Abstract
Paternal uniparental disomy 14 (UPD(14)pat) results in a unique constellation of clinical features, and a similar phenotypic constellation is also caused by microdeletions involving the DLK1-MEG3 intergenic differentially methylated region (IG-DMR) and/or the MEG3-DMR and by epimutations (hypermethylations) affecting the DMRs. However, relative frequency of such underlying genetic causes remains to be clarified, as well as that of underlying mechanisms of UPD(14)pat, that is, trisomy rescue (TR), gamete complementation (GC), monosomy rescue (MR), and post-fertilization mitotic error (PE). To examine this matter, we sequentially performed methylation analysis, microsatellite analysis, fluorescence in situ hybridization, and array-based comparative genomic hybridization in 26 patients with UPD(14)pat-like phenotype. Consequently, we identified UPD(14)pat in 17 patients (65.4%), microdeletions of different patterns in 5 patients (19.2%), and epimutations in 4 patients (15.4%). Furthermore, UPD(14)pat was found to be generated through TR or GC in 5 patients (29.4%), MR or PE in 11 patients (64.7%), and PE in 1 patient (5.9%). Advanced maternal age at childbirth (≥35 years) was predominantly observed in the MR/PE subtype. The results imply that the relative frequency of underlying genetic causes for the development of UPD(14)pat-like phenotype is different from that of other imprinting disorders, and that advanced maternal age at childbirth as a predisposing factor for the generation of nullisomic oocytes through non-disjunction at meiosis 1 may be involved in the development of MR-mediated UPD(14)pat.
Keywords: genetic cause, maternal age effect, monosomy rescue, UPD(14)pat subtype
Introduction
Human chromosome 14q32.2 carries a ∼1.2 Mb imprinted region with the germline-derived primary DLK1-MEG3 intergenic differentially methylated region (IG-DMR) and the post-fertilization-derived secondary MEG3-DMR, together with multiple imprinted genes.1, 2 Both DMRs are methylated after paternal transmission and unmethylated after maternal transmission in the body, whereas in the placenta the IG-DMR alone remains as a DMR and the MEG3-DMR is rather hypomethylated irrespective of the parental origin.2, 3 Furthermore, it has been shown that the unmethylated IG-DMR and MEG3-DMR of maternal origin function as the imprinting centers in the placenta and the body, respectively, and that the IG-DMR acts as an upstream regulator for the methylation pattern of the MEG3-DMR in the body but not in the placenta.3
As a result of the presence of the imprinted region, paternal uniparental disomy 14 (UPD(14)pat) (OMIM #608149) causes a unique constellation of body and placental phenotypes such as characteristic face, bell-shaped small thorax, abdominal wall defect, polyhydramnios, and placentomegaly.2, 4, 5 Furthermore, consistent with the essential role of the DMRs in the imprinting regulation, microdeletions and epimutations affecting the IG-DMR or both DMRs of maternal origin result in UPD(14)pat-like phenotype in both the body and the placenta, whereas a microdeletion involving the maternally inherited MEG3-DMR alone leads to UPD(14)pat-like phenotype in the body, but not in the placenta.2, 3
Of the three underlying genetic causes for UPD(14)pat-like phenotype (UPD(14)pat, microdeletions, and epimutations), UPD(14)pat is primarily generated by four mechanisms, that is, trisomy rescue (TR), gamete complementation (GC), monosomy rescue (MR), and post-fertilization mitotic error (PE).6 TR refers to a condition in which chromosome 14 of maternal origin is lost from a zygote with trisomy 14 formed by fertilization between a disomic sperm and a normal oocyte. GC results from fertilization of a disomic sperm with a nullisomic oocyte. MR refers to a condition in which chromosome 14 of paternal origin is replicated in a zygote with monosomy 14 formed by fertilization between a normal sperm and a nullisomic oocyte. PE is an event after formation of a normal zygote. In this regard, a nullisomic oocyte specific to GC and MR is produced by non-disjunction at meiosis 1 (M1) or meiosis 2 (M2), and non-disjunction at M1 is known to increase with maternal age, probably because of a long-term (10–50 years) meiotic arrest at prophase 1.7
However, relative frequency of the genetic causes for UPD(14)pat-like phenotype remains to be determined, as well as that of underlying mechanisms for the generation of UPD(14)pat. Here, we report our data on this matter, and discuss the difference in the relative frequency among imprinted disorders and the possible maternal age effect on the relative frequency.
