magnitude to ~105 M−1s−1 [86].
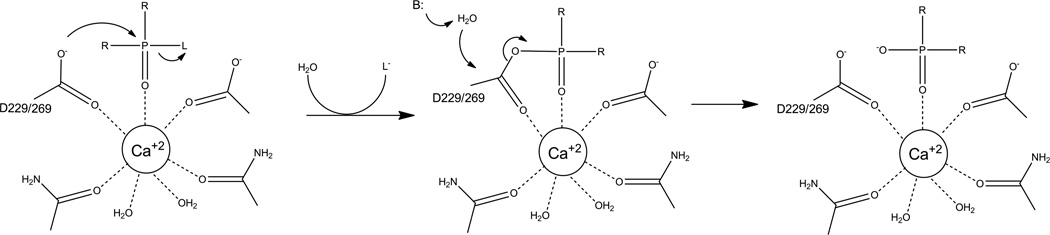
The proposed mechanism for phosphotriester cleavage by PON1 and DFPase shown in Figure 12 is unique among the phosphotriesterases in that only a single active site metal is employed and the mechanism proceeds through a covalent enzyme bound intermediate [21,29,31]. The mechanistic work that has been done on DFPase and PON1 is limited in scope, and the proposed mechanism is mainly based on the structures of the enzymes. pH profiles for both enzymes indicate that a single group must be deprotonated with a pKa of ~7 [23,94]. The initial supposition was for a simple attack by an activated water or hydroxide as observed in PTE. This notion was supported by the finding of a presumptive catalytic base H287 in DFPase and a catalytic diad H115 and H134 in PON1 [23,95]. It was proposed that the catalytic base deprotonated a water allowing nucleophilic attack to occur. While this mechanism appears to be correct for the lactonase and esterase activity of PON1 [92,93], in both DFPase and PON1, the presumed catalytic base is not required for phosphotriesterase activity [23,28,92,93]. In DFPase, substitution of H287 for a leucine or a phenylalanine resulted in a less than two-fold loss of activity while in PON1 substitution of H115W resulted in a 2-fold improvement in activity against paraoxon. The crystal structure of DFPase in complex with a slow substrate analog revealed that the leaving group was oriented toward the presumptive base while the phosphorus center was aligned for a backside attack by the carboxyl group of residue D229 [29]. Aspartate-229 is one of the calcium binding residues, but a role as the attacking nucleophile generating a high energy phosphoacyl intermediate was unexpected. Evidence for the phosphoacyl enzyme intermediate has been provided by 18O labeling experiments with DFPase under single turnover conditions [29,31]. These experiments have demonstrated that the oxygen found in the product comes from the carboxyl group of D229 rather than solvent. The carboxyl oxygen is found to be exchanged with a solvent oxygen during the course of the reaction indicating that the hydrolysis of the enzyme-bound intermediate comes via attack at the carboxylate rather than at the phosphorus center. In PON1 the catalytic aspartate is D269, and numerous molecular modeling studies have shown that substrates align well for nucleophilic attack by the carboxylate oxygen [29,82]. The relatively high turnover numbers for DFPase and PON1 with fluoride leaving groups both indicate that an activated water is needed to cleave the intermediate, but current structural information does not indicate how the water is activated. Brønsted analysis for PON1 using paraoxon analogs give a βLG value of −1.6 for leaving groups with a pKa > 7 [16]. The large negative value indicates that cleavage of the leaving group is fully rate limiting for these substrates and that the transition state is very product-like. Current evidence indicates that the initial cleavage of the leaving group is rate limiting and that the cleavage of the acyl intermediate is relatively fast, though how that is achieved is yet to be determined [21,29,31].
Figure 12.
Proposed catalytic mechanism for phosphotriesterase reaction catalyzed by PON1 and DFPase. R is ester linked alcohol group or a methyl group. L is leaving group which is fluoride for DFPase or for PON1 a fluoride, a phenol, or a thiol.
6. Concluding Comments
The organophosphates encompass a very broad group of structurally related compounds. The toxicity of these compounds has fueled a search for enzymes that are able to hydrolyze them [1,2]. It is somewhat surprising to find enzymes in nature that can hydrolyze these relatively new, synthetic compounds, but the phosphotriesterase activity is found in widely varied organisms, many of which would not be expected to encounter organophosphates in their native environments [4–12]. While the enzymes that posses phosphotriesterase activity are widely varied and utilize completely different protein scaffolds and even different catalytic mechanisms, they are all metal dependent hydrolases with hydrophobic active sites [18–23,29–31].
