Abstract
Performance of two supervised cluster analysis (SVCA) algorithms for extracting reference tissue curves was evaluated to improve quantification of dynamic (R)-[11C]PK11195 brain positron emission tomography (PET) studies. Reference tissues were extracted from images using both a manually defined cerebellum and SVCA algorithms based on either four (SVCA4) or six (SVCA6) kinetic classes. Data from controls, mild cognitive impairment patients, and patients with Alzheimer's disease were analyzed using various kinetic models including plasma input, the simplified reference tissue model (RPM) and RPM with vascular correction (RPMVb). In all subject groups, SVCA-based reference tissue curves showed lower blood volume fractions (Vb) and volume of distributions than those based on cerebellum time-activity curve. Probably resulting from the presence of specific signal from the vessel walls that contains in normal condition a significant concentration of the 18 kDa translocation protein. Best contrast between subject groups was seen using SVCA4-based reference tissues as the result of a lower number of kinetic classes and the prior removal of extracerebral tissues. In addition, incorporation of Vb in RPM improved both parametric images and binding potential contrast between groups. Incorporation of Vb within RPM, together with SVCA4, appears to be the method of choice for analyzing cerebral (R)-[11C]PK11195 neurodegeneration studies.
Keywords: clustering, parametric analysis, (R)-[11C]PK11195, reference tissue
Introduction
(R)-[11C]PK11195 is a widely used positron emission tomography (PET) ligand for the imaging of activated microglia in the brain and provides means to study neuroinflammation in brain disorders such as Alzheimer's disease (AD), schizophrenia, neurotrauma, and normal aging among others (Cagnin et al, 2001; Gerhard et al, 2003; van Berckel et al, 2008; Schuitemaker et al, 2010). Quantification of (R)-[11C]PK11195 uptake is also important for monitoring the effects of antiinflammatory therapy (Dodel et al, 2010).
The gold standard for analysis of dynamic (R)-[11C]PK11195 studies is the two tissue reversible plasma input model (2T4k) (Gunn et al, 2001). The 2T4k model requires accurate arterial blood sampling during the scan, which is invasive, complicates logistics, and is prone to errors. Reference tissue models, in contrast, do not require arterial blood sampling and are therefore better suited for routine clinical studies. However, a reference tissue model needs to be validated before routine clinical use.
In a previous study, Kropholler et al (2005) evaluated both the plasma input model (2T4k) and the simplified reference tissue model (SRTM; Lammertsma and Hume, 1996) for analysis of dynamic (R)-[11C]PK11195 studies. The binding potentials (BPND) estimated using both models showed good correlation. Nevertheless, SRTM-derived BPND can be biased due to (specific) binding in the reference region; for example, the cerebellum, a typical reference region of choice, is surrounded by venous sinuses that have significant concentration of TSPO in endothelial tissue and smooth muscle (Turkheimer et al, 2007). Besides, the pattern of microglial activation is generally unknown and the choice of an anatomical region as reference may be challenging without appropriate pathological information. Therefore, automatic approaches to extract reference tissue kinetics of (R)-[11C]PK11195 would be appealing as long as sensitive and robust.
Turkheimer et al (2007) applied a new clustering method for extracting reference tissue curves, called supervised cluster analysis (SVCA6). The supervised clustering algorithm segments PET voxels based on differences in time-activity curves (TACs). The method assumes that each TAC is a linear sum of six predefined kinetic classes associated with gray matter with specific binding, gray matter without specific binding, white matter, blood, bone, and soft tissue regions. This algorithm selects reference tissue voxels primarily from gray-matter tissue without specific binding avoiding binding in blood vessels. It was shown that the use of SVCA6 extracted reference tissues curves improved the agreement between reference tissue and plasma input models (Turkheimer et al, 2007).
Boellaard et al (2008) modified SVCA6 by removing bone and soft tissue regions from the PET scan before SVCA analysis. The modified method (SVCA4) only requires four kinetic classes; however, it also requires a brain mask which usually can be obtained from a magnetic resonance image (MRI) of the subject. This improved precision, because of the reduction in fit parameters and the elimination of noise and errors from extracerebral signal coming from bone and soft tissue regions. The SVCA4 provided more accurate reference tissues than cerebellum and SVCA6. However, it has also been recently shown that variations in vascular binding, typical for example of aging and neurodegenerative disorders, may contribute significantly to the estimation of BPND. Tomasi et al (2008) proposed a modified SRTM method that corrects for vascular signal in the target region based on an image-derived blood TAC. This method uses a basis function implementation of SRTM (i.e., RPM; Gunn et al, 1997) with blood volume fraction (Vb) correction (RPMVb). They reported that the use of RPMVb together with SVCA6 resulted in better differentiation of AD from controls. Additionally, a group difference in estimated Vb between healthy controls and AD patients was found, which is in line with known pathological changes in AD. The purpose of the present study was to compare different methods for extracting reference tissue for kinetic analysis of (R)-[11C]PK11195 studies. To this end, two assessments were performed:
The extracted reference tissue curves were analyzed using the plasma input model with additional parameter for blood volume fraction (2T4k) for the assessment of volume of distribution (VT) and blood volume fraction (Vb).
The extracted reference tissue curves were used as input in various reference tissue algorithms for the assessment of the specific binding (BPND) in the target region (=thalamus).
The second aim was to replicate the result of Tomasi et al (2008), in a new cohort of AD patients as further validation of the RPMVb modeling approach.
Reference TACs were extracted using SVCA4, SVCA6, and manual definition of cerebellum.
Kinetic analysis of the target region was performed using 2T4k, SRTM, RPM, and RPMVb. In all assessments, data were used from young controls (YC), old controls (OC), patients with mild cognitive impairment (MCI), and patients with AD.
Materials and methods
Scanning Protocol
Clinical data were derived from patient studies, approved by the Medical Ethics Review Committee of VU University Medical Center, and consisted of 34 subjects (9 YC, 8 OC, 9 MCI, and 8 AD). The age range was 19 to 56 (average 27±11), 64 to 78 (average 72±5), 64 to 79 (average 72±6), and 61 to 81 (average 72±6) for YC, OC, MCI, and AD, respectively. Each subject gave written informed consent before inclusion in the study.