Patients and methods
Patients
This study comprised 26 patients with UPD(14)pat-like phenotype (9 male patients and 17 female patients) (Table 1). Of the 26 patients, 18 patients have been reported previously; they consisted of nine sporadic patients with full UPD(14)pat,4, 5 one sporadic patient with segmental UPD(14)pat,4 the proband of sibling cases and four sporadic patients with different patterns of microdeletions involving the unmethylated DMRs of maternal origin,2, 3 and three patients with epimutations (hypermethylations) of the two normally unmethylated DMRs of maternal origin.2 The remaining eight patients were new sporadic cases.
Table 1. Summary of patients examined in this study.
| Patient | Genetic cause | UPD(14)pat subtype | Maternal age at childbirth (years) | Paternal age at childbirth (years) | Remark | Reference |
|---|---|---|---|---|---|---|
| 1 | UPD(14)pat | TR/GC [M1] | 31 | 35 | 5 | |
| 2 | UPD(14)pat | TR/GC [M1] | 28 | 29 | 5 | |
| 3 | UPD(14)pat | TR/GC [M1] | 29 | 38 | This report | |
| 4 | UPD(14)pat | TR/GC [M1] | 36 | 41 | 5 | |
| 5 | UPD(14)pat | TR/GC [M2] | 30 | 30 | This report | |
| 6 | UPD(14)pat | MR/PE | 42 | Unknown | 4,5 | |
| 7 | UPD(14)pat | MR/PE | 31 | 28 | 5 | |
| 8 | UPD(14)pat | MR/PE | 32 | 33 | 5 | |
| 9 | UPD(14)pat | MR/PE | 26 | 35 | 5 | |
| 10 | UPD(14)pat | MR/PE | 38 | 38 | This report | |
| 11 | UPD(14)pat | MR/PE | 26 | 32 | This report | |
| 12 | UPD(14)pat | MR/PE | 41 | 36 | This report | |
| 13 | UPD(14)pat | MR/PE | 30 | 28 | This report | |
| 14 | UPD(14)pat | MR/PE | 39 | 34 | This report | |
| 15 | UPD(14)pat | MR/PE | 42 | 37 | Born after IVF-ET | 5 |
| 16 | UPD(14)pat | MR/PE | 36 | 36 | 4,5 | |
| 17 | UPD(14)pat-seg. | PE | 27 | 24 | Segmental isodisomy | 4,5 |
| 18 | Microdeletion | 31 | 34 | 2 | ||
| 19 | Microdeletion | 33 | 36 | 2 | ||
| 20 | Microdeletion | 28 | 27 | 2 | ||
| 21 | Microdeletion | 27 | 37 | IG-DMR alone | 3 | |
| 22 | Microdeletion | 25 | 25 | MEG3-DMR alone | 3 | |
| 23 | Epimutation | 35 | 36 | 2 | ||
| 24 | Epimutation | 28 | 26 | 2 | ||
| 25 | Epimutation | 27 | 30 | 2 | ||
| 26 | Epimutation | 33 | 33 | This report |
Abbreviation: IVF-ET, in vivo fertilization-embryo transfer using parental gametes.
The microdeletions in patients 18–22 are different in size.
Phenotypic findings of the 26 patients are summarized in Supplementary Table 1; detailed clinical features of patients 6 and 16–25 are as described previously,2, 3, 4 and those of the eight new patients 3, 5, 10–14, and 26 are shown in Supplementary Table 2, together with those of patients 1, 2, 4, 7–9, and 15 in whom detailed phenotypes were not described in the previous report.5 All the 26 patients were identified shortly after birth because of the unique bell-shaped thorax with coat-hanger appearance of the ribs on roentgenograms obtained because of asphyxia. Subsequent clinical analysis revealed that 25 of the 26 patients exhibited both body and placental UPD(14)pat-like phenotype, whereas the remaining one previously reported patient (patient 22) manifested body, but not placental, UPD(14)pat-like phenotype.3 The karyotype was found to be normal in 25 patients, although cytogenetic analysis was not performed in one previously reported patient who died of respiratory failure at 2 h of age (patient 6).4 One patient (patient 15) was conceived by in vitro fertilization-embryo transfer.5 This study was approved by the Institute Review Board Committee at the National Center for Child Health and Development, and performed after obtaining written informed consent.
