Abstract
The pathogenesis of Alzheimer’s disease (AD) is associated with proteolytic processing of the amyloid precursor protein (APP) to an amyloidogenic peptide termed Aβ. Although mutations in APP and the secretase enzymes that mediate its processing are known to result in familial forms of AD, the mechanisms underlying the more common sporadic forms of the disease are still unclear. Evidence suggests that the susceptibility of APP to amyloidogenic processing is related to its intracellular localization, and that secretase-independent degradation may prevent the formation of cytotoxic peptide fragments. Recently, single nucleotide polymorphisms in the UBQLN1 gene have been linked to late-onset AD, and its protein product, ubiquilin-1, may regulate the maturation of full-length APP. Here we show that ubiquilin-1 inhibits the maturation of APP by sequestering it in the early secretory pathway, primarily within the Golgi apparatus. This sequestration significantly delayed the proteolytic processing of APP by secretases and the proteasome. These effects were mediated by ubiquilin-1–stimulated K63-linked polyubiquitination of lysine 688 in the APP intracellular domain. Our results reveal the mechanistic basis by which ubiquilin-1 regulates APP maturation, with important consequences for the pathogenesis of late-onset AD.
Keywords: trafficking, neurodegeneration, ubiquitin-associated domain, ubiquitin-like domain
Amyloid precursor protein (APP) is a type I transmembrane protein that matures in the secretory pathway, where it undergoes classical N- and O-linked glycosylation during transit through the endoplasmic reticulum (ER) and Golgi, respectively (1, 2). Though most APP is degraded by lysosomes (3), a small portion reaches the cell surface to be cleaved by the secretases (4). Amyloidogenic processing of APP occurs by sequential processing by β- and γ-secretase to release Aβ peptide and the APP intracellular domain (AICD) (5). APP half-life is short, and thus the amount of APP detected at the cell surface is very low (6). The rapid internalization rate of APP is due to the presence of the highly conserved YENPTY (single amino acid code) sequence in the APP cytoplasmic domain, which contains a canonical NPxY internalization signal for clathrin-mediated endocytosis (7). Proteolytic processing of APP has been shown to occur in various sites throughout the secretory and endocytic pathways. However, amyloidogenic processing primarily occurs after transition through the Golgi apparatus (8) and in endosomal compartments, where acidic conditions promote optimal activity of the beta-site APP cleaving enzyme (BACE) or β-secretase (9). Deletion or mutation of the YENPTY internalization signal leads to an increase in plasma membrane-associated APP and a significant decrease in Aβ production, underscoring the importance of endocytic recycling for Aβ generation (5, 7, 10). Various cytoplasmic adaptor proteins have been shown to interact with the YENPTY motif and regulate sorting of APP to different cell compartments and thus regulate its proteolytic processing. The protein SorLA or LR11 (11) acts as a sorting receptor that traps APP in the Golgi, reducing the amount of APP that reaches the cell surface for processing. SorLA also shuttles APP from the early endosomes back to the Golgi, further reducing Aβ production in late endosomes (3, 12). Clathrin coat proteins and adaptor proteins JIP1b, Fe65, retromer, nexin, homer2, homer3, Numb, and X11 family members also regulate sorting of APP through the endocytic pathway by interacting with the intracellular tail of APP (3, 6, 9, 13).
Production of Aβ may also be suppressed by factors that target full-length APP or its C-terminal fragments to the proteasome or lysosome, resulting in γ-secretase–independent degradation (14–16). Recent studies have suggested a role for ubiquilin-1 in the regulation of APP trafficking and processing (17). Specifically, reduction in proteins levels of ubiquilin-1 accelerated the maturation of APP through the secretory pathway, which was associated with increased secretion of APPs and increased production of Aβ (17). Here we show that ubiquilin-1 can sequester APP in the Golgi apparatus and delay its access to the cell surface and subsequent proteolytic processing. We found that ubiquilin-1 suppresses APP maturation and proteasomal degradation by stimulating APP lysine 63 (K63)-linked polyubiquitination. We also found that mutation of lysine 688 arginine (K688R) in the APP intracellular domain abrogated ubiquitination and Golgi sequestration, suggesting that nondegradative ubiquitination of APP by ubiquilin-1 is a critical sorting signal limiting APP amyloidogenic processing. Combined with our previous observations that ubiquilin-1 functions as a molecular chaperone and its levels are reduced in Alzheimer’s disease (AD) brains (18), these results indicate that ubiquilin-1 is a critical quality-control molecule regulating APP biosynthesis, trafficking, and degradation.
Results
Ubiquilin-1 Sequesters APP in the Early Secretory Pathway.