Highlights.
The role of binuclear metal clusters in phosphotriester hydrolysis.
The mechanism of action for the hydrolysis of oganophosphate triesters.
Variations in protein structure for various phosphotriesterases.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Raushel FM. Bacterial detoxification of organophosphate nerve agents. Curr. Opin. Microbiol. 2002;5:288–295. doi: 10.1016/s1369-5274(02)00314-4. [DOI] [PubMed] [Google Scholar]
- 2.Musilek K, Dolezal M, Gunn-Moore F, Kuca K. Design, evaluation and structure---Activity relationship studies of the AChE reactivators against organophosphorus pesticides. Med. Res Rev. 2011;31:548–575. doi: 10.1002/med.20192. [DOI] [PubMed] [Google Scholar]
- 3.Munro NB, Ambrose KR, Watson AP. Toxicity of the organophosphate chemical warfare agents GA, GB, and VX: Implications for public protection. Environ. Health Perspect. 1994;102:18–38. doi: 10.1289/ehp.9410218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harper LL, McDaniel CS, Miller CE, Wild JR. Dissimilar plasmids isolated from Pseudomonas diminuta MG and a Flavobacterium sp. (ATCC 27551) contain identical opd genes. Appl. Environ. Microbiol. 1988;54:2586–2589. doi: 10.1128/aem.54.10.2586-2589.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun L, Dong Y, Zhou Y, Yang M, Zhang C, Rao Z, Zhang X. Crystalization and preliminary X-ray studies of methyl parathion hydrolase from Pseudomonas sp. WBC-3. Acta Crystallogr. D Biol. Crystallogr. 2004;60:954–956. doi: 10.1107/S0907444904005669. [DOI] [PubMed] [Google Scholar]
- 6.Zhongli C, Shunpeng L, Guoping F. Isolation of methyl parathion-degrading strain M6 and cloning of the methyl parathion hydrolase gene. Appl. Environ. Microbiol. 2001;67:4922–4925. doi: 10.1128/AEM.67.10.4922-4925.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rani NL, Lalithakumari D. Degradation of methyl parathion by Pseudomonas putida. Can. J. Microbiol. 1994;40:1000–1006. doi: 10.1139/m94-160. [DOI] [PubMed] [Google Scholar]
- 8.DeFrank JJ, Cheng TC. Purification and properties of an organophophorus acid anhydrase from a halophilic bacterial isolate. J. Bacteriol. 1991;173:1938–1943. doi: 10.1128/jb.173.6.1938-1943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng TC, Liu L, Wang B, Wu J, DeFrank JJ, Anderson DM, Rastogi VK, Hamilton AB. Nucleotide sequence of a gene encoding an organophosphorus nerve agent degrading enzyme from Alteromonas haloplanktis. J. Ind. Microbiol. Biotechnol. 1997;18:49–55. doi: 10.1038/sj.jim.2900358. [DOI] [PubMed] [Google Scholar]
- 10.Horne I, Sutherland TD, Harcourt RL, Russell RJ, Oakeshott JG. Identification of an opd (organophosphate degradation) gene in an Agrobacterium isolate. Appl. Environ. Microbiol. 2002;68:3371–3376. doi: 10.1128/AEM.68.7.3371-3376.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoskin FCG, Long RJ. Purification of a DFP-hydrolyzing enzyme from squid head ganglion. Arch. Biochem. Biophys. 1972;150:548–555. doi: 10.1016/0003-9861(72)90073-2. [DOI] [PubMed] [Google Scholar]
- 12.Furlong CE, Richter RJ, Chapline C, Crabb JW. Purification of rabbit and human serum paraoxonase. Biochemistry. 1991;30:10133–10140. doi: 10.1021/bi00106a009. [DOI] [PubMed] [Google Scholar]
- 13.Elias M, Dupuy J, Merone L, Mandrich L, Porzio E, Moniot S, Rochu D, Lecomte C, Rossi M, Masson P, Manco G, Chabriere E. Structural basis for natural lactonase and promiscuous phosphotriesterase activities. J. Mol. Biol. 2008;379:1017–1028. doi: 10.1016/j.jmb.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 14.Roodveldt C, Tawfik DS. Shared promiscuous activities and evolutionary features in various members of the amidohydrolase superfamily. Biochemistry. 2005;44:12728–12736. doi: 10.1021/bi051021e. [DOI] [PubMed] [Google Scholar]