After bolus injection of 370 MBq (R)-[11C]PK11195, a dynamic emission scan was acquired in 3D mode using an ECAT EXACT HR+ (CTI/Siemens, Knoxville, TN, USA). In addition, a 10-minute 2D transmission scan was acquired to correct for tissue attenuation. The dynamic emission scan consisted of 22 frames with a total scan duration of 60.5 minutes (1 × 30, 1 × 15, 1 × 5, 1 × 10, 2 × 15, 2 × 30, 3 × 60, 4 × 150, 5 × 300, and 2 × 600 seconds). The first frame has a relatively large duration because it is used as a background frame to verify running of the scanner and blood sampling device. Next, the tracer is injected during the first 10 seconds of the second frame. The later frames have shorter frame durations to be able to accurately obtain the shape of the TACs during the initial uptake phase. Frames were reconstructed using FORE (Defrise et al, 1997) + 2D Filtered Back Projection with a Hanning filter at a cutoff of 0.5 times the Nyquist frequency. A matrix size of 256 × 256 and a zoom factor of two were applied during reconstruction, resulting in an image pixel size of 1.2 × 1.2 mm and a slice thickness of 2.5 mm. The reconstruction settings resulted in an image resolution of 7 mm full width at half maximum. Reconstructions included all usual corrections, such as detector normalization, and decay, dead time, attenuation, randoms, and scatter corrections.
The arterial input function was measured using an online continuous blood sampling device (Boellaard et al, 2001). At discrete times (∼3, 6, 10, 20, 30, 40, and 60 minutes after injection), additional manual samples were obtained for online calibration of the measured whole blood input function, determination of plasma/whole blood ratios, and measurement of metabolite fractions.
The protocol included a T1-weighted MRI scan (1T IMPACT, Siemens Medical Solutions, Erlangen, Germany) to facilitate the definition of anatomical regions of interest (ROIs).
Regions of Interest Definitions
First, the T1-weighted MRI scan was coregistered (West et al, 1997; Maes et al, 1997) with the dynamic (R)-[11C]PK11195 scan summed over 1 to 8 minutes, where the latter summed image approximates a flow image with a fair contrast between gray and white matter. Next, the coregistered T1-weighted MRI was segmented into gray matter, white matter, and cerebrospinal fluid (Zhang et al, 2001). Next, anatomical ROIs for thalamus and cerebellum were drawn manually on the coregistered T1-weighted MRI. Finally, these ROIs were projected onto reconstructed dynamic (R)-[11C]PK11195 images to extract regional TACs.
The thalamus was selected as target region because it is known to have a relatively high level of specific binding of (R)-[11C]PK11195 in AD (Schuitemaker et al, 2007). The cerebellum ROI consisted of both gray and white matter (Schuitemaker et al, 2007) and was used to extract a reference tissue TAC for comparison with reference tissue TACs extracted with the supervised cluster algorithms.
Supervised Cluster Analysis
Two SVCA algorithms were used to automatically extract reference tissue TACs from dynamic PET scans. Predefined kinetic classes were available from a previous study using data from seven healthy controls and seven patients with traumatic brain injury (Boellaard et al, 2008). Details concerning this procedure and cluster analysis were reported in Turkheimer et al (2007); however, a summary of cluster analysis is as follows.
The original method (SVCA6) uses six kinetic classes normal gray matter, normal white matter, blood, bone, and soft tissue regions, and gray matter with specific binding. The first five classes were defined on a separate set of normal controls while the last one, corresponding to gray matter with high microglia density, was obtained from the brain injury patients.
To extract the reference, the dynamic PET scan is first normalized as described by Turkheimer et al (2007): each voxel value is reduced by the frame average and divided by the standard deviation. Therefore, the normalization is affected by the size of the reconstructed field of view; for this study, both definition of kinetic classes and application cluster analysis were preformed on scans acquired from the same scanner using similar scanning protocol. However, in case of using SVCA4, normalization was done on voxels that correspond with brain tissue only (based on MRI-derived coregistered gray- and white-matter segmentations). Thereby, this method also avoids the effects of differences in field of view between different scanners.
Next, each voxel TAC of this scan is analyzed using the set of predefined kinetic classes to find the scaling coefficient of each kinetic class, so that the total TAC is equal to the sum of these scaled kinetic classes. As the kinetic classes are not orthogonal, a nonnegative least squares algorithm (Turkheimer et al, 2007) is used for finding the scaling coefficients. Scaling coefficients of each kinetic class are stored in coefficient maps showing their spatial distribution.
Finally, to extract the reference tissue curve, the coefficient map from the (normal) gray-matter kinetic class is used to calculate the weighted average, as follows:
 |
where, N is the number of voxels, TACNS(t) the resulting reference tissue TAC, TACiVoxel(t) the TAC from voxel i of the (nonnormalized) dynamic PET scan, and wiGray the gray-matter kinetic class scaling coefficient estimated for voxel i.
The modified supervised cluster analysis method (SVCA4) (Boellaard et al, 2008) is similar to SVCA6, except that only four kinetic classes are used: gray matter with specific (R)-[11C]PK11195 binding, gray matter without specific binding, white matter, and blood. This modified method uses the mentioned coregistered segmented MRI scans to exclude skull and soft tissue parts from each frame of the PET scan before performing cluster analysis, same as mentioned above but now with only four kinetic classes. Removal of skull and soft tissue was simply done by setting voxel values to zero for nonbrain structures.
Kinetic Analysis of Clinical Data
Each of extracted reference tissue TACs was analyzed using a two tissue reversible plasma input model with additional parameter for blood volume fraction (2T4k) (Gunn et al, 2001) for the assessment of volume of distribution (VT) and blood volume fraction (Vb). As mentioned before, reference tissue TACs were based on manual definition of cerebellum or extracted using cluster analysis algorithms.
Next, extracted reference tissue curves were used as input in four different reference tissue algorithms for the assessment of the specific binding (BPND) in the target region (e.g., thalamus). The reference tissue analysis was performed using 2T4k, SRTM (Lammertsma and Hume 1996), the basis function approach of SRTM (RPM) (Gunn et al, 1997), and the basis function approach of SRTM with additional correction for blood volume fraction Vb (RPMVb) (Tomasi et al, 2008). Note that in this step the 2T4k was used to calculate the BPND indirectly, i.e., reference tissue based, BPND was calculated using the thalamus to reference tissue volume of distribution ratio DVR (=VT_thalamus/VT_reference_tissue=DVR=BPND+1).
