Abstract
DNA from Epstein–Barr virus-transformed lymphocyte cell lines (LCLs) has proven useful for studies of genetic sequence polymorphisms. Whether LCL DNA is suitable for methylation studies is less clear. We conduct a genome-wide methylation investigation using an array set with 45 million probes to investigate the methylome of LCL DNA and technical duplicates of WB DNA from the same 10 individuals. We focus specifically on methylation sites that show variation between individuals and, therefore, are potentially useful as biomarkers. The sample correlations for the methylation variable probes ranged from 0.69 to 0.78 for the WB duplicates and from 0.27 to 0.72 for WB vs LCL. To compare the pattern of the methylation signals, we grouped adjacent probes based on their inter-correlations. These analyses showed ∼29 000 and ∼14 000 blocks in WB and LCL, respectively. Merely 31% of the methylated regions detected in WB were detectable in LCLs. Furthermore, we observed significant differences in mean difference between WB and LCL as compared with duplicates of WB (P-value =2.2 × 10−16). Our study shows that there are substantial differences in the DNA methylation patterns between LCL and WB. Thus, LCL DNA should not be used as a proxy for WB DNA in methylome-wide studies.
Keywords: DNA methylation, methylome, inter-individual variation, biomarkers
Introduction
It is common to extract DNA from Epstein–Barr virus (EBV)-transformed lymphocyte cell lines (LCLs). This provides an almost unlimited amount of DNA, which has proven very useful for studies of genetic sequence polymorphisms.1, 2, 3, 4 Although LCL DNA has been used to investigate associations between methylation markers and phenotypes,5 it is less clear whether it is equally suitable for methylation investigations.6, 7, 8
A number of targeted studies indicate that EBV transformation affects the methylation pattern,9, 10 and a recent study comparing DNA from primary B-lymphocytes and their corresponding LCLs, using >27 000 markers mainly located in CpG-rich regions, has shown that gene regulation is changed during EBV transformation.6 Other studies using similar protocols to investigate CpG-rich regions have shown that the correlations between the methylation patterns in DNA from WB and LCL are high (r>0.9).7, 8
In this article, we focus specifically on methylation sites that show variation between individuals and, therefore, are potentially useful as biomarkers in disease studies. As such sites may not be in CpG-rich regions,11 we use a genome-wide (tiling array) approach, which is not limited to pre-selected regions of interest. Similar approaches have been used to study the methylome of, for example, human brain samples12 and Arabidopsis.13 In total, we investigate 45 million probes per sample in 30 methylomes from 10 individuals. To identify the sites that reliably measure inter-individual variation in methylation in WB, we use technical duplicates of WB DNA from the 10 individuals. Next, we compare the methylation pattern from the variable methylation regions in WB DNA with LCL DNA extracted from the same individuals.
Materials and methods
Study sample
We obtained 10 individuals from the National Institute of Mental Health (Site 150) for whom WB DNA and LCL DNA were available. All subjects gave their informed consent. The sample consisted of five males and five females of European American descent. The participants' ages ranged from 22 to 80, with a mean age of 45.8 years and SD=20.2. Details on DNA extraction and LCL are presented in the Supplementary Material.
DNA methylation profiling
Following the manufacturers' protocols, DNA was fragmented with MseI to a median size of 500 bp, and the methylome was enriched for using MethylMiner (Invitrogen, Carlsbad, CA, USA) and further amplified using the Sigma WGA2 kit (Sigma-Aldrich, St Louis, MO, USA). Amplified methylomes were fragmented using the Affymetrix (Santa Clara, CA, USA) 6.0 fragment reagents and labeled using the Affymetrix WT-ds DNA Terminal labeling kit followed by hybridization to the Affymetrix GeneChip Human Tiling 2.0R array set. The arrays were washed and scanned according to the manufacturers' protocol.
Data analyses
The data were background-corrected using the robust multi-array procedure,14 followed by quantile normalization.15 The variable methylation sites were identified by calculating the probe correlation, that is, the correlation between the technical duplicates of WB from the 10 individuals for each probe.