Analysis of underlying genetic causes in patients with UPD(14)pat-like phenotype
We sequentially performed methylation analysis, microsatellite analysis, and fluorescence in situ hybridization (FISH), using leukocyte genomic DNA samples and lymphocyte metaphase spreads of all the 26 patients with UPD(14)pat-like phenotype. The detailed methods were as reported previously.2, 3 In brief, methylation analysis was performed for the IG-DMR (CG4 and CG6) and the MEG3-DMR (CG7 and the CTCF-biding sites C and D) by combined bisulfite restriction analysis and bisulfite sequencing. Microsatellite analysis was performed for multiple loci on chromosome 14, by determining the sizes of PCR products obtained with fluorescently labeled forward primers and unlabeled reverse primers. FISH analysis was carried out for the IG-DMR and the MEG3-DMR using 5104-bp and 5182-bp long PCR products, respectively, together with the RP11-566I2 probe for 14q12 utilized as an internal control.
In this study, furthermore, oligonucleotide array-based comparative genomic hybridization (CGH) was also performed for the imprinted region of non-UPD(14)pat patients, using a custom-build oligo-microarray containing 12 600 probes for 14q32.2–q32.3 encompassing the imprinted region and ∼10 000 reference probes for other chromosomal region (4 × 180K format, Design ID 032112) (Agilent Technologies, Palo Alto, CA, USA). The procedure was as described in the manufacturer's instructions.
Analysis of subtypes in patients with UPD(14)pat
UPD(14)pat subtype was determined by microsatellite analysis.8, 9 In brief, heterodisomy for at least one locus was regarded as indicative of TR- or GC-mediated UPD(14)pat (TR/GC subtype), whereas isodisomy for all the informative microsatellite loci was interpreted as indicative of MR- or PE-mediated UPD(14)pat (MR/PE subtype) (for details, see Supplementary Figure S1). Here, while heterodisomy and isodisomy for a pericentromeric region in the TR/GC subtype imply a disomic sperm generation through M1 and M2 non-disjunction respectively,9 such discrimination between M1 and M2 non-disjunctions is impossible for the development of a nullisomic oocyte. Furthermore, it is usually impossible to discriminate between TR and GC, although the presence of trisomic cells is specific to TR. Similarly, it is also usually impossible to discriminate between MR and PE, although identification of segmental isodisomy or mosaicism is unique to PE (PE subtype).
Analysis of parental ages
We examined parental ages at childbirth in patients of different underlying causes and different UPD(14)pat subtypes. Statistical significance of the relative frequency was examined by the Fisher's exact probability test, and that of the median age by the Mann–Whitney's U-test. P<0.05 was considered significant.
Results
Analysis of underlying causes in patients with UPD(14)pat-like phenotype
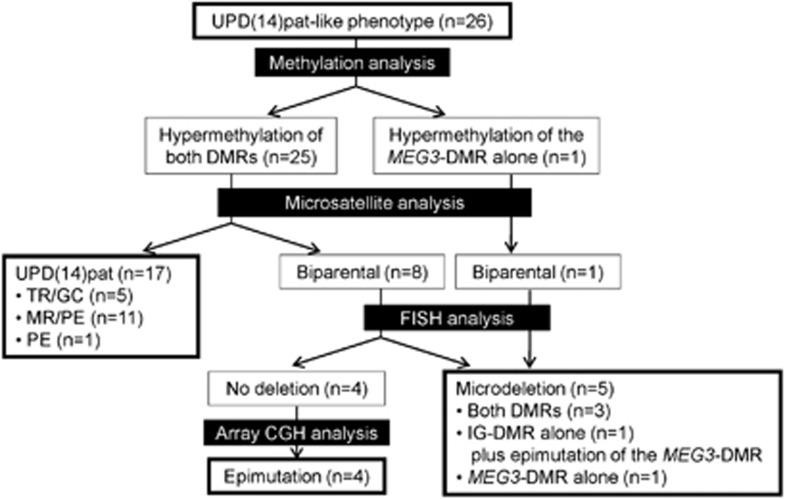
For the eight new sporadic patients, methylation analysis invariably revealed hypermethylation of both DMRs, and microsatellite analysis showed UPD(14)pat in seven patients and biparentally inherited homologs of chromosome 14 in the remaining one patient (patient 26). FISH analysis for patient 26 identified two signals for the two DMRs, and subsequently performed array CGH analysis showed no evidence for genomic rearrangements (Supplementary Figure S2). Thus, patient 26 was assessed to have an epimutation affecting the two DMRs. Furthermore, the results of array CGH analysis confirmed the presence of microdeletions in patients 18–21 and the absence of a discernible microdeletion in patients 23–25 (Supplementary Figure S2) (array CGH analysis was not performed in patient 22 with a 4303-bp microdeletion3 because of the lack of DNA sample available). Thus, together with our previous data, all the 26 patients with UPD(14)pat-like phenotype had genetic alteration involving the imprinted region on chromosome14q32.2.