The SRTM, RPM, and RPMVb were mainly used to calculate the BPND. The RPMVb requires an image-derived blood curve to correct for Vb in the target TAC. The fractional blood volume for the reference input TAC was fixed to 5% in RPMVb that equals to the original implementation by Tomasi et al (2008). The image-derived blood curve was defined as the average TAC from 10 voxels showing the highest uptake during the initial period (<1 minutes) (Tomasi et al, 2008). Optimization of image quality and quantification required empirical assessment of the basis function parameters/range in case of using RPM and RPMVb. The RPM was run using number of basis functions of 30, θmin of 0.04/minute and θmax of 1.0/minute (Gunn et al, 1997). The RPMVb was run using number of basis functions of 30, θmin of 0.01/minute, and θmax of 0.3/minute.
Both RPM and RPMVb methods are faster and more robust against high noise levels than nonlinear methods (i.e., SRTM and 2T4k); therefore, RPM and RPMVb were used to calculate volumetric images showing the binding potential for each image voxel (=parametric BPND images). Next, the manually defined ROI on segmented MRI was projected onto parametric BPND images to calculate regionally averaged BPND.
Results
Extraction of Reference Tissues
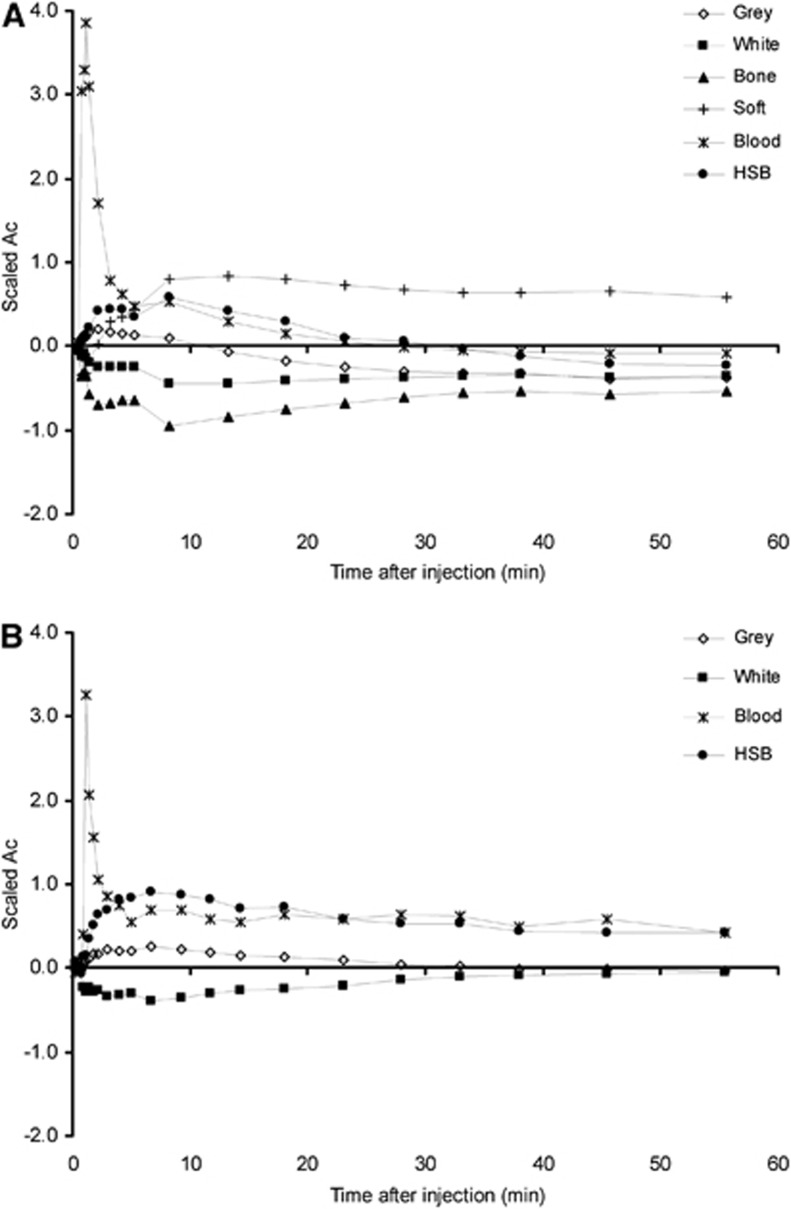
Kinetic classes used in SVCA6 and SVCA4 are shown in Figure 1. The coefficient maps for each kinetic class showed expected distributions (data not shown): the blood pool was clearly visible and healthy gray-matter maps (used for extraction of reference tissue curve) are well identified. Coefficient maps representing high specific binding showed high uptake in the thalamus region and also uptake (unexpectedly) in a region just outside the brain.
Figure 1.
Typical time-activity curves (TACs) for the kinetic classes used in SVCA6 (A) and SVCA4 (B). TACs were scaled by subtracting from each time point the frame average and dividing by the frame standard deviation. SVCA4 uses only four kinetic classes and the curves are different because scaling is performed after brain extraction. HSB stands for high specific binding (i.e., binding to activated microglia). SVCA, supervised cluster analysis.
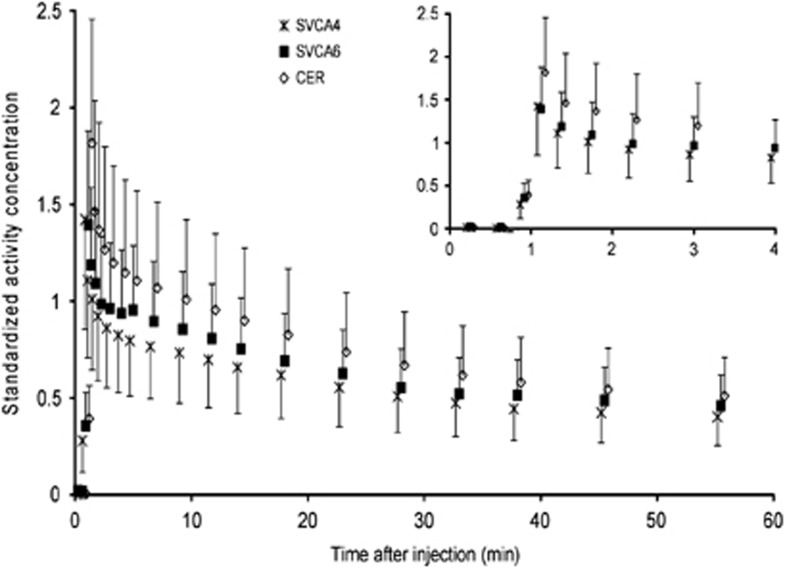
Figure 2 shows population average reference tissue TACs for all eight AD subjects. In general, the highest uptake was seen in the cerebellum TAC. The SVCA4 and SVCA6 TACs were slightly different in shape, and SVCA6 showed overall higher uptakes than SVCA4. Similar uptake differences between the reference tissue curves were seen for the other clinical subject groups (YC, OC, and MCI, not shown here).
