Abstract
Cat eye syndrome (CES) is caused by a gain of the proximal part of chromosome 22. Usually, a supernumerary marker chromosome is present, containing two extra copies of the chromosome 22q11.1q11.21 region. More sporadically, the gain is present intrachromosomally. The critical region for CES is currently estimated to be about 2.1 Mb and to contain at least 14 RefSeq genes. Gain of this region may cause ocular coloboma, preauricular, anorectal, urogenital and congenital heart malformations. We describe a family in which a 600 kb intrachromosomal triplication is present in at least three generations. The copy number alteration was detected using MLPA and further characterized with interphase and metaphase FISH and SNP-array. The amplified fragment is located in the distal part of the CES region. The family members show anal atresia and preauricular tags or pits, matching part of the phenotype of this syndrome. This finding suggests that amplification of the genes CECR2, SLC25A18 and ATP6V1E1, mapping within the critical region for CES, may be responsible for anorectal, renal and preauricular anomalies in patients with CES.
Keywords: cat eye syndrome, 22q11, intrachromosomal gain, triplication, anal atresia, preauricular anomalies
Introduction
Cat eye syndrome (CES) has a broad spectrum of clinical features. Most frequently found clinical features in diagnosed CES patients are preauricular tags or pits (81–87%), anorectal malformations (atresia, anterior placement) (73–81%), urogenital malformations (eg, unilateral absence of kidney) (71%), ocular coloboma (55–61%) and congenital heart anomalies (eg, anomalous pulmonary venous return, tetralogy of Fallot, persistence of the left superior vena cava, valve anomalies) (50–63%),1 but none are necessarily present.2, 3 Mental development is generally normal or near-normal2, 3, 4, 5 and many other anomalies are described in a smaller number of patients. Only 41% of the patients show all three main visible anomalies (coloboma, anal anomalies and preauricular anomalies), which may indicate that CES is underdiagnosed.2, 3
CES is caused by duplication or triplication of a part of chromosome 22. In most cases, a bisatellited small dicentric supernumerary chromosome is found, consisting of twice the chromosome 22pter to proximal 22q11.21 fragment, cytogenetically written as inv dup(22)(q11.21).4, 5 Intrachromosomal duplication or triplication of the proximal 22q fragment can also give a similar phenotype with a partial or full spectrum of the clinical characteristics. No significant difference in severity of the syndrome is seen between patients with the partial trisomy variant, resulting from a duplication, or the partial tetrasomy variant, resulting from a triplication.6, 7, 8, 9
Mosaicism of the supernumerary inv dup(22) chromosome has been reported several times, but the degree of mosaicism could not be correlated to severity of the phenotype.2 Also inheritance of the supernumerary chromosome has been published.5, 10 In several of these cases, family members have common phenotypic characteristics, but in other cases also family members show phenotypic variance ranging from milder to more severe phenotypes. Therefore, recurrence risk and phenotypic severity for offspring of a CES patient is difficult to predict. Considerable overlap can exist between CES and other syndromes with anal, auricular, heart and urogenital anomalies, such as Townes–Brocks syndrome (anal, auricular, heart and urogenital anomalies), caused by mutations in SALL1, oculoauriculovertebral spectrum/Goldenhar syndrome (ophthalmological, auricular, heart and renal anomalies) or VACTERL association (anal, heart and renal anomalies). These syndromes are also highly variable.11
In this study, we report a SALL1-mutation negative, three-generation family in which four members carry a 600 kb intrachromosomal partial triplication of the CES region, causing a partial tetrasomy of the CES critical region. The amplified region is smaller than the currently accepted critical region. There is phenotypic variation in this family, but all carrier members show phenotypic overlap with CES. The influence of the amplified and non-amplified genes of the CES critical region to the phenotype in the reported patients is discussed. The presented data may contribute to the knowledge of gene-specific causality of CES features.