Consequently, the 26 patients with UPD(14)pat-like phenotype were classified as follows: (1) 16 sporadic patients with full UPD(14)pat and 1 sporadic patient with segmental UPD(14)pat (UPD(14)pat group); (2) the proband of the sibling cases and two sporadic patients with different patterns of microdeletions involving the two DMRs, one sporadic patient with a microdeletion involving the IG-DMR alone in whom the MEG3-DMR was epimutated, and one patient with a microdeletion involving the MEG3-DMR alone (deletion group); and (3) four patients with epimutations (hypermethylations) of both DMRs (epimutation group) (Figure 1 and Table 1).
Figure 1.
Classification of 26 patients with UPD(14)pat-like phenotype.
Analysis of subtypes in patients with UPD(14)pat
Heterozygosity for at least one locus indicative of TR/GC subtype was identified in five patients (patients 1–5), and the disomic pattern of pericentromeric region indicated M1 non-disjunction in patients 1–4 and M2 non-disjunction in patient 5. Full isodisomy consistent with MR/PE subtype was detected in 11 patients (patients 6–16), and segmental isodisomy unique to PE subtype was revealed in 1 patient (patient 17) (Table 1, Figure 1, and Supplementary Figure S3).
Analysis of parental ages
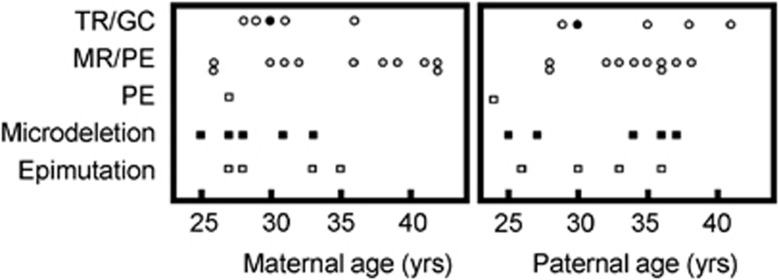
The distribution of parental ages at childbirth is shown in Figure 2. The advanced maternal age at childbirth (≥35 years) was predominantly observed in the MR/PE subtype of UPD(14)pat. Furthermore, while the relative frequency of aged mothers (≥35 years) did not show a significant difference between the MR/PE subtype of UPD(14)pat (6/11) and (i) other subtypes of UPD(14)pat (1/6) (P=0.159), (ii) deletion group (0/5) (P=0.057), and (iii) epimutation group (1/4) (P=0.338), it was significantly different between the MR/PE subtype and the sum of other subtypes of UPD(14)pat, deletion group, and epimutation group (2/15) (P=0.034). Similarly, while the median maternal age did not show a significant difference between the MR/PE subtype of UPD(14)pat (36 years) vs (i) other subtypes of UPD(14)pat (29.5 years) (P=0.118), (ii) deletion type (28 years) (P=0.088), and (iii) epimutation type (30.5 years) (P=0.295), it was significantly different between the MR/PE subtype of UPD(14)pat and the sum of other subtypes of UPD(14)pat, deletion group, and epimutation group (29 years) (P=0.045).
Figure 2.
The distribution of parental ages at childbirth according to the underlying genetic causes for the development of UPD(14)pat-like phenotype and UPD(14)pat subtypes. Of the five plots for the TR/GC subtype, open and black circles indicate the TR/GC subtype due to non-disjunction at paternal M1 and M2, respectively.