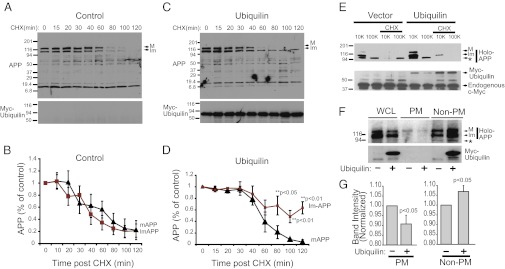
To investigate the possible effects of exogenous expression of ubiquilin-1 on APP degradation, we monitored the degradation rate of endogenous APP and APP fragments in PC12 cells treated with the protein synthesis inhibitor cycloheximide (CHX). Trafficking through the Golgi was monitored by the transition of APP from an immature form, which migrates faster on SDS/PAGE, to a mature more slowly migrating form that has undergone O-linked glycosylation (1, 8). CHX treatment resulted in a time-dependent loss of immature APP and mature APP levels with a half-life of ∼40 min (Fig. 1 A and B). In cells overexpressing ubiquilin-1, the mature form of APP is degraded at approximately similar rates; however, the immature form of APP persists throughout the time course (Fig. 1 C and D). These results suggest that ubiquilin-1 may be modulating APP maturation by sequestering immature APP in early secretory compartments. To explore this possibility, subcellular fractionation experiments were performed to examine the compartmentalization of mature and immature APP in the presence and absence of ubiquilin-1 overexpression. PC12 cells were treated with CHX or vehicle for 2 h before collection. The lysates were subjected to differential centrifugation and separated into 10,000 × g pellets (containing heavy membranes such as rough ER, plasma membrane, and high-density Golgi) and 100,000 × g pellets (containing light membranes such as smooth ER, low-density Golgi, and small vesicles). In vector-only transfected cells, the 10,000 × g pellets contained immature, and a small amount of the mature form of, endogenous APP (Fig. 1E). In control cells, addition of CHX for 2 h resulted in the loss of mature and immature APP in both fractions. In contrast, in PC12 cells overexpressing ubiquilin-1, there was in increased amount of mature APP in the 10,000 × g pellet. Furthermore, the immature form of APP persisted in the 10,000 × g pellet after CHX treatment, suggesting that ubiquilin-1 may stabilize APP in light membrane compartments such as the Golgi. An additional band with slightly faster mobility (marked by an asterisk) was observed in fractionation experiments but not in whole-cell lysates, which may correspond to APP before N-linked glycosylation.
Fig. 1.
Ubiquilin-1 inhibits APP maturation. (A) Degradation rate of mature (M) and immature (Im) levels of endogenous APP in PC12 cells. Cells were treated with CHX or vehicle and collected at the indicated time points (in minutes). (B) Quantification of three separate experiments as in A represented as the mean ± SEM. (C) Degradation of APP in cells expressing ubiquilin-1 as in A. (D) Quantification from three separate experiments as in B. P values are shown. In A and C, ubiquilin expression was determined by blotting for the myc epitope present on the N terminus of the protein. (E) Subcellular fractionation and endogenous APP distribution in PC12 cells overexpressing ubiquilin-1 or empty-vector transfected cells. Cells were treated with CHX for 2 h, and lysates were prepared and subjected to differential centrifugation. Mature and immature APP bands are identified with an arrow. The asterisk (*) indicates an unidentified reactive band that has faster mobility than immature APP, possibly full-length APP that has not undergone N-linked glycosylation. 10K, 10,000 × g pellet; 100K, 100,000 × g pellet. Equal protein loading was determined by examining endogenous levels of c-Myc (Lower). (F) Biotinylation experiments to examine cell-surface expression of endogenous APP in cells expressing ubiquilin-1 or transfected with empty vector. WCL, whole-cell lysate; PM, plasma membrane; non-PM, nonplasma membrane. (G) Quantification of PM and non-PM APP levels pooled from four experiments.
Sequestration of APP in the secretory pathway would prevent trafficking of APP to the cell surface. To explore this possibility, cell-surface biotinylation experiments were performed to determine if ubiquilin-1 overexpression was associated with decreased plasma membrane expression of APP (Fig. 1 F and G). We observed that ubiquilin-1 overexpression caused a significant decrease in plasma membrane-associated APP, and this decrease was concomitant with an enrichment of both mature and immature APP in the nonplasma membrane fraction.
Ubiquilin-1 Delays the Proteolytic Processing of APP.
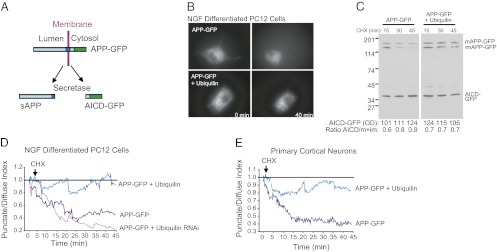
To study trafficking and degradation of APP in real-time, we used live-cell imaging of PC12 cells expressing an APP–GFP fusion construct consisting of a GFP moiety fused to the C terminus of full-length APP (Fig. 2A). Upon APP processing by the secretases, the AICD–GFP fusion is released from the membrane, and the fluorescence pattern changes from punctate to diffuse. Before addition of CHX, a punctate pattern of GFP fluorescence was observed in cells expressing APP–GFP (Fig. 2B, Upper Left), corresponding to a predominantly vesicular localization of APP–GFP, as expected. After addition of CHX, this punctate fluorescence was gradually replaced by diffuse cytosolic fluorescence over time, reflecting secretase processing (Fig. 2B, Upper Right and Movie S1). In contrast, in cells coexpressing APP–GFP and ubiquilin-1, the punctate pattern of fluorescence persisted for much longer after the addition of CHX (Fig. 2B, Lower and