- 15.Xiang DF, Kolb P, Fedorov AA, Meier MM, Fedorov LV, Nguyen TT, Sterner R, Almo SC, Shoichet BK, Raushel FM. Functional annotation and three-dimensional structure of Dr0930 from Deinococcus radiodurans, a close relative of phosphotriesterase in the amidohydrolase superfamily. Biochemistry. 2009;48:2237–2247. doi: 10.1021/bi802274f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khersonsky O, Tawfik DS. Structure-reactivity studies of serum paraoxonase PON1 suggest that its native activity is lactonase. Biochemistry. 2005;44:6371–6382. doi: 10.1021/bi047440d. [DOI] [PubMed] [Google Scholar]
- 17.Wilce MCJ, Bond CS, Dixon NE, Freeman HC, Guss JM, Lilley PE, Wilce JA. Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3472–3477. doi: 10.1073/pnas.95.7.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benning MM, Kuo JM, Raushel FM, Holden HM. Three-dimensional structure of the binuclear metal center of phosphotriesterase. Biochemistry. 1995;34:7973–7978. doi: 10.1021/bi00025a002. [DOI] [PubMed] [Google Scholar]
- 19.Dong YJ, Bartlam M, Sun L, Zhou YF, Zhang ZP, Zhang CG, Rao Z, Zhang XE. Crystal structure of methyl parathion hydrolase from Pseudomonas sp. WBC-3. J. Mol. Biol. 2005;353:655–663. doi: 10.1016/j.jmb.2005.08.057. [DOI] [PubMed] [Google Scholar]
- 20.Koepke J, Scharff EI, Lucke C, Ruterjans H, Fritzsch G. Statistical analysis of crystallographic data obtained from squid ganglion DFPase at 0.85 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1744–1754. doi: 10.1107/s0907444903016135. [DOI] [PubMed] [Google Scholar]
- 21.Blum MM, Mustyakimov M, Ruterjans H, Kehe K, Schoenborn BP, Langan P, Chen JCH. Rapid determination of hydrogen positions and protonation states of diisopropyl fluorophosphatase by joint neutron and X-ray diffraction refinement. Proc. Natl. Acad. Sci. USA. 2009;106:713–718. doi: 10.1073/pnas.0807842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vyas NK, Nickitenko A, Rastogi VK, Shah SS, Quiocho FA. Structural insights into the dual activities of the nerve agent degrading organophosphate anhydrolase/prolidase. Biochemistry. 2010;49:547–559. doi: 10.1021/bi9011989. [DOI] [PubMed] [Google Scholar]
- 23.Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RBG, McCarthy A, Toker L, Silman I, Sussman JL, Tawfik DS. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Biol. 2004;11:412–419. doi: 10.1038/nsmb767. [DOI] [PubMed] [Google Scholar]
- 24.Omburo GA, Kuo JM, Mullins LS, Raushel FM. Characterization of the zinc binding site of bacterial phosphotriesterase. J. Biol. Chem. 1992;267:13278–13283. [PubMed] [Google Scholar]
- 25.Hartleib J, Geschwindner S, Scharff EI, Ruterjans H. Role of calcium ions in the structure and function of the di-isopropylfluorophosphatase from Loligo vulgaris. Biochem. J. 2001;353:579–589. doi: 10.1042/0264-6021:3530579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen-Goodspeed M, Sogorb MA, Wu F, Hong SB, Raushel FM. Stuctural determinants of the substrate and stereochemical specificity of phosphotriesterase. Biochemistry. 2001;40:1325–1331. doi: 10.1021/bi001548l. [DOI] [PubMed] [Google Scholar]
- 27.Hu X, Jiang X, Lenz DE, Cerasoli DM, Wallqvist A. In silico analyses of substrate interactions with human serum paraoxonase 1. Proteins. 2009;75:486–498. doi: 10.1002/prot.22264. [DOI] [PubMed] [Google Scholar]