Materials and methods
Clinical evaluation of patient and family
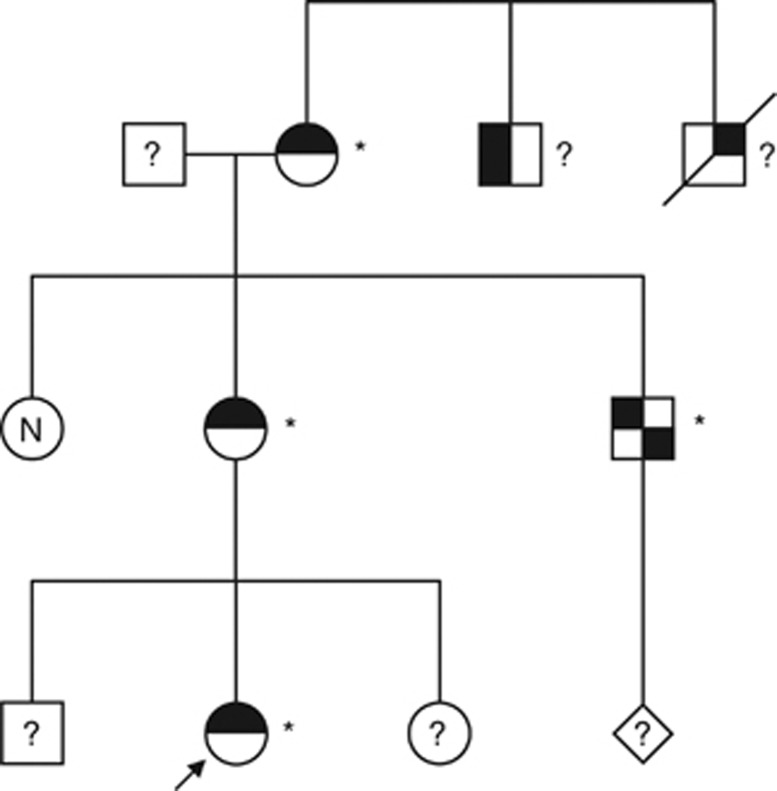
The index patient was a newborn girl with vestibular anus and a preauricular ear pit at the left. Both the mother and the maternal grandmother had surgery for anal atresia and preauricular ear tags. The latter was also found in a brother of the mother who was known with and treated for idiopathic growth hormone deficiency and who had a unilateral kidney agenesis. A brother of the grandmother was known with ‘Goldenhar' syndrome with deafness, facial asymmetry, microtia, absence of one auditory canal and was described as ‘simple'. Another brother died neonatally and probably also had an anal anomaly (Figure 1).
Figure 1.
Family pedigree, arrow indicates index patient. Carriers of the 22q11.1q11.21 triplication are indicated with an asterisk (*). Untested family members are indicated with a question mark and ‘N' stands for ‘not a carrier' of the triplication. Individuals with a non-filled symbol have no phenotypic characteristics of CES. A black upper-right quadrant stands for anal malformation, the black upper-left quadrant stands for preauricular anomalies, the black lower-right quadrant stands for renal anomalies and the black lower left quadrant stands for deafness. The uncle of the index patient also had idiopathic growth hormone deficiency; this is not indicated in this figure.
None of the affected had congenital cardiac anomalies or was known with eye abnormalities. Ophthalmological examination was performed in the proband, her two brothers, her mother and the maternal grandmother. The granduncle with Goldenhar syndrome was not available for examination.
Cytogenetic and molecular evaluations
Chromosome analysis using GTG banding was performed on stimulated peripheral blood cultures of the index patient using standard methods. Extraction of DNA from peripheral blood cells of the investigated family members was performed according to standard techniques. MLPA was performed using the commercially available SALSA MLPA kit P250-B1 (MRC-Holland, Amsterdam, The Netherlands), containing probes specific to genomic regions that are most probably involved in Velocardiofacial syndrome, DiGeorge syndrome, 22q11.2 duplication syndrome, distal 22q11.2 deletion syndrome, 22q13.3 deletion syndrome and CES. The MLPA experiments were performed according to the manufacturer's instructions.