Figure 2.
Average standardized reference tissue time-activity curves (TACs) over all eight AD subjects. Curves represent manually drawn gray+white-matter cerebellum (CER), and those extracted automatically using SVCA4 and SVCA6. The insert highlights the first 4 minutes. TACs were normalized for injected dose and patient weight before calculating averages (symbols) and one standard deviation (error bars).
Analysis of the Reference Tissue Curves Using Plasma Input
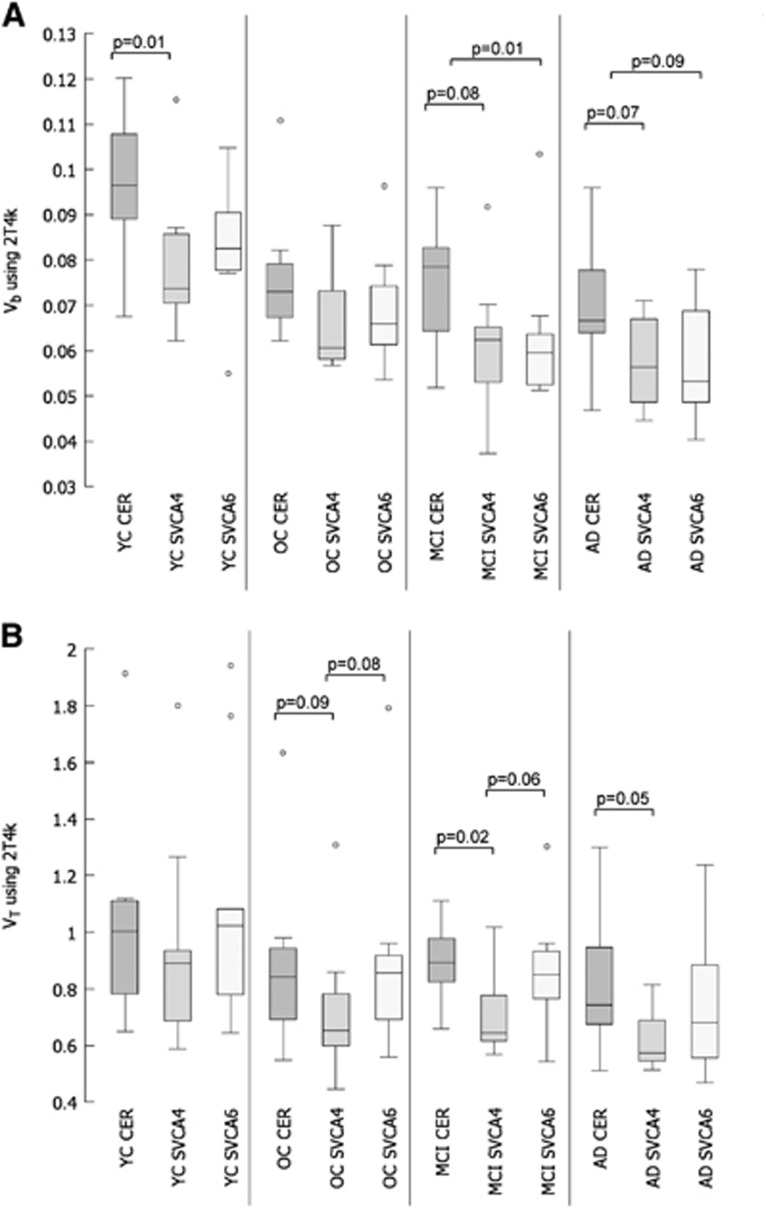
Reference tissue Vb values obtained using plasma input analysis (2T4k) are shown in Figure 3A. The number of outliers shown in the figure (defined as interquartile range × 1.5 outside first and third quartiles) seems to be related to a clinical subject group rather than to the reference tissue extraction method, i.e., largest number of outliers was seen for YC probably due to a lower specific signal in this group. Note that all T-test analyses were performed after removal of the shown outliers in the figures using a significance level of P=0.05. Compared with cerebellum, there are indications for lower Vb for both the SVCA4- and SVCA6-derived reference tissues (see two-tailed significance P in the figure). However, the differences between the clinical subject groups were similar for all methods, i.e., for all methods group differences in Vb were only significant between YC versus OC, YC versus MCI, and YC versus AD (T-test data not shown).
Figure 3.
Comparison of reference tissue Vb (A) and VT (B) using box and whisker plots, including two-tailed T-test analysis for comparison between methods. Reference tissue time-activity curves (TACs) were based on manually drawn gray+white-matter cerebellum (CER), and those extracted automatically using SVCA4 and SVCA6. Data were taken from young controls (YC), old controls (OC), and patients with mild cognitive impairment (MCI) and Alzheimer's disease (AD). Note that all two-tailed T-tests were performed after removing the outliers (marked with open circles) shown in the figure.
Reference tissue VT values obtained using plasma input analysis (2T4k) are shown in Figure 3B. Again, the number of outliers shown in the figure seems to be related to a clinical subject group rather than to the reference tissue extraction method. There are indications for lower VT for SVCA4-derived reference tissue compared with VT for cerebellum-based reference tissue. Furthermore, only SVCA4 showed a significantly different VT between YC versus MCI, and YC versus AD, all other group comparisons for each method were not significant (T-test data not shown).
Estimation of Specific Binding in Thalamus Using Plasma Input
VT values of the target region (=thalamus) estimated using plasma input analysis did not show any significant differences among the groups (t-test data not shown). Significant differences in VT between target and reference tissue input were found for OC, MCI, and AD, however, only in case of using SVCA4.