- 28.Katsemi V, Lucke C, Koepke J, Lohr F, Maurer S, Fritzsch G, Ruterjans H. Mutational and structural studies of the diisopropylfluorophosphatase from Loligo vulgaris shed new light on the catalytic mechanism of the enzyme. Biochemistry. 2005;44:9022–9033. doi: 10.1021/bi0500675. [DOI] [PubMed] [Google Scholar]
- 29.Blum MM, Lohr F, Richardt A, Ruterjans H, Chen JCH. Binding of a designed substrate analogue to diisopropyl fluorophosphatase: Implications for the phosphotriesterase mechanism. J. Am. Chem. Soc. 2006;128:12750–12757. doi: 10.1021/ja061887n. [DOI] [PubMed] [Google Scholar]
- 30.Aubert SD, Li Y, Raushel FM. Mechanism for the hydrolysis of organophosphates by the bacterial phosphotriesterase. Biochemistry. 2004;43:5707–5715. doi: 10.1021/bi0497805. [DOI] [PubMed] [Google Scholar]
- 31.Chen JCH, Mustyakimov M, Schoenborn BP, Langan P, Blum MM. Neutron structure and mechanistic studies of diisopropyl fluorophosphatase (DFPase) Acta Crystallogr. D Biol. Crystallogr. 2010;66:1131–1138. doi: 10.1107/S0907444910034013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serdar CM, Gibson DT, Munnecke DM, Lancaster JH. Plasmid involvement in parathion hydrolysis by Pseudomonas diminuta. Appl. Environ. Microbiol. 1982;44:246–249. doi: 10.1128/aem.44.1.246-249.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulbry WW, Karns JS, Kearney PC, Nelson JO, McDaniel CS, Wild JR. Identification of a plasmid-borne parathion hydrolase gene from Flavobacterium sp. by Southern hybridization with opd form Pseudomonas diminuta. Appl. Environ. Microbiol. 1986;51:926–930. doi: 10.1128/aem.51.5.926-930.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Somara S, Sidavattam D. Plasmid mediated organophosphate pesticide degradation by Flavobacterium balustinum. Biochem. Mol. Biol. Int. 1995;36:627–631. [PubMed] [Google Scholar]
- 35.Yang H, Carr PD, McLoughlin SY, Liu JW, Horne I, Qui X, Jeffries CMJ, Russell RJ, Oakeshott JG, Ollis DL. Evolution of an organophosphate-degrading enzyme: a comparison of natural and directed evolution. Protein Eng. 2003;16:135–145. doi: 10.1093/proeng/gzg013. [DOI] [PubMed] [Google Scholar]
- 36.Vanhooke JL, Benning MM, Raushel FM, Holden HM. Three-dimensional structure of the zinc-containing phosphotriesterase with the bound substrate analong diethyl 4-methylbenzylphosphonate. Biochemistry. 1996;35:6020–6025. doi: 10.1021/bi960325l. [DOI] [PubMed] [Google Scholar]
- 37.Kim J, Tsai PC, Chen SL, Himo F, Almo SC, Raushel FM. Structure of diethyl phosphate bound to the binuclear metal center of phosphotriesterase. Biochemisty. 2008;47:9497–9504. doi: 10.1021/bi800971v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson C, Kim HK, Carr PD, Liu JW, Ollis DL. The structure of an enzyme-product complex reveals the critical role of a terminal hydroxide nucleophile in the bacterial phosphotriesterase mechanism. Biochim. Biophys. Acta. 2005;1752:56–64. doi: 10.1016/j.bbapap.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Jackson CJ, Foo JL, Kim HK, Carr PD, Liu JW, Salem G, Ollis DL. In crystallo capture of a Mchaelis complex and product-binding modes of a bacterial phosphotriesterase. J. Mol. Biol. 2008;375:1189–1196. doi: 10.1016/j.jmb.2007.10.061. [DOI] [PubMed] [Google Scholar]
- 40.Benning MM, Shim H, Raushel FM, Holden HM. High resolution X-ray structures of different metal-substituted forms of phosphotriesterase from Pseudomonas diminuta. Biochemistry. 2001;40:2712–2722. doi: 10.1021/bi002661e. [DOI] [PubMed] [Google Scholar]