FISH analysis of the CES region was performed in the index patient with BAC clones RP11-155N18 and RP11-958H20, located in 22q11.1 and 22q11.1q11.21, respectively, and together covering about 470 kb of the CES critical region,12, 13 including the genes CECR1, CECR2, CECR4, CECR5, CECR6 and IL17RA. FISH experiments were performed using standard procedures14 with alphoid probe p190.22 as a control probe for centromere 22.15
Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA) was performed in the index patient according to the manufacturer's protocols. Analysis of the data was performed using the Affymetrix software packages Genotyping Console (GTC) and Chromosome Analysis Suite (ChAS). The result was compared in silico with pooled data of 94 reference samples, consisting of in-house generated normal control samples.
Results
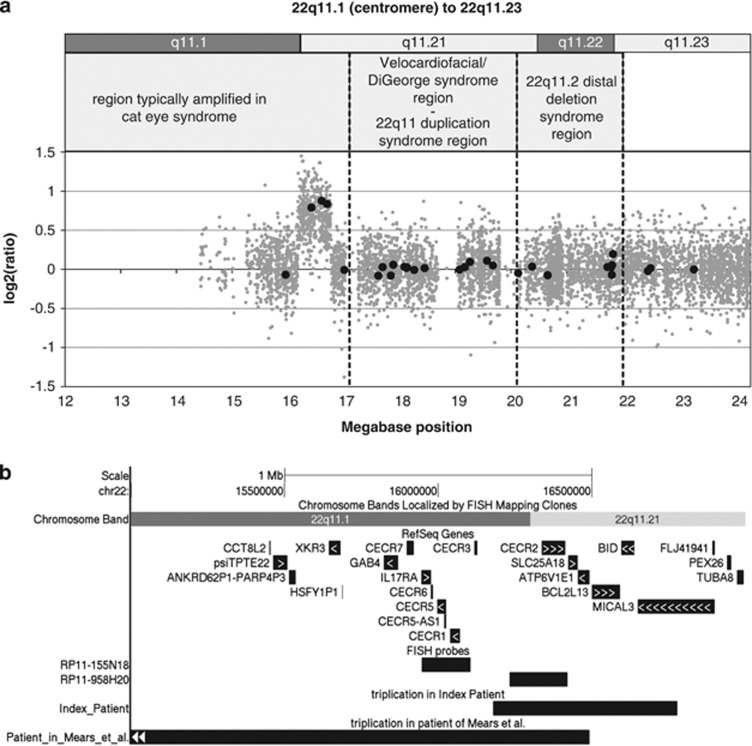
Based on a clinical suspicion for a possible rearrangement of the CES region, MLPA screening of the index patient showed a partial gain of this 22q11 region. Three of the five probes specific for the area (ie, SLC25A18, BID and MICAL3) showed a gain of signal ratio, suggesting a copy number of four. The most proximal probe for the CES region in the kit (IL17RA) showed a normal ratio, as did the most distal probe for the region (USP18) and all other chromosome 22 probes distal to the three amplified probes (Figure 2a).
Figure 2.
(a) MLPA result (black closed circles) and SNP array result (grey closed circles) of the 22q11.1 to 22q11.23 region in the index patient. The different syndromes, including CES, linked to this chromosomal region are displayed above. (b) Overview of the genes in the CES critical region and the locations of the BAC probes RP11-155N18 and RP11-958H20 used for FISH, together with the location of the triplicated region in the index patient and the triplication of the patient of Mears et al. Arrows within the genes indicate the transcriptional direction.
Standard karyotyping followed the MLPA analysis to investigate whether the partial chromosome 22 amplification was caused by the presence of a supernumerary marker chromosome. GTG banding revealed a microscopically normal female karyotype in 10 metaphases.
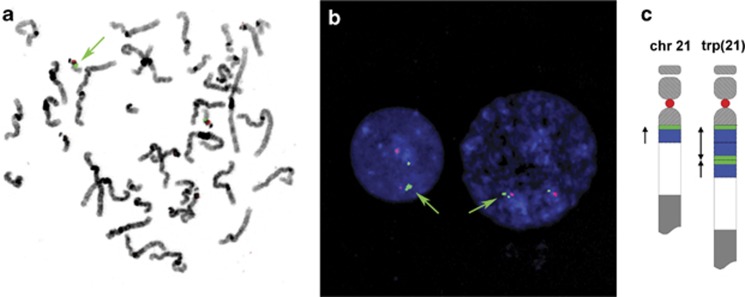
Metaphase FISH analysis using two probes specific for the CES region showed specific signals only in band q11 of both chromosomes 22. In some of the metaphases, the RP11-958H20 signal seemed larger on one chromosome 22 than on the other homolog, but signals were not split (Figure 3b). RP11-155N18 showed no clear difference in signal strength between both homologues.