The paternal ages were similar irrespective of the genetic causes and the UPD(14)pat subtypes. In addition, the median paternal age was comparable between the TR/GC subtype of UPD(14)pat that postulates the production of a disomic sperm (35.0 years) and the sum of other subtypes of UPD(14)pat, deletion group, and epimutation group that assumes the production of a normal sperm (33.5 years) (P=0.322).
Discussion
This study revealed that the UPD(14)pat-like phenotype was caused by UPD(14)pat in 65.4% of patients, by microdeletions in 19.2% of patients, and by epimutations in 15.4% of patients. Although the relative frequency of underlying genetic factors for the development of UPD(14)pat-like phenotype has been reported previously,10 most data are derived from our previous publications. Thus, the present results are regarded as the updated and extended data on the relative frequency. For the relative frequency, it is notable that 25 of the 26 patients were confirmed to have normal karyotype, although chromosome analysis was not performed in patient 6. Thus, while Robertsonian translocations involving chromosome 14 is known to be a predisposing factor for the occurrence of UPD(14)pat,11, 12, 13, 14, 15, 16 such a possible chromosomal effect has been excluded in nearly all patients examined in this study.
The relative frequency of underlying causes has also been reported in other imprinting disorders.8, 17, 18, 19 The data are summarized in Table 2 (a similar summary has also been reported recently by Hoffmann et al).10 In particular, the results in patients with normal karyotype are available in Prader–Willi syndrome (PWS).8 Furthermore, PWS is also known to be caused by UPD, microdeletions, and epimutations affecting a single imprinting region,8, 19 although Silver–Russell syndrome and Beckwith–Wiedemann syndrome (BWS) can result from perturbation of at least two imprinted regions,17, 18 and BWS and Angelman syndrome can occur as a single gene disorder.17, 19 Thus, it is notable that the relative frequency of underlying causes is quite different between patients with UPD(14)pat-like phenotype and those with PWS.8, 19 This would primarily be due to the presence of low copy repeats flanking the imprinted region on chromosome 15, because chromosomal deletions are prone to occur in regions harboring such repeat sequences.20 Indeed, two types of microdeletions mediated by such low copy repeats account for a vast majority of microdeletions in patients with PWS,21 whereas the microdeletions identified in patients with UPD(14)pat-like phenotype are different to each other. This would explain why microdeletions are less frequent and UPD and epimutations are more frequent in patients with UPD(14)pat-like phenotype than in those with PWS.
Table 2. Relative frequency of genetic mechanisms in imprinting disorders.
| UPD(14)pat-like phenotype | BWS | SRS | AS | PWS | |
|---|---|---|---|---|---|
| Uniparental disomy | 65.4% | 16% | 10% | 3–5% | 25% (25%) |
| UPD(14)pat | UPD(11)pat (mosaic) | UPD(7)mat | UPD(15)pat | UPD(15)mat | |
| Cryptic deletion | 19.2% | Rare | — | 70% | 70% (72%) |
| Cryptic duplication | — | — | Rare | — | — |
| Epimutation | |||||
| Hypermethylation | 15.4% | 9% | — | — | 2–5% (2%) |
| Affected DMR | IG-DMR/MEG3-DMR | H19-DMR | — | — | SNRPN-DMR |
| Hypomethylation | — | 44% | >38% | 2–5% | — |
| Affected DMR | KvDMR1 | H19-DMR | SNRPN-DMR | ||
| Gene mutation | — | 5% | — | 10-15% | — |
| Mutated gene | CDKN1C | UBE3A | |||
| Unknown | 25% | >40% | 10% | ||
| Reference | This study | 17 | 18 | 19 | 8, 19 |
Abbreviations: AS, Angelman syndrome; BWS, Beckwith–Wiedemann syndrome; PWS, Prader–Willi syndrome; SRS, Silver–Russell syndrome.
Patients with abnormal karyotypes are included in BWS and AS, and not included in SRS. In PWS, the data including patients with abnormal karyotypes are shown, and those from patients with normal karyotype alone are depicted in parentheses.