- 41.Omburo GA, Mullins LS, Raushel FM. Structural characterization of the divalent cation sites of bacterial phosphotriesterase by 113Cd NMR Spectroscopy. Biochemistry. 1993;32:9148–9155. doi: 10.1021/bi00086a021. [DOI] [PubMed] [Google Scholar]
- 42.Caldwell SR, Newcomb JR, Schlecht KA, Raushel FM. Limits of diffusion in the hydrolysis of substrates by the phosphotriesterase from Pseudomonas diminuta. Biochemistry. 1991;30:7438–7444. doi: 10.1021/bi00244a010. [DOI] [PubMed] [Google Scholar]
- 43.Nowlan C, Li Y, Hermann JC, Evans T, Carpenter J, Ghanem E, Shoichet BK, Raushel FM. Resolution of chiral phosphate, phosphonate, and phosphineate esters by an enantioselective enzyme library. J. Am. Chem. Soc. 2006;128:15892–15902. doi: 10.1021/ja0658618. [DOI] [PubMed] [Google Scholar]
- 44.Watkins LM, Mahoney HJ, McCulloch JK, Raushel FM. Augmented hydrolysis of diisopropyl fluorophosphate in engineered mutants of phosphotriesterase. J. Biol. Chem. 1997;272:25596–25601. doi: 10.1074/jbc.272.41.25596. [DOI] [PubMed] [Google Scholar]
- 45.Chae MY, Postula JF, Raushel FM. Stereospecific enzymatic hydrolysis of phosphorus-sulfur bonds in chiral organophosphate triesters. Bioorg. Med. Chem. Lett. 1994;4:1473–1478. [Google Scholar]
- 46.Lai K, Stolowich NJ, Wild JR. Characterization of P-S bond hydrolysis in organophosphorothioate pesticides by organophosphorus hydrolase. Arch. Biochem. Biophys. 1995;318:59–64. doi: 10.1006/abbi.1995.1204. [DOI] [PubMed] [Google Scholar]
- 47.Rastogi VK, DeFrank JJ, Cheng T, Wild JR. Enzymatic hydrolysis of Russian-VX by organophosphorus hydrolase. Biochem. Biophys. Res. Commun. 1997;241:294–296. doi: 10.1006/bbrc.1997.7569. [DOI] [PubMed] [Google Scholar]
- 48.Lai K, Grimsley JK, Kuhlmann BD, Scapozza L, Harvey SP, DeFrank JJ, Kolakowski JE, Wild JR. Rational enzyme design: Computer modeling and site-directed mutagenesis for the modification of catalytic specificity in organophosphorus hydrolase. Chimia. 1996;50:430–431. [Google Scholar]
- 49.Shim H, Hong SB, Raushel FM. Hydrolysis of phosphodiesters through transformation of the bacterial phosphotriesterase. J. Biol. Chem. 1998;273:17445–17450. doi: 10.1074/jbc.273.28.17445. [DOI] [PubMed] [Google Scholar]
- 50.Cho CMH, Mulchandani A, Chen W. Altering the substrate specificity of organophosphorus hydrolase for enhanced hydrolysis of chlorpyrifos. Appl. Environ, Microbiol. 2004;70:4681–4685. doi: 10.1128/AEM.70.8.4681-4685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho CMH, Mulchandani A, Chen W. Functional analysis of organophosphorus hydrolase variants with high degradation activity towards organophosphate pesticides. Protein Eng. Des. Sel. 2006;19:99–105. doi: 10.1093/protein/gzj007. [DOI] [PubMed] [Google Scholar]
- 52.Schofield DA, DiNovo AA. Generation of a mutagenized organophosphorus hydrolase for the biodegradation of the organophosphate pesticides malathion and demeton-S. J. Appl. Microbiol. 2010;109:548–557. doi: 10.1111/j.1365-2672.2010.04672.x. [DOI] [PubMed] [Google Scholar]
- 53.Hong SB, Raushel FM. Metal-substrate interactions facilitate the catalytic activity of the bacterial phosphotriesterase. Biochemistry. 1996;35:10904–10912. doi: 10.1021/bi960663m. [DOI] [PubMed] [Google Scholar]
- 54.Hong SB, Raushel FM. Stereochmical constraints on the substrate specificity of phosphotriesterase. Biochemistry. 1999;38:1159–1165. doi: 10.1021/bi982204m. [DOI] [PubMed] [Google Scholar]