Figure 4 shows box plots of thalamus BPND (derived from DVR-1) obtained using plasma input analysis (2T4k) of both the reference input and thalamus TAC, in this case, T-test significance levels are given for comparison between groups for each method. Overall, use of SVCA4 generated higher specific binding in thalamus than the other reference tissue extraction methods. Differences between cerebellum input and SVCA4 were significant for most clinical subject groups (data not shown). Also, the largest variability was seen for SVCA4 values in MCI and AD. Nevertheless, thalamus BPND estimates using SVCA methods were in most cases significantly higher for subjects with increasing pathology (Figure 4).
Figure 4.
Comparison of thalamus-specific binding potential (BPND) using box and whisker plots, including two-tailed T-test analysis for comparison between groups. BPND (=DVR-1) was calculated using thalamus to reference tissue VT ratio's.
Estimation of Specific Binding in Thalamus without Plasma Input
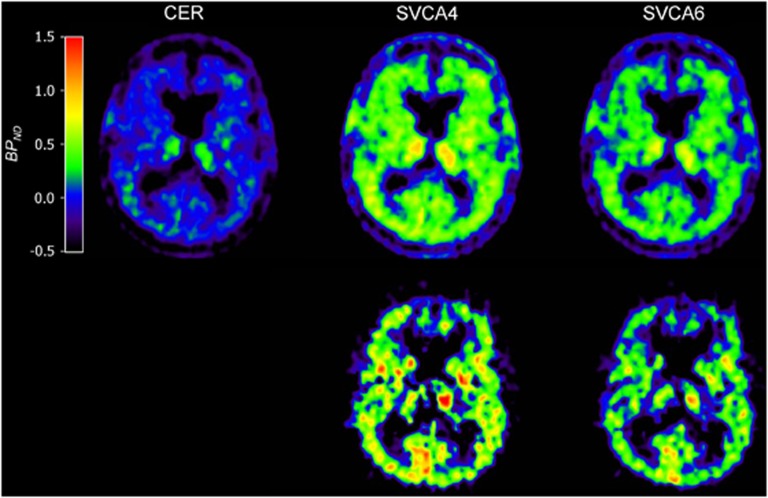
In Figure 5, several typical parametric BPND images (RPM and RPMVb) are shown. In general, use of both SVCA methods showed large contrast between gray- and white-matter areas compared with the use of the cerebellum as reference input. In addition, use of RPMVb showed higher contrast in the brain tissues and less (R)-[11C]PK11195 binding in extracerebral tissues (in or near the skull) than when using RPM.
Figure 5.
Parametric binding potential (BPND) images generated using reference tissue model (RPM) (top row) and RPM with vascular correction (RPMVb) (bottom row). Reference tissue inputs were taken from cerebellum (CER), SVCA4, and SVCA6. Note the higher contrast when using RPMVb (bottom row). Data were taken from an AD subject.
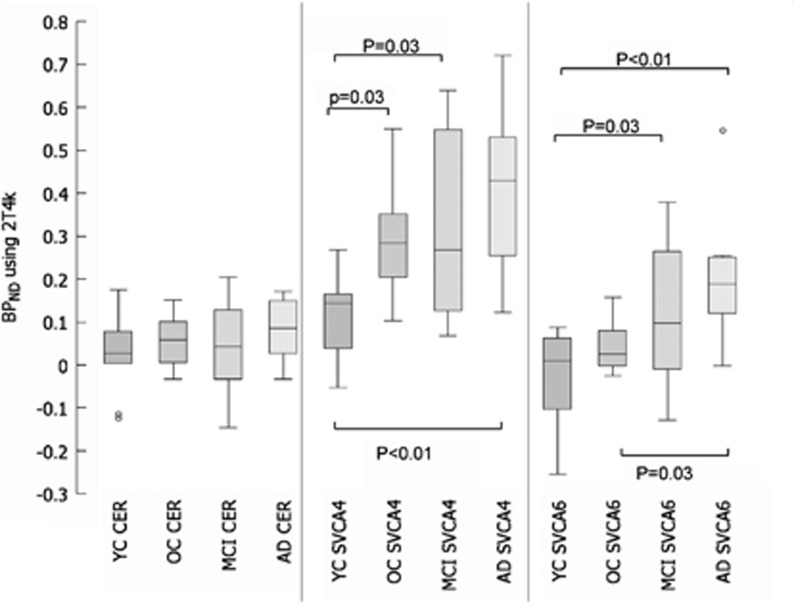
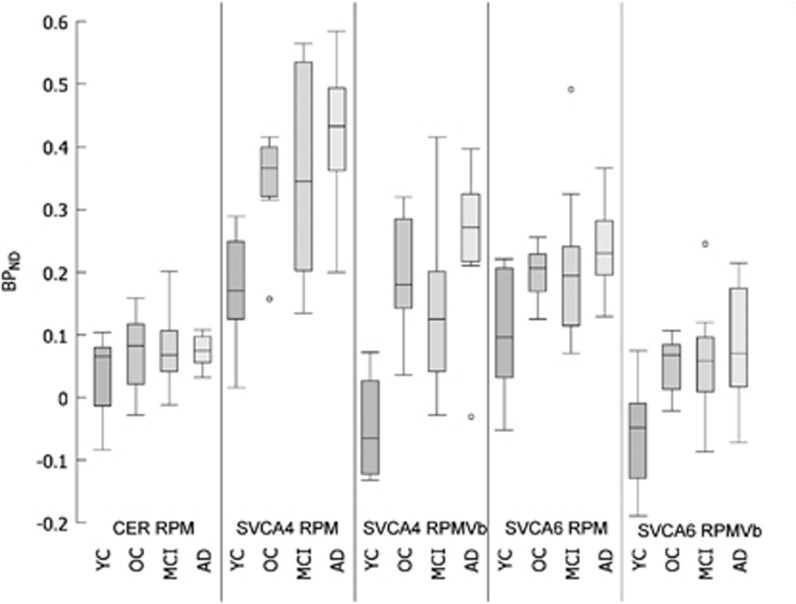
Regional thalamus BPND values obtained using SRTM were almost identical to corresponding RPM results (data not shown). Therefore, SRTM results are not shown. Figure 6 shows box plots for thalamus BPND estimated using either RPM or RPMVb in combination with cerebellum, SVCA4, or SVCA6 reference tissue curves. Table 1 shows multiple two-tailed t-test analyses performed on these data (after removing outliers shown in the figure) to investigate the differences between each method (Table 1) and for each method the differences between clinical subject groups (Table 1). Again, BPND based on cerebellum reference tissue was not significantly different between subject groups (Figure 6; Table 1), whereas SVCA-based BPND showed more contrast (Figure 6; Table 1). By comparing methods, it was observed that there are larger differences between cerebellum and SVCA4 input than between cerebellum and SVCA6 methods (Table 1). Furthermore, results are significantly changed when applying Vb correction in SVCA methods. Finally, only use of RPMVb with SVCA4-based reference tissue input showed significant differences between all groups (Table 1) except OC versus MCI.