We used two statistical measures based on correlation to compare WB DNA with LCL DNA. The first is the calculation of the sample correlation that reflects whether the level, that is, rank, of the methylation signals remains unchanged after EBV transformation. The sample correlation is preferably calculated using only the variable methylation sites (probe correlation >0.75). However, in order to be able to compare our work with previously reported studies, we also report the sample correlation including all 45 million probes. The second statistical measure was used to investigate if the pattern of the methylation signal, regardless of its level, is conserved after the EBV transformation by creating blocks of adjacent probes based on their inter-correlations. That is, we investigate if methylation sites (probes) that are correlated in WB DNA are also correlated in LCL DNA (for details see Supplementary Material).
In addition to the measures of correlation, we also investigated the similarity between the duplicate WB samples and between each WB vs LCLs for each of the 439 K probes by calculating the mean individual difference between the sample types for a given probe. Furthermore, we are also calculating the mean difference divided by a pooled estimate of the SD (also know as Cohen's D). To investigate if the differences between WB vs LCL were equally similar as the duplicates of WB, we used a t-test.
Results
Comparison of methylation levels and methylation patterns
The probe correlation from the technical duplicates indicated that 439 264 probes (439 K) showed variation in WB between individuals. The sample correlation, that is, the correlation between samples from the same individual, calculated using the 439 K probes, ranged from 0.69 to 0.78 for the duplicates of WB (mean=0.74, SD=0.03). In contrast, for WB vs LCL, the sample correlations were highly variable and ranged from 0.27 to 0.72 (mean≤0.62) with a dramatically increased SD (SD≥0.12) as compared with WB. Sample correlation calculated using all 45 million probes (45 M) range 0.90–0.96 and did not show the difference between WB and LCL (Table 1).
Table 1. Sample correlation detected for all probes (45 M) and for the methylation variable probes (439 K).
| Samples | Probe set | Sample correlation range | Mean | SD |
|---|---|---|---|---|
| Blood 1 vs Blood 2 | 45 Ma | 0.92–0.96 | 0.95 | 0.01 |
| Blood 1 vs LCL | 45 Ma | 0.91–0.96 | 0.94 | 0.02 |
| Blood 2 vs LCL | 45 Ma | 0.90–0.96 | 0.94 | 0.02 |
| Blood 1 vs Blood 2 | 439 K | 0.69–0.78 | 0.74 | 0.03 |
| Blood 1 vs LCL | 439 K | 0.30–0.72 | 0.60 | 0.12 |
| Blood 2 vs LCL | 439 K | 0.27–0.71 | 0.62 | 0.13 |
For computational reasons, the sample correlations for the 45 million probes are calculated for each of the seven arrays in the array set. The average sample correlations of all arrays are reported.
When grouping probes into blocks based on their inter-correlation with closely located variable probes (439 K), we detected ∼29 000 blocks in each of the WB (Table 2) and ∼14 000 blocks in LCL. These blocks, including 88 296 probes, indicate regions in the human genome where the methylation pattern appears to be different among individuals. These regions vary in length from 26 to 7905 bp, with a median size of 229–232 bp in WB and 205 bp in LCL. While the size and the distribution of the blocks are fairly similar in WB and LCL, the number of blocks differs. The lower number of blocks observed in LCLs as compared with WB indicates that less than half of the regions which show inter-individual variation in WB show a similar methylation pattern in LCL. The location of the blocks detected in the two blood samples are similar with an 89% overlap. Furthermore, ∼71% of the blocks detected in the LCLs overlap with the blocks detected in WB. However, as the number of blocks detected in LCL is approximately half of what is detected in WB only ∼31% of the methylated regions detected in WB were detectable in LCLs (Table 2).