- 55.Tsai PC, Bigley A, Li Y, Ghanem E, Cadieux CL, Kasten SA, Reeves TE, Cerasoli DM, Raushel FM. Selective hydrolysis of organophosphate nerve agents by the bacterial phosphotriesterase. Biochemistry. 2010;49:7978–7987. doi: 10.1021/bi101056m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li WS, Lum KT, Chen-Goodspeed M, Sogorb MA, Raushel FM. Stereoselective detoxification of chiral sarin and soman analogues by phosphotriesterase. Bioorg. Med. Chem. 2001;9:2083–2091. doi: 10.1016/s0968-0896(01)00113-4. [DOI] [PubMed] [Google Scholar]
- 57.Chen-Goodspeed M, Sogorb MA, Wu F, Raushel FM. Enhancement, relaxation, and reversal of the stereoselectivity for phosphotriesterase by rational evolution of active site residues. Biochemistry. 2001;40:1332–1339. doi: 10.1021/bi001549d. [DOI] [PubMed] [Google Scholar]
- 58.Li WS, Li Y, Hill CM, Lum KT, Raushel FM. Enzymatic synthesis of chiral organophosphothioates from prochiral precursors. J. Am. Chem. Soc. 2002;124:3498–3499. doi: 10.1021/ja017840d. [DOI] [PubMed] [Google Scholar]
- 59.Li Y, Aubert SD, Raushel FM. Operational control of stereoselectivity during the enzymatic hydrolysis of racemic organophosphorus compounds. J. Am. Chem. Soc. 2003;125:7526–7527. doi: 10.1021/ja035625m. [DOI] [PubMed] [Google Scholar]
- 60.Hill CM, Li WS, Thoden JB, Holden HM, Raushel FM. Enhanced degradation of chemical warfare agents through molecular engineering of the phosphotriesterase active site. J. Am. Chem. Soc. 2003;125:8990–8991. doi: 10.1021/ja0358798. [DOI] [PubMed] [Google Scholar]
- 61.Li Y, Aubert SD, Maes EG, Raushel FM. Enzymatic resolution of chiral phosphinate esters. J. Am. Chem. Soc. 2004;126:8888–8889. doi: 10.1021/ja048457m. [DOI] [PubMed] [Google Scholar]
- 62.Reeves TE, Wales ME, Grimsley JK, Li P, Cerasoli DM, Wild JR. Balancing the stability and the catalytic specificities of OP hydrolase with enhanced V-agent activities. Protein Eng. Des. Sel. 2008;21:405–412. doi: 10.1093/protein/gzn019. [DOI] [PubMed] [Google Scholar]
- 63.Lewis VE, Donarski WJ, Wild JR, Raushel FM. Mechanism and stereochemical course at the phosphorus of the reaction catalyzed by a bacterial phosphotriesterase. Biochemistry. 1988;27:1591–1597. doi: 10.1021/bi00405a030. [DOI] [PubMed] [Google Scholar]
- 64.Samples CR, Howard T, Raushel FM, DeRose VJ. Protonation of the binuclear metal center within the active site of phosphotriesterase. Biochemistry. 2005;44:11005–11013. doi: 10.1021/bi0506270. [DOI] [PubMed] [Google Scholar]
- 65.Ely F, Hadler KS, Mitic N, Gahan LR, Ollis DL, Plugis NM, Russo MT, Larrabee JA, Schenk G. Electronic and geometric structures of the organophosphate-degrading enzyme from Agrobacterium radiobacter (OpdA) J. Biol. Inorg. Chem. 2011;16:777–787. doi: 10.1007/s00775-011-0779-6. [DOI] [PubMed] [Google Scholar]
- 66.Ely F, Hadler KS, Gahan LR, Guddat LW, Ollis DL, Schenk F. The organophosphate-degrading enzyme from Agrobacterium radiobacter displays mechanistic flexibility for catalysis. Biochem. J. 2010;432:565–573. doi: 10.1042/BJ20101054. [DOI] [PubMed] [Google Scholar]
- 67.Samples CR, Raushel FM, DeRose VJ. Activation of the binuclear metal center through formation of phosphotriesterase-inhibitor complexes. Biochemistry. 2007;46:3435–3442. doi: 10.1021/bi061951d. [DOI] [PubMed] [Google Scholar]
- 68.Caldwell SR, Raushel FM, Weiss PM, Cleland WW. Transition-state structures for enzymatic and alkaline phosphotriester hydrolysis. Biochemistry. 1991;30:7444–7450. doi: 10.1021/bi00244a011. [DOI] [PubMed] [Google Scholar]
- 69.Chen SL, Fang WH, Himo F. Theoretical study of the phosphotriesterase reaction mechanism. J. Phys. Chem. B. 2007;111:1253–1255. doi: 10.1021/jp068500n. [DOI] [PubMed] [Google Scholar]
- 70.Wong KY, Gao J. The reaction mechanism of paraoxon hydrolysis by phosphotriesterase from combined QM/MM simulations. Biochemistry. 2007;46:13352–13369. doi: 10.1021/bi700460c. [DOI] [PubMed] [Google Scholar]