Figure 6.
Comparison of thalamus-specific binding (BPND) using box and whisker plots.
Table 1. BPND estimates were compared using two-tailed t-test: (a) for each reference tissue method are given the significance levels for testing differences in the clinical subject groups; (b) for each clinical subject group are given the significance levels for testing differences in the reference tissue methods.
| Test | RPM cerebellum | RPM SVCA4 | RPMVb SVCA4 | RPM SVCA6 | RPMVb SVCA6 |
|---|---|---|---|---|---|
| (a) | |||||
| YC versus OC | 0.266 | 0.000 | 0.000 | 0.025 | 0.004 |
| YC versus MCI | 0.167 | 0.019 | 0.004 | 0.142 | 0.017 |
| YC versus AD | 0.137 | 0.000 | 0.000 | 0.007 | 0.005 |
| OC versus MCI | 0.779 | 0.690 | 0.452 | 0.519 | 0.581 |
| OC versus AD | 0.911 | 0.295 | 0.050 | 0.209 | 0.435 |
| MCI versus AD | 0.809 | 0.299 | 0.039 | 0.144 | 0.286 |
| Test | YC | OC | MCI | AD |
|---|---|---|---|---|
| (b) | ||||
| RPM CER versus RPM SVCA4 | 0.002 | 0.000 | 0.001 | 0.000 |
| RPM CER versus RPMVb SVCA4 | 0.030 | 0.008 | 0.241 | 0.000 |
| RPM CER versus RPM SVCA6 | 0.078 | 0.000 | 0.021 | 0.000 |
| RPM CER versus RPMVb SVCA6 | 0.013 | 0.499 | 0.195 | 0.803 |
| RPM SVCA4 versus RPMVb SVCA4 | 0.000 | 0.001 | 0.022 | 0.021 |
| RPM SVCA4 versus RPM SVCA6 | 0.153 | 0.000 | 0.026 | 0.002 |
| RPM SVCA6 versus RPMVb SVCA6 | 0.001 | 0.000 | 0.003 | 0.003 |
| RPMVb SVCA4 versus RPMVb SVCA6 | 0.628 | 0.002 | 0.075 | 0.000 |
BPND, binding potential; CER, cerebellum; RPM, reference tissue model; RPMVb, RPM with vascular correction; SVCA, supervised cluster analysis.
Data were taken from young controls (YC), old controls (OC), and patients with mild cognitive impairment (MCI) and Alzheimer's disease (AD). Significant P values are indicated with bold fonts.
Discussion
Reference Tissue
During the extraction of reference tissues the coefficient maps for each kinetic class showed expected distributions (data not shown). However, coefficient maps representing high specific binding also showed some uptake (unexpectedly) in a region just outside the brain. This does not affect the reference tissue extraction process because the coefficient maps for the (healthy) gray matter (that was used to extract the reference input curves) were well identified without high probability values outside the brain.
Reference tissue TACs extracted from cerebellum and the two cluster analysis algorithms did not show similar uptake (Figure 2). Similar results were found for different clinical subject groups. Reference tissue curves extracted using SVCA4 always showed lower uptake than the two other methods (Figure 2). Increased uptake at the peak of the cerebellum TAC hints toward contributions from the vascular component; at first, the vascular contribution will reflect radiotracer in blood and the increased uptake at the latter part of the TAC is likely to reflect binding in endothelium and smooth muscles (Figure 2).
The fractional blood volume of the reference tissue curves was estimated using the 2T4k model with correction for the blood volume fraction. In general, for all reference tissue extraction methods, analysis of the reference tissue curves showed a decreasing Vb with increasing probability of pathology in the brain (Figure 3A), the exact reason for this decrease is unknown. In theory, both SVCA algorithms should produce reference tissue TACs that are free of blood (i.e., Vb=0), as they have a separate kinetic class for blood. For both methods, however, reference tissue Vb was only slightly lower than that for the manually defined cerebellum TAC (Figure 3A). This indicates that cluster analysis is only partly capable of correcting for the intravascular contribution in the reference tissue. There can be several reasons for this observation. First, ROIs defined to obtain the gray-matter kinetic class may also contain intravascular activity and second, partial volume effects due to the low resolution of PET images may compromise reference tissue TACs. Third, the SVCA used only one kinetic class to describe the tracer kinetics in blood. However, a significant arterial-venous gradient of the activity concentration of (R)-[11C]PK11195 due to its high extraction exists; therefore, the vasculature may not be completely captured by that single blood class.
The volume of distribution, VT, was estimated using the 2T4k model with correction for the blood volume fraction. The VT should give an estimate of uptake of the tracer in tissue. For all reference tissue extraction methods, analysis of the reference tissue curves themselves using the two tissue reversible plasma input model showed a decreasing VT with increasing probability of pathology in the brain (Figure 3B). In all subject groups, SVCA4 showed the lowest VT values. Furthermore, only SVCA4 showed a significantly different VT between YC versus MCI, and YC versus AD. All other group comparisons did not show significant differences in VT. The lower VT with SVCA4 resulted possibly from less binding in the extracted reference tissue or binding in blood vessel walls.
Specific Binding
VT values of the target region (=thalamus) estimated using plasma input analysis did not show any significant differences among the groups. However, VT is not only reflecting specific binding, but also includes a large (and intersubject variable) contribution of nonspecific binding. Therefore, VT is not very sensitive to pick up differences in specific binding. Significant differences in VT between target and reference tissue input were found for OC, MCI, and AD, however, only in case of using SVCA4. The latter differences are as expected because the VT target to VT reference tissue ratio (plasma input-based DVR=BPND+1) is an estimate of specific binding and therefore a more suitable parameter to evaluate tracer binding.