Table 2. Description of blocks with a variable methylation pattern.
| Sample | Minimum block size | Median block size | Mean block size | Maximum block size | Number of blocks | Total coverage |
|---|---|---|---|---|---|---|
| LCL | 27 | 205 | 258.0 | 7705 | 14 042 | 3 623 509 |
| Blood 1 | 26 | 229 | 287.8 | 7705 | 29 032 | 8 354 475 |
| Blood 2 | 26 | 232 | 288.4 | 7905 | 29 079 | 8 385 979 |
Values are given in base pairs.
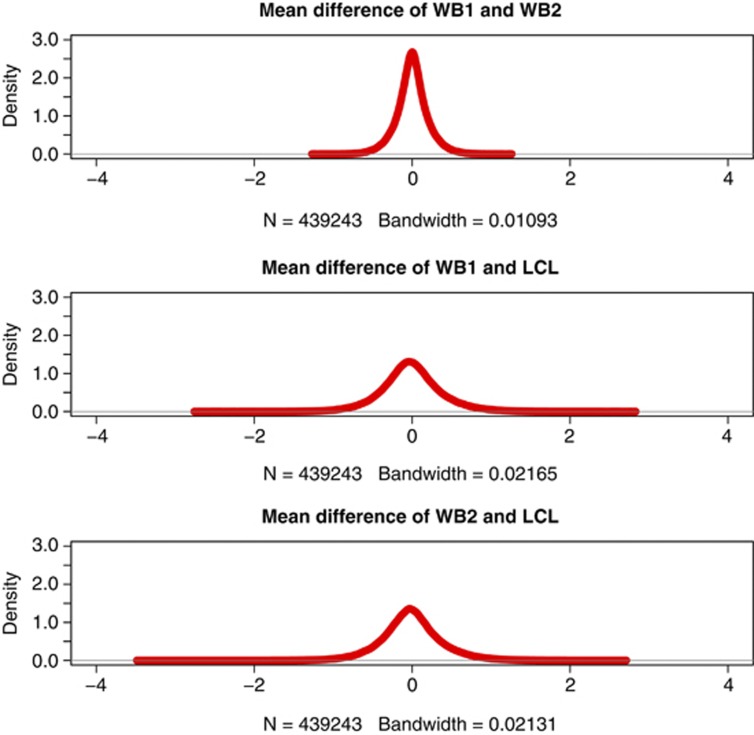
The distributions of the difference in mean between the sample combinations are plotted in Figure 1 (the distribution of Cohen's D is shown in Supplementary Figure S1). These figures show that the majority of probes show small differences between the technical duplicates of WB while much bigger differences are observed for the comparisons with WB vs LCL DNA. When exploring the differences in mean and the SD's in mean (Cohen's D), we noticed a significant difference (P-value =2.2 × 10−16 for each of the two measures of difference) between WB vs LCL as compared with the duplicates of WB.
Figure 1.
The distribution of the mean difference between the duplicates of WB DNA (top) and each of the WB samples vs LCLs (middle and bottom, respectively) are shown. Probes with complete data (no missing data) from all samples that showed inter-individuals variation are included.
Discussion
Focusing on variable methylation sites, we show that the sample correlations between WB and LCL from the same individuals are, on average, lower and their SD is higher than what is observed between the technical duplicates of WB. In addition, fewer blocks could be constructed in LCL and a limited number of blocks detected in WB overlap in LCL. These observations suggest that both the levels of methylation and the methylation patterns are different in LCLs as compared with WB. Furthermore, we observed highly significant differences in the mean difference between WB and LCL as compared with duplicates of WB.
There are several potential reasons for the observed differences. Comparisons of primary B-cells and their corresponding LCLs,6 and studies showing altered methylation pattern in cells that have undergone a large number of passages7 suggest that transformation itself may affect the methylation pattern. In our case, all samples have been prepared the same way and are from a low passage number. However, even thought a high passage number is likely to have a higher effect, it is possible that the transformation itself already after one or a few passages affect the methylation pattern differently in different samples. It is also important to note that DNA from WB represents the methylation pattern from all cells present in WB while LCL DNA represents a subpopulation of B-lymphocytes only. Furthermore, differences in the hemograms as well as in the number of B-cells actually transforming, and their growth rate, could partly explain the high SD observed between LCL and WB.
As reported previously,7, 8 the sample correlation of all probes (45 M) between DNA extracted from whole blood and LCL is high. This high correlation can be explained by that the majority of probes are in regions that are not at all methylated. Unmethylated regions will not show any variation between WB and LCL methylation signals, which would cause the sample correlation to be artificially high. For a proper comparison between WB and LCL it is, therefore, important to confine the analysis to methylated probes or, if the focus is on potential biomarkers for disease, on methylation variables sites.
In conclusion, our study shows that many samples have extensive differences in the methylation pattern between LCL and WB derived from the same subjects. Thus, LCL should not be used as a proxy for WB in methylation studies.
Data availability
Raw data and background-corrected normalized data are made available though the Gene Expression Omnibus (GEO) database http://www.ncbi.nlm.nih.gov/geo/ with accession number GSE35204. Scripts for block construction as well as the pre-constructed blocks are made available as Supplementary Material.
Acknowledgments
The lab technical work was conducted at the Vanderbilt Functional Genomics shared resource, Vanderbilt University, TN, USA. DNA was obtained from National Institute of Mental Health Schizophrenia Genetics Initiative (NIMH-GI) (full acknowledgement is given in the Supplementary Material).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Adkins DE, Aberg K, McClay JL, et al. A genomewide association study of citalopram response in major depressive disorder-a psychometric approach. Biol Psychiatry. 2010;68:e25–e27. doi: 10.1016/j.biopsych.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Lin D, Tzeng JY, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ijzendoorn MH, Caspers K, Bakermans-Kranenburg MJ, Beach SR, Philibert R. Methylation matters: interaction between methylation density and serotonin transporter genotype predicts unresolved loss or trauma. Biol Psychiatry. 2010;68:405–407. doi: 10.1016/j.biopsych.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliskan M, Cusanovich DA, Ober C, Gilad Y. The effects of EBV transformation on gene expression levels and methylation profiles. Hum Mol Genet. 2011;20:1642–1652. doi: 10.1093/hmg/ddr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafodatskaya D, Choufani S, Ferreira JC, et al. EBV transformation and cell culturing destabilizes DNA methylation in human lymphoblastoid cell lines. Genomics. 2010;95:73–83. doi: 10.1016/j.ygeno.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Sun YV, Turner ST, Smith JA, et al. Comparison of the DNA methylation profiles of human peripheral blood cells and transformed B-lymphocytes. Hum Genet. 2010;127:651–658. doi: 10.1007/s00439-010-0810-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan EP, Ehrich M, Brazil DP, et al. Comparative analysis of DNA methylation profiles in peripheral blood leukocytes versus lymphoblastoid cell lines. Epigenetics. 2009;4:159–164. doi: 10.4161/epi.4.3.8793. [DOI] [PubMed] [Google Scholar]
- Saferali A, Grundberg E, Berlivet S, et al. Cell culture-induced aberrant methylation of the imprinted IG DMR in human lymphoblastoid cell lines. Epigenetics. 2010;5:50–60. doi: 10.4161/epi.5.1.10436. [DOI] [PubMed] [Google Scholar]
- Bock C, Walter J, Paulsen M, Lengauer T. Inter-individual variation of DNA methylation and its implications for large-scale epigenome mapping. Nucleic Acids Res. 2008;36:e55. doi: 10.1093/nar/gkn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morahan JM, Yu B, Trent RJ, Pamphlett R. A genome-wide analysis of brain DNA methylation identifies new candidate genes for sporadic amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2009;10:418–429. doi: 10.3109/17482960802635397. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data and background-corrected normalized data are made available though the Gene Expression Omnibus (GEO) database http://www.ncbi.nlm.nih.gov/geo/ with accession number GSE35204. Scripts for block construction as well as the pre-constructed blocks are made available as Supplementary Material.