- 71.Zhang X, Wu R, Song L, Lin Y, Lin M, Cao Z, Wu W, Mo Y. Molecular dynamics simulations of the detoxification of paraoxon catalyzed by phosphotriesterase. J. Comput. Chem. 2009;30:2388–2401. doi: 10.1002/jcc.21238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jao SC, Huang LF, Tao YS, Li WS. Hydrolysis of organophosphate triesters by Escherichia coli aminopeptidase P. J. Mol. Catal. B Enzym. 2004;27:7–12. [Google Scholar]
- 73.Yoshimoto T, Orawski AT, Simmons WH. Substrate specificity of aminopeptidase P from Escherichia coli: Comparison with membrane-bound forms from rat and bovine lung. Arch. Biochem. Biophys. 1994;311:28–34. doi: 10.1006/abbi.1994.1204. [DOI] [PubMed] [Google Scholar]
- 74.Graham SC, Lilley PE, Lee M, Schaeffer PM, Kralicek AV, Dixon NE, Guss JM. Kinetic and crystallographic analyis of mutant Escherichia coli aminopeptidase P: Insites into substrate recognition and mechanism of catalysis. Biochemistry. 2006;45:964–975. doi: 10.1021/bi0518904. [DOI] [PubMed] [Google Scholar]
- 75.Hill CM, Wu F, Cheng TC, DeFrank JJ, Raushel FM. Substrate and stereochemical specificity of the organophosphorus acid anhydrolase from Alteromonas sp. JD6.5 toward p-nitrophenyl phosphotriesters. Bioorg. Med. Chem. Lett. 2000;10:1285–1288. doi: 10.1016/s0960-894x(00)00213-4. [DOI] [PubMed] [Google Scholar]
- 76.Hill CM, Li WS, Cheng TC, DeFrank JJ, Raushel FM. Stereochemical Specificity of organophorus acid anhydrolase toward p-nitrophenyl analogs of soman and sarin. Bioorg. Chem. 2001;29:27–35. doi: 10.1006/bioo.2000.1189. [DOI] [PubMed] [Google Scholar]
- 77.Huang LF, Su B, Jao SC, Liu KT, Li WS. Aminopeptidase P mediated detoxification of organophosphonate analogues of sarin: mechanistic and stereochemical study at the phosphorus atom of the substrate. Chembiochem. 2006;7:506–514. doi: 10.1002/cbic.200500412. [DOI] [PubMed] [Google Scholar]
- 78.Aharoni A, Gaidukov L, Yagur S, Toker L, Silman I, Tawfik DS. Directed evolution of mammalian paraoxonases PON1 and PON3 for bacterial expression and catalytic specialization. Proc. Natl. Acad. Sci. U.S.A. 2004;101:482–487. doi: 10.1073/pnas.2536901100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harel M, Brumshtein B, Meged R, Dvir H, Ravelli RBG, McCarthy A, Toker L, Silman I, Sussman JL. 3-D structure of serum paraoxonase 1 sheds light on its activity, stability, soulubility and crystallizability. Arh Hig. Rada. Toksikol. 2007;58:347–353. doi: 10.2478/v10004-007-0028-0. [DOI] [PubMed] [Google Scholar]
- 80.Kuo CL, La Du BN. Calcium binding by human and rabbit serum paraoxonases. Structural stability and enzymatic activity. Drug Metab. Dispos. 1998;26:653–660. [PubMed] [Google Scholar]
- 81.Hartleib J, Ruterjans H. High-yield expression, purification, and characterization of the recombinant diisopropylfluorophosphatase from Loligo vulgaris. Protein Expr. Purif. 2001;21:210–219. doi: 10.1006/prep.2000.1360. [DOI] [PubMed] [Google Scholar]
- 82.Blum MM, Timperley CM, Williams GR, Thiermann H, Worek F. Inhibitory potency against human acetylcholinesterase and enzymatic hydrolysis of fluorogenic nerve agent mimics by human paraoxonase 1 and squid diisopropyl fluorophosphatase. Biochemistry. 2008;47:5216–5224. doi: 10.1021/bi702222x. [DOI] [PubMed] [Google Scholar]
- 83.Camps J, Marsillach J, Joven J. The paraoxonases: role in human diseases and methodological difficulties in measurement. Crit. Rev. Clin. Lab. Sci. 2009;46:83–106. doi: 10.1080/10408360802610878. [DOI] [PubMed] [Google Scholar]