The specific binding (BPND) was estimated using reference tissue methods with or without plasma input. The results of BPND using different reference tissue TACs were analyzed for accuracy. Although accuracy is difficult to estimate for clinical data because the underlying true parameters are not known, both box plots and (expected) differences between subject groups can be used as an index of performance. Therefore, here the focus was on evaluating tracer binding in the thalamus because this area was found to show differences in binding in several other studies (Banati et al, 2000; Schuitemaker et al, 2010).
Although the plasma input-based specific binding is also corrected for the differences in Vb between the target and the reference region, still similar outcomes (Figure 4) were seen in the nonplasma input-based reference tissue methods (Figure 6). In general, one could conclude that SVCA4 showed higher binding in all subject groups than all other methods (Figures 4 and 6). However, expected differences between subject groups were only significant when using SVCA method (Table 1). Furthermore, parametric images based on SVCA reference tissue input functions showed higher contrast and appeared less noisy (Figure 5). Clearly, cerebellum-based reference tissue input curves were less suitable than those of SVCA when using a reference tissue model for analyzing (R)-[11C]PK11195 data.
Finally, incorporation of Vb in the model improved both gray- and white-matter contrast in parametric images (Figure 5) and BPND contrast between groups (Table 1). A possible explanation of this observation could be that (R)-[11C]PK11195 binds specifically in the blood vessel walls, which then needs to be corrected for by the model (Tomasi et al, 2008). The RPMVb algorithm effectively corrects for this effect in both reference tissue and target region without the need of arterial sampling.
Possible Limitations of Cluster Analysis
Cluster analysis requires a set of predefined kinetic classes, which are generated from manually defined ROIs on selected (R)-[11C]PK11195 scans (Turkheimer et al, 2007; Boellaard et al, 2008; Hinz et al, 2008). Next, dynamic (R)-[11C]PK11195 PET scans are analyzed using this set of predefined kinetic classes. After normalization of the kinetic classes, only relative shape differences between the kinetic classes are important (Figure 1). The shape of the various kinetic classes depends on both scanning protocol (e.g., speed of bolus injection) and characteristics of the PET scanner and image reconstruction (Boellaard et al, 2008). As a consequence, kinetic classes may have to be derived at, or validated for, each site, at least per type of PET scanner or when using different imaging procedures. This may limit the use of SVCA. However, recently another study applied a similar approach with predefined kinetic classes to enhance the visualization of tumors imaged with [18F]FLT (Gray et al, 2010) showing the feasibility of using comparable supervised cluster methods for other applications. Yet, further research is needed to explore the applicability of these approaches to other tracers and scanners.
In our study, the used kinetic classes were redefined for our setting (scanner and imaging procedure) during a previous study (Boellaard et al, 2008), using dynamic (R)-[11C]PK11195 brain PET studies from seven young healthy controls and seven patients with traumatic brain injury. The latter studies were only used to obtain the kinetic class representing high specific binding.
The SVCA4 needs additional preprocessed PET data for which the skull and soft tissues outside the brain are automatically removed using a coregistered T1-weighted MRI scan. Use of smaller number of kinetic classes (SVCA4 versus SVCA6) could result in reduced sensitivity for noise because the clustering algorithm requires fewer fit parameters (Boellaard et al, 2008). However, accurate coregistration and gray–white matter versus cerebrospinal fluid segmentation are required to correctly remove nonbrain tissues from the PET study. Small misalignments could result in incorrect removal of brain tissue or residual nonbrain voxels in the processed PET images. Therefore, in our study all preprocessed PET data were first inspected visually.
Conclusion
The SVCA4 provided reference tissue input curves with lower volumes of distribution and blood volume fractions than those obtained using a cerebellum ROI or SVCA6. Moreover, the use of cerebellum as reference region was not able to show differences in specific binding between subject groups, while use of SVCA (4 or 6) did show group differences in specific binding. Based on the ability to show differences in binding between subject groups as well as showing lowest reference VT and Vb, SVCA4 in combination with RPMVb might be the method of choice for analyzing dynamic (R)-[11C]PK11195 brain PET studies.
The authors declare no conflict of interest.
Footnotes
The research leading to these results has received funding from the European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement n° HEALTH-F2-2011-278850 (INMiND), The Netherlands Organisation of Scientific Research (VIDI grant 016.066.309), the Dutch Brain Foundation (grant 9F01.21) and Federico E. Turkheimer was supported by PET Methodology Programme Grant (Medical Research Council UK).