Advanced maternal age at childbirth was predominantly observed in the MR/PE subtype. This may imply the relevance of advanced maternal age to the development of MR-mediated UPD(14)pat, because the generation of nullisomic oocytes through M1 non-disjunction is a maternal age-dependent phenomenon.22 Although no paternal age effect was observed, this is consistent with the previous data indicating no association of advanced paternal age with a meiotic error.23 For the maternal age effect, however, several matters should be pointed out: (1) the number of analyzed patients is small, although it is very difficult to collect a large number of patients in this extremely rare disorder; (2) of the MR/PE subtype, the advanced maternal age is a risk factor for the generation of MR-mediated UPD(14)pat, but not for the development of PE-mediated UPD(14)pat; (3) it is impossible to discriminate between maternal age-dependent M1 non-disjunction and maternal age-independent M2 non-disjunction in the MR and GC subtypes (however, GC must be extremely rare, because it requires the concomitant occurrence of a nullisomic oocyte and a disomic sperm); (4) of the TR/GC subtype, the advanced maternal age is a risk factor for the generation of GC-mediated UPD(14)pat, but not for the development of TR-mediated UPD(14)pat; and (5) if a cryptic recombination(s) might remain undetected in some patients with apparently full isodisomy, this argues that such patients actually have TR- or GC-mediated UPD(14)pat rather than MR- or PE-mediated UPD(14)pat. Thus, further studies are required to examine the maternal age effect on the generation of MR-mediated UPD(14)pat. In addition, while a relationship is unlikely to exist between advanced maternal age and microdeletions and epimutations, this notion would also await further investigations.
Such a maternal age effect is also expected in the TR/GC subtype maternal UPDs after M1 non-disjunction, because the generation of disomic oocytes through M1 non-disjunction is also a maternal age-dependent phenomenon.7 Indeed, such a maternal age effect has been shown for PWS patients with normal karyotype; the maternal age at childbirth was significantly higher in patients with heterodisomy for a very pericentromeric region indicative of TR/GC subtype UPD(15)mat after M1 non-disjunction than in those with other genetic causes.8, 9 For various chromosomes other than chromosome 15, furthermore, since maternal age at childbirth is higher in patients with maternal heterodisomy than in those with maternal isodisomy,24 this would also argue for maternal age effect on the development of maternal UPDs. However, in the previous studies on maternal UPDs other than UPD(15)mat, the available data are quite insufficient to assess the maternal age effect. For example, although a relatively large number of patients with UPD(14)mat phenotype have been reported in the literature (reviewed in reference Hoffmann et al),10 we could identify only six UPD(14)mat patients with normal karyotype in whom maternal age at childbirth was documented and microsatellite analysis was performed.25, 26, 27, 28, 29, 30 Furthermore, the microsatellite data are insufficient to identify the subtype of UPD(14)mat and to distinguish between M1 and M2 non-disjunction in the TR/GC subtype. Thus, while the maternal age at childbirth may be advanced in five patients with apparently TR/GC-mediated UPD(14)mat (27, 35, 37, 41, and 44 years)25, 26, 27, 29, 30 (the maternal age at childbirth in the remaining one patient with apparently MR/PE-mediated UPD(14)mat is 40 years),28 the notion of a maternal age effect awaits further investigations for UPD(14)mat.
Finally, it appears to be worth pointing out that methylation analysis invariably revealed hypermethylated DMR(s) in all the 26 patients who were initially ascertained because of bell-shaped thorax with coat-hanger appearance of the ribs. This indicates that methylation analysis of the DMRs can be utilized for a screening of this condition, and that the constellation of clinical features in the UPD(14)pat-like phenotype, especially the bell-shaped thorax with coat-hanger appearance of the ribs, is highly unique to patients with UPD(14)pat-like phenotype.
In summary, this study confirms the relative frequency of underlying genetic causes for the UPD(14)pat phenotype and reveals the relative frequency of UPD(14)pat subtypes. Furthermore, the results emphasize the difference in the relative frequency of underlying genetic causes among imprinted disorders, and may support a possible maternal age effect on the generation of the nullisomic oocyte mediated UPD(14)pat. Further studies will permit a more precise assessment on these matters.