- 84.Josse D, Xie W, Renault F, Rochu D, Schopfer LM, Masson P, Lockridge O. Identification of residues essential for human paraoxonase (PON1) arylesterase/organophosphatase activities. Biochemistry. 1999;38:2816–2825. doi: 10.1021/bi982281h. [DOI] [PubMed] [Google Scholar]
- 85.Amitai G, Gaidukov L, Adani R, Yishay S, Yacov G, Kushnir M, Teitlboim S, Lindenbaum M, Bel P, Khersonsky O, Tawfik DS, Meshulam H. Enhanced stereoselective hydrolysis of toxic organophosphates by directly evolved variants of mammalian serum paraoxonase. FEBS J. 2006;273:1906–1919. doi: 10.1111/j.1742-4658.2006.05198.x. [DOI] [PubMed] [Google Scholar]
- 86.Gupta RD, Goldsmith M, Ashani Y, Simo Y, Mullokandov G, Bar H, Ben-David M, Leader H, Margalit R, Silman I, Sussman JL, Tawfik DS. Directed evolution of hydrolases for prevention of G-type nerve agent intoxication. Nat. Chem. Biol. 2011;7:120–125. doi: 10.1038/nchembio.510. [DOI] [PubMed] [Google Scholar]
- 87.Valiyaveettil M, Alamneh Y, Biggemann L, Soojhawon I, Doctor BP, Nambiar MP. Efficient hydrolysis of the chemical warfare nerve agent tabun by recombinant and purified human and rabbit serum paraoxonase 1. Biochem. Biophys. Res. Commun. 2010;403:97–102. doi: 10.1016/j.bbrc.2010.10.125. [DOI] [PubMed] [Google Scholar]
- 88.Peterson MW, Fairchild SZ, Otto TC, Mohtashemi M, Cerasoli DM, Chang WE. VX hydrolysis by human serum paraoxonase 1: A comparison of experimental and computational results. PLoS One. 2011;6:e20335. doi: 10.1371/journal.pone.0020335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Davies HG, Richter RJ, Keifer M, Broomfield CA, Sowalla J, Furlong CE. The effect of the human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nat. Genet. 1996;14:334–336. doi: 10.1038/ng1196-334. [DOI] [PubMed] [Google Scholar]
- 90.Melzer M, Chen JCH, Heidenreich A, Gab J, Koller M, Kehe K, Blum MM. Reversed enantioselectivity of diisopropyl fluorophosphatase against organophosphorus nerve agents by rational design. J. Am. Chem. Soc. 2009;131:17226–17232. doi: 10.1021/ja905444g. [DOI] [PubMed] [Google Scholar]
- 91.Yeung DT, Josse D, Nicholson JD, Khanal A, McAndrew CW, Bahnson BJ, Lenz DE, Cerasoli DM. Structure/function analysis of human serum paraoxonase (HuPON1) mutants designed from a DFPase-like homology model. Biochim. Biophys. Acta. 2004;1702:67–77. doi: 10.1016/j.bbapap.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 92.Khersonsky O, Tawfik DS. The histidine 115-histidine 134 dyad mediates the lactonase activity of mammalian serum paraoxonases. J. Biol. Chem. 2006;281:7649–7656. doi: 10.1074/jbc.M512594200. [DOI] [PubMed] [Google Scholar]
- 93.Rosenblat M, Gaidukov L, Khersonsky O, Vaya J, Oren R, Tawfik DS, Aviram M. The catalytic histidine dyad of high density lipoprotein-associated serum paraxonase-1 (PON1) is essential for PON1-mediated inhibition of low density lipoprotein oxidation and stimulation of macrophage cholesterol efflux. J. Biol. Chem. 281:7657–7665. doi: 10.1074/jbc.M512595200. [DOI] [PubMed] [Google Scholar]
- 94.Hartleib J, Ruterjans H. Insites into the reaction mechanism of the diisopropyl fluorophosphatase from Loligo vulgaris by means of kinetic studies, chemical modification and site-directed mutagenesis. Biochim. Biophys. Acta. 2001;1546:312–324. doi: 10.1016/s0167-4838(01)00153-4. [DOI] [PubMed] [Google Scholar]
- 95.Scharff EI, Koepke J, Fritzsch G, Lucke C, Ruterjans H. Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris. Structure. 2001;9:493–502. doi: 10.1016/s0969-2126(01)00610-4. [DOI] [PubMed] [Google Scholar]