References
- Banati RB, Newcombe J, Gunn RN, Cagnin A, Turkheimer F, Heppner F, Price G, Wegner F, Giovannoni G, Miller DH, Perkin GD, Smith T, Hewson AK, Bydder G, Kreutzberg GW, Jones T, Cuzner ML, Myers R. The peripheral benzodiazepine binding site in the brain in multiple sclerosis: quantitative in vivo imaging of microglia as a measure of disease activity. Brain. 2000;123 (Pt 11:2321–2337. doi: 10.1093/brain/123.11.2321. [DOI] [PubMed] [Google Scholar]
- Boellaard R, Turkheimer FE, Hinz R, Schuitemaker A, Scheltens P, van Berckel BNM, Lammertsma AA.2008. Performance of a modified supervised cluster algorithm for extracting reference region input functions from (R)-[11C]PK11195 brain PET studies. Nuclear Science Symposium Conference Record, 2008. NSS ‘08. IEEE, 2008, pp 5400–5402. doi 10.1109/NSSMIC.2008.4774453 [DOI]
- Boellaard R, van Lingen A, van Balen SCM, Hoving BG, Lammertsma AA. Characteristics of a new fully programmable blood sampling device for monitoring blood radioactivity during PET. Eur J Nucl Med. 2001;28:81–89. doi: 10.1007/s002590000405. [DOI] [PubMed] [Google Scholar]
- Cagnin A, Brooks DJ, Kennedy AM, Gunn RN, Myers R, Turkheimer FE, Jones T, Banati RB. In-vivo measurement of activated microglia in dementia. Lancet. 2001;358:461–467. doi: 10.1016/S0140-6736(01)05625-2. [DOI] [PubMed] [Google Scholar]
- Defrise M, Kinahan PE, Townsend DW, Michel C, Sibomana M, Newport DF. Exact and approximate rebinning algorithms for 3-D PET data. IEEE Trans Med Imaging. 1997;16:145–158. doi: 10.1109/42.563660. [DOI] [PubMed] [Google Scholar]
- Dodel R, Spottke A, Gerhard A, Reuss A, Reinecker S, Schimke N, Trenkwalder C, Sixel-Doring F, Herting B, Kamm C, Gasser T, Sawires M, Geser F, Kollensperger M, Seppi K, Kloss M, Krause M, Daniels C, Deuschl G, Bottger S, Naumann M, Lipp A, Gruber D, Kupsch A, Du Y, Turkheimer F, Brooks DJ, Klockgether T, Poewe W, Wenning G, Schade-Brittinger C, Oertel WH, Eggert K. Minocycline 1-year therapy in multiple-system-atrophy: effect on clinical symptoms and [(11)C] (R)-PK11195 PET (MEMSA-trial) Mov Disord. 2010;25:97–107. doi: 10.1002/mds.22732. [DOI] [PubMed] [Google Scholar]
- Gerhard A, Banati RB, Goerres GB, Cagnin A, Myers R, Gunn RN, Turkheimer F, Good CD, Mathias CJ, Quinn N, Schwarz J, Brooks DJ. [11C](R)-PK11195 PET imaging of microglial activation in multiple system atrophy. Neurology. 2003;61:686–689. doi: 10.1212/01.wnl.0000078192.95645.e6. [DOI] [PubMed] [Google Scholar]
- Gray KR, Contractor KB, Kenny LM, Al-Nahhas A, Shousha S, Stebbing J, Wasan HS, Coombes RC, Aboagye EO, Turkheimer FE, Rosso L. Kinetic filtering of [(18)F]Fluorothymidine in positron emission tomography studies. Phys Med Biol. 2010;55:695–709. doi: 10.1088/0031-9155/55/3/010. [DOI] [PubMed] [Google Scholar]
- Gunn RN, Gunn SR, Cunningham VJ. Positron emission tomography compartmental models. J Cereb Blood Flow Metab. 2001;21:635–652. doi: 10.1097/00004647-200106000-00002. [DOI] [PubMed] [Google Scholar]
- Gunn RN, Lammertsma AA, Hume SP, Cunningham VJ. Parametric imaging of ligand-receptor binding in PET using a simplified reference region model. Neuroimage. 1997;6:279–287. doi: 10.1006/nimg.1997.0303. [DOI] [PubMed] [Google Scholar]
- Hinz R, Jones M, Bloomfield PM, Boellaard R, Turkheimer FE, Tyrrell PJ. Reference tissue kinetics extraction from [11C]-(R)-PK11195 scans on the High Resolution Research Tomograph (HRRT) [abstract] Neuroimage. 2008;41 (Suppl 2:T65. [Google Scholar]
- Kropholler MA, Boellaard R, Schuitemaker A, van Berckel BN, Luurtsema G, Windhorst AD, Lammertsma AA. Development of a tracer kinetic plasma input model for (R)-[11C]PK11195 brain studies. J Cereb Blood Flow Metab. 2005;25:842–851. doi: 10.1038/sj.jcbfm.9600092. [DOI] [PubMed] [Google Scholar]
- Lammertsma AA, Hume SP. Simplified reference tissue model for PET receptor studies. Neuroimage. 1996;4:153–158. doi: 10.1006/nimg.1996.0066. [DOI] [PubMed] [Google Scholar]
- Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE Trans Med Imaging. 1997;16:187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- Schuitemaker A, van Berckel BNM, Kropholler MA, Veltman DJ, Scheltens P, Jonker C, Lammertsma AA, Boellaard R. SPM analysis of parametric (R)-[C-11]PK11195 binding images: plasma input versus reference tissue parametric methods. Neuroimage. 2007;35:1473–1479. doi: 10.1016/j.neuroimage.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Schuitemaker A, van der Doef TF, Boellaard R, van der Flier WM, Yaqub M, Windhorst AD, Barkhof F, Jonker C, Kloet RW, Lammertsma AA, Scheltens P, van Berckel BN. Microglial activation in healthy aging. Neurobiol Aging. 2010;5:6–9. doi: 10.1016/j.neurobiolaging.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Tomasi G, Edison P, Bertoldo A, Roncaroli F, Singh P, Gerhard A, Cobelli C, Brooks DJ, Turkheimer FE. Novel reference region model reveals increased microglial and reduced vascular binding of 11C-(R)-PK11195 in patients with Alzheimer′s disease. J Nucl Med. 2008;49:1249–1256. doi: 10.2967/jnumed.108.050583. [DOI] [PubMed] [Google Scholar]
- Turkheimer FE, Edison P, Pavese N, Roncaroli F, Anderson AN, Hammers A, Gerhard A, Hinz R, Tai YF, Brooks DJ. Reference and target region modeling of [11C]-(R)-PK11195 brain studies. J Nucl Med. 2007;48:158–167. [PubMed] [Google Scholar]
- van Berckel BN, Bossong MG, Boellaard R, Kloet R, Schuitemaker A, Caspers E, Luurtsema G, Windhorst AD, Cahn W, Lammertsma AA, Kahn RS. Microglia activation in recent-onset schizophrenia: a quantitative (R)-[11C]PK11195 positron emission tomography study. Biol Psychiatry. 2008;64:820–822. doi: 10.1016/j.biopsych.2008.04.025. [DOI] [PubMed] [Google Scholar]
- West J, Fitzpatrick JM, Wang MY, Dawant BM, Maurer CR, Kessler RM, Maciunas RJ, Barillot C, Lemoine D, Collignon A, Maes F, Suetens P, Vandermeulen D, vandenElsen PA, Napel S, Sumanaweera TS, Harkness B, Hemler PF, Hill DLG, Hawkes DJ, Studholme C, Maintz JBA, Viergever MA, Malandain G, Pennec X, Noz ME, Maguire GQ, Pollack M, Pelizzari CA, Robb RA, Hanson D, Woods RP. Comparison and evaluation of retrospective intermodality brain image registration techniques. J Comput Assist Tomogr. 1997;21:554–566. doi: 10.1097/00004728-199707000-00007. [DOI] [PubMed] [Google Scholar]
- Zhang YY, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]