Acknowledgments
We thank Drs Kenji Kurosawa, Michiko Hayashidani, Toshio Takeuchi, Shinya Tanaka, Mika Noguch, Kouji Masumoto, Takeshi Utsunomiya, Yumiko Komatsu, Hirofumi Ohashi, Maureen J O'Sullivan, Andrew J Green, Yoshiyuki Watabe, Tsuyako Iwai, Hitoshi Kawato, Miho Torikai, Akiko Yamamoto, Nobuhiro Suzumori, Makoto Kuwajima, Hiroshi Yoshihashi, Yoriko Watanabe, and Jin Nishimura for material sampling and phenotype assessment. This work was supported by Grants for Research on Intractable Diseases (H22-161) and for Health Research on Children, Youth and Families (H21-005) from the Ministry of Health, Labor and Welfare, by Grants-in-Aid for Scientific Research (A) (22249010) and (B) (21028026) from the Japan Society for the Promotion of Science (JSPS), by Grants from Takeda Science Foundation and from Kanehara Foundation, and by the Grant for National Center for Child Health and Development (23A-1).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Kagami M, Sekita Y, Nishimura G, et al. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet. 2008;40:237–242. doi: 10.1038/ng.2007.56. [DOI] [PubMed] [Google Scholar]
- Kagami M, O'Sullivan MJ, Green AJ, et al. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet. 2010;6:e1000992. doi: 10.1371/journal.pgen.1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami M, Nishimura G, Okuyama T, et al. Segmental and full paternal isodisomy for chromosome 14 in three patients: narrowing the critical region and implication for the clinical features. Am J Med Genet A. 2005;138A:127–132. doi: 10.1002/ajmg.a.30941. [DOI] [PubMed] [Google Scholar]
- Kagami M, Yamazawa K, Matsubara K, Matsuo N, Ogata T. Placentomegaly in paternal uniparental disomy for human chromosome 14. Placenta. 2008;29:760–761. doi: 10.1016/j.placenta.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Agan N, Goldberg JD, Ledbetter DH, Longshore JW, Cassidy SB. American College of Medical Genetics statement of diagnostic testing for uniparental disomy. Genet Med. 2001;3:206–211. doi: 10.1097/00125817-200105000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KT. Meiosis in oocytes: predisposition to aneuploidy and its increased incidence with age. Hum Reprod Update. 2008;14:143–158. doi: 10.1093/humupd/dmm043. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Murakami N, Nagai T, Ogata T. Maternal age effect on the development of Prader-Willi syndrome resulting from upd(15)mat through meiosis 1 errors. J Hum Genet. 2011;56:566–571. doi: 10.1038/jhg.2011.59. [DOI] [PubMed] [Google Scholar]
- Robinson WP, Christian SL, Kuchinka BD, et al. Somatic segregation errors predominantly contribute to the gain or loss of a paternal chromosome leading to uniparental disomy for chromosome 15. Clin Genet. 2000;57:349–358. doi: 10.1034/j.1399-0004.2000.570505.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann K, Heller R. Uniparental disomies 7 and 14. Best Pract Res Clin Endocrinol Metab. 2011;25:77–100. doi: 10.1016/j.beem.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Wang JC, Passage MB, Yen PH, Shapiro LJ, Mohandas TK. Uniparental heterodisomy for chromosome 14 in a phenotypically abnormal familial balanced 13/14 Robertsonian translocation carrier. Am J Hum Genet. 1991;48:1069–1074. [PMC free article] [PubMed] [Google Scholar]
- Papenhausen PR, Mueller OT, Johnson VP, Sutcliffe M, Diamond TM, Kousseff BG. Uniparental isodisomy of chromosome 14 in two cases: an abnormal child and a normal adult. Am J Med Genet. 1995;59:271–275. doi: 10.1002/ajmg.1320590302. [DOI] [PubMed] [Google Scholar]
- Cotter PD, Kaffe S, McCurdy LD, Jhaveri M, Willner JP, Hirschhorn K. Paternal uniparental disomy for chromosome 14: a case report and review. Am J Med Genet. 1997;70:74–79. doi: 10.1002/(sici)1096-8628(19970502)70:1<74::aid-ajmg14>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Yano S, Li L, Owen S, Wu S, Tran T. A further delineation of the paternal uniparental disomy (UPD14): the fifth reported liveborn case. Am J Hum Genet. 2001;69 (Suppl:A739. [Google Scholar]