Figure 3.
(a) Metaphase spread and (b) interphase nuclei, counterstained with DAPI, after hybridization with RP11-958H20 (green) and p190.22 (red, specific for the centromeric region of chromosome 22). Green arrows show a larger signal for RP11-958H20 in the metaphase chromosomes and a split signal in the interphase nuclei, in which the signal is split in a larger and a smaller signal. (c) Schematic representation of the proposed arrangement of the triplication in chromosome 22 with in red and green the location of the FISH probes used in panels (a, b).
Interphase FISH analysis, however, showed two copies of RP11-155N18 with different intensity in each nucleus, whereas 12 out of 30 nuclei showed a split signal of the larger of two signals for RP11-958H20 (Figure 3b), resulting in three signals in the interphase cell. In the cells showing three signals, additional intensity difference between the signals was observed. The resulting karyotype was communicated as 46,XX.ish 22q11.1q11.21(RP11-958H20,RP11-155N18)x2[10].nuc ish 22q11.1(RP11-155N18x2)[29],22q11.21(RP11-958H20x3)[12/30].
Subsequently, SNP array analysis was performed with targeted analysis of the 22q11 region for determination of the exact amplification size. The array showed an amplification of 421 probes in a region of 598 kb, starting at 16 179 398 bp in band q11.1 and ending at 16 777 897 bp in band q11.21 (genome build NCBI36/hg18. Genome build GRCh37/hg19: position 17 799 398 to 18 397 897 bp) with an estimated copy number of 4, in concordance with the MLPA result (Figure 2a). The amplification involved the genes CECR2, SLC25A18, ATP6V1E1, BCL2L13, BID, all completely and MICAL3 partially (Figure 2b).
Evaluation of other family members with phenotypic characteristics of CES using the 22q11 MLPA test showed inheritance of the amplification in four family members and three generations (Figure 1), with similar size and signal ratio.
Discussion
We describe four family members from three generations in whom an atypical gain of a part of the CES critical region was found using MLPA. The signal ratio of the aberrant probes indicated a tetrasomy of this part of chromosome 22 (Figure 2a). Standard karyotyping excluded the presence of a supernumerary marker chromosome, usually found in CES. Metaphase FISH analysis excluded an insertion of the amplified fragment into another chromosome or chromosome region. Consequently, we concluded that there is a local amplification in one of the chromosomes 22. This was supported by interphase FISH results, because the larger of the two signals for probe RP11-958H20 was split in two in 12 out of 30 nuclei (Figure 3b).
Using high-resolution SNP Array, we found that the amplification was confined to the distal part of the CES critical region and proved to be about 600 kb (Figure 2a). The four signals, as could be expected in interphase FISH based on the MLPA and array results, were not observed, probably because of close physical proximity of the different copies on the aberrant chromosome 22. In about half of the investigated nuclei, three signals for BAC probe RP11-958H20 were observed. There was a clear difference in intensity of the three signals, as shown in Figure 3b. BAC probe RP11-958H20 is located at the proximal end of the 600 kb amplified fragment. Therefore, we conclude not only that three copies of the fragment are present in the same locus of the aberrant chromosome 22, but also that the middle fragment is in inverted orientation compared with the flanking two, as proposed in Figure 3c. Consequently, this will lead to one separated signal and one combined, double signal. The fact that three copies of the 600 kb fragment are present on the same chromosome is also supported by the (autosomal dominant) inheritance pattern in the family (Figure 1).
The currently accepted CES critical region is based on one patient with all cardinal features of CES caused by an atypical supernumerary ring chromosome 22.13 This atypical supernumerary marker chromosome did not involve amplification of the directly distally located genes BCL2L13, BID and MICAL3 (Figure 2b). However, these genes are amplified in the more frequently occurring standard type supernumerary marker chromosome in patients with CES. As the original patient described by Mears et al13 died at 17 days of life, a contribution to physical and mental development of these distally located genes BCL2L13, BID and MICAL3 cannot be ruled out.12
In the amplification found in the current family, six genes, CECR2, SLC25A18, ATP6V1E1, BCL2L13, BID and MICAL3 (latter only partially amplified), are involved. The family members carrying the intrachromosomal amplification have anal atresia (3 out of 4) and/or preauricular pits or tags (4 out of 4). As these congenital anomalies are frequently found in CES and as only three of the genes involved in our family overlap with the genes involved in the amplification reported by Mears et al13 (CECR2, SLC25A18 and ATP6V1E1) (Figure 2b), anal atresia and preauricular pits or tags may be caused by amplification and/or overexpression of one or more of these three genes. The same genes might also be responsible for the growth hormone deficiency and the unilateral kidney agenesis, as was seen in the uncle of the index patient, because short stature and kidney anomalies are also part of the phenotypic spectrum of CES. A CES patient with low serum growth hormone and hypogonadotrophic hypogonadism was reported before.16 Additionally, we cannot rule out that even the deafness and the congenital facial anomalies in the granduncle of the index patient, earlier clinically diagnosed as Goldenhar syndrome, is caused by the same familial triplication. Unfortunately, the granduncle of the index patient refused further testing for possible carrier status. These genes could also be involved in other phenotypic features of CES that may be non-penetrant in this family.
Mutations in the gene CECR2 trigger dysregulation of mesenchymal or ectodermal transcription factors and the protein is described to contribute to neurogenesis and inner ear development.17, 18 SLC25A18 catalyzes unidirectional transport of glutamate in the inner membranes of mitochondria.19 ATP6V1E1 codes for the regulatory accessory subunit E of the vacuolar-type H(+)-ATPase, which is responsible for acidification of intracellular organelles and involved in hydrogen ion transport across the plasma membrane. Mutations in other subunits of the V-ATPase complex have shown to be involved in distal renal tubular acidosis and sensorineural deafness.20
In conclusion, CECR2, SLC25A18 and ATP6V1E1 are strong candidate genes in causing anal atresia, preauricular pits or tags and kidney anomalies in the described family members and consequently in causing these frequent components of the CES phenotype spectrum.
The authors declare no conflict of interest.
References
- Belangero SI, Bellucco FT, Cernach MC, Hacker AM, Emanuel BS, Melaragno MI.Interrupted aortic arch type B in A patient with cat eye syndrome Arq Bras Cardiol 200992e29–e31.e56-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berends MJ, Tan-Sindhunata G, Leegte B, van Essen AJ. Phenotypic variability of cat-eye syndrome. Genet Couns. 2001;12:23–34. [PubMed] [Google Scholar]
- Rosias PR, Sijstermans JM, Theunissen PM, et al. Phenotypic variability of the cat eye syndrome. Case report and review of the literature. Genet Couns. 2001;12:273–282. [PubMed] [Google Scholar]
- Buhler EM, Mehes K, Muller H, Stalder GR. Cat-eye syndrome, a partial trisomy 22. Humangenetik. 1972;15:150–162. doi: 10.1007/BF00295742. [DOI] [PubMed] [Google Scholar]
- Schinzel A, Schmid W, Fraccaro M, et al. The ‘cat eye syndrome': dicentric small marker chromosome probably derived from a no.22 (tetrasomy 22pter to q11) associated with a characteristic phenotype. Report of 11 patients and delineation of the clinical picture. Hum Genet. 1981;57:148–158. doi: 10.1007/BF00282012. [DOI] [PubMed] [Google Scholar]
- Reiss JA, Weleber RG, Brown MG, Bangs CD, Lovrien EW, Magenis RE. Tandem duplication of proximal 22q: a cause of cat-eye syndrome. Am J Med Genet. 1985;20:165–171. doi: 10.1002/ajmg.1320200120. [DOI] [PubMed] [Google Scholar]
- Knoll JH, Asamoah A, Pletcher BA, Wagstaff J. Interstitial duplication of proximal 22q: phenotypic overlap with cat eye syndrome. Am J Med Genet. 1995;55:221–224. doi: 10.1002/ajmg.1320550214. [DOI] [PubMed] [Google Scholar]
- Meins M, Burfeind P, Motsch S, et al. Partial trisomy of chromosome 22 resulting from an interstitial duplication of 22q11.2 in a child with typical cat eye syndrome. J Med Genet. 2003;40:e62. doi: 10.1136/jmg.40.5.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay EA, Shaffer LG, Carrozzo R, Greenberg F, Baldini A. De novo tandem duplication of chromosome segment 22q11-q12: clinical, cytogenetic, and molecular characterization. Am J Med Genet. 1995;56:296–299. doi: 10.1002/ajmg.1320560316. [DOI] [PubMed] [Google Scholar]
- Luleci G, Bagci G, Kivran M, Luleci E, Bektas S, Basaran S. A hereditary bisatellite-dicentric supernumerary chromosome in a case of cat-eye syndrome. Hereditas. 1989;111:7–10. doi: 10.1111/j.1601-5223.1989.tb00369.x. [DOI] [PubMed] [Google Scholar]
- Rosa RF, Mombach R, Zen PR, Graziadio C, Paskulin GA. Clinical characteristics of a sample of patients with cat eye syndrome. Rev Assoc Med Bras. 2010;56:462–465. doi: 10.1590/s0104-42302010000400021. [DOI] [PubMed] [Google Scholar]
- Footz TK, Brinkman-Mills P, Banting GS, et al. Analysis of the cat eye syndrome critical region in humans and the region of conserved synteny in mice: a search for candidate genes at or near the human chromosome 22 pericentromere. Genome Res. 2001;11:1053–1070. doi: 10.1101/gr.154901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears AJ, el-Shanti H, Murray JC, McDermid HE, Patil SR. Minute supernumerary ring chromosome 22 associated with cat eye syndrome: further delineation of the critical region. Am J Hum Genet. 1995;57:667–673. [PMC free article] [PubMed] [Google Scholar]
- van der Veken LT, Dieleman MM, Douben H, et al. Low grade mosaic for a complex supernumerary ring chromosome 18 in an adult patient with multiple congenital anomalies. Mol Cytogenet. 2010;3:13. doi: 10.1186/1755-8166-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi M, Archidiacono N, Antonacci R, et al. Cloning and comparative mapping of recently evolved human chromosome 22-specific alpha satellite DNA. Somat Cell Mol Genet. 1994;20:443–448. doi: 10.1007/BF02257462. [DOI] [PubMed] [Google Scholar]
- Masukawa H, Ozaki T, Nogimori T. Cat eye syndrome with hypogonadotropic hypogonadism. Internal Med. 1998;37:853–856. doi: 10.2169/internalmedicine.37.853. [DOI] [PubMed] [Google Scholar]
- Fairbridge NA, Dawe CE, Niri FH, Kooistra MK, King-Jones K, McDermid HE. Cecr2 mutations causing exencephaly trigger misregulation of mesenchymal/ectodermal transcription factors. Birth Defects Res A Clin Mol Teratol. 2010;88:619–625. doi: 10.1002/bdra.20695. [DOI] [PubMed] [Google Scholar]
- Dawe CE, Kooistra MK, Fairbridge NA, Pisio AC, McDermid HE. Role of chromatin remodeling gene Cecr2 in neurulation and inner ear development. Dev Dyn. 2011;240:372–383. doi: 10.1002/dvdy.22547. [DOI] [PubMed] [Google Scholar]
- Fiermonte G, Palmieri L, Todisco S, Agrimi G, Palmieri F, Walker JE. Identification of the mitochondrial glutamate transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J Biol Chem. 2002;277:19289–19294. doi: 10.1074/jbc.M201572200. [DOI] [PubMed] [Google Scholar]
- Stover EH, Borthwick KJ, Bavalia C, et al. Novel ATP6V1B1 and ATP6V0A4 mutations in autosomal recessive distal renal tubular acidosis with new evidence for hearing loss. J Med Genet. 2002;39:796–803. doi: 10.1136/jmg.39.11.796. [DOI] [PMC free article] [PubMed] [Google Scholar]