- Kurosawa K, Sasaki H, Sato Y, et al. Paternal UPD14 is responsible for a distinctive malformation complex. Am J Med Genet A. 2002;110:268–272. doi: 10.1002/ajmg.10404. [DOI] [PubMed] [Google Scholar]
- McGowan KD, Weiser JJ, Horwitz J, et al. The importance of investigating for uniparental disomy in prenatally identified balanced acrocentric rearrangements. Prenat Diagn. 2002;22:141–143. [PubMed] [Google Scholar]
- Sasaki K, Soejima H, Higashimoto K, et al. Japanese and North American/European patients with Beckwith-Wiedemann syndrome have different frequencies of some epigenetic and genetic alterations. Eur J Hum Genet. 2007;15:1205–1210. doi: 10.1038/sj.ejhg.5201912. [DOI] [PubMed] [Google Scholar]
- Eggermann T. Epigenetic regulation of growth: lessons from Silver-Russell syndrome. Endocr Dev. 2009;14:10–19. doi: 10.1159/000207472. [DOI] [PubMed] [Google Scholar]
- Gurrieri F, Accadia M. Genetic imprinting: the paradigm of Prader-Willi and Angelman syndromes. Endocr Dev. 2009;14:20–28. doi: 10.1159/000207473. [DOI] [PubMed] [Google Scholar]
- Pujana MA, Nadal M, Guitart M, Armengol L, Gratacos M, Estivill X. Human chromosome 15q11-q14 regions of rearrangements contain clusters of LCR15 duplicons. Eur J Hum Genet. 2002;10:26–35. doi: 10.1038/sj.ejhg.5200760. [DOI] [PubMed] [Google Scholar]
- Varela MC, Kok F, Setian N, Kim CA, Koiffmann CP. Impact of molecular mechanisms, including deletion size, on Prader-Willi syndrome phenotype: study of 75 patients. Clin Genet. 2005;67:47–52. doi: 10.1111/j.1399-0004.2005.00377.x. [DOI] [PubMed] [Google Scholar]
- Pellestor F, Andreo B, Anahory T, Hamamah S. The occurrence of aneuploidy in human: lessons from the cytogenetic studies of human oocytes. Eur J Med Genet. 2006;49:103–116. doi: 10.1016/j.ejmg.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Sloter E, Nath J, Eskenazi B, Wyrobek AJ. Effects of male age on the frequencies of germinal and heritable chromosomal abnormalities in humans and rodents. Fertil Steril. 2004;81:925–943. doi: 10.1016/j.fertnstert.2003.07.043. [DOI] [PubMed] [Google Scholar]
- Kotzot D. Advanced parental age in maternal uniparental disomy (UPD): implications for the mechanism of formation. Eur J Hum Genet. 2004;12:343–346. doi: 10.1038/sj.ejhg.5201158. [DOI] [PubMed] [Google Scholar]
- Fokstuen S, Ginsburg C, Zachmann M, Schinzel A. Maternal uniparental disomy 14 as a cause of intrauterine growth retardation and early onset of puberty. J Pediatr. 1999;134:689–695. doi: 10.1016/s0022-3476(99)70282-9. [DOI] [PubMed] [Google Scholar]
- Hordijk R, Wierenga H, Scheffer H, Leegte B, Hofstra RM, Stolte-Dijkstra I. Maternal uniparental disomy for chromosome 14 in a boy with a normal karyotype. J Med Genet. 1999;36:782–785. doi: 10.1136/jmg.36.10.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanlaville D, Aubry MC, Dumez Y, et al. Maternal uniparental heterodisomy of chromosome 14: chromosomal mechanism and clinical follow up. J Med Genet. 2000;37:525–528. doi: 10.1136/jmg.37.7.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner DR, Shaffer LG, Yang SP, Walgenbach DD. Confined placental mosaicism for trisomy 14 and maternal uniparental disomy in association with elevated second trimester maternal serum human chorionic gonadotrophin and third trimester fetal growth restriction. Prenat Diagn. 2001;21:395–398. doi: 10.1002/pd.75. [DOI] [PubMed] [Google Scholar]
- Aretz S, Raff R, Woelfle J, et al. Maternal uniparental disomy 14 in a 15-year-old boy with normal karyotype and no evidence of precocious puberty. Am J Med Genet A. 2005;135:336–338. doi: 10.1002/ajmg.a.30755. [DOI] [PubMed] [Google Scholar]
- Mitter D, Buiting K, von Eggeling F, et al. Is there a higher incidence of maternal uniparental disomy 14 [upd(14)mat]? Detection of 10 new patients by methylation-specific PCR. Am J Med Genet A. 2006;140:2039–2049. doi: 10.1002/ajmg.a.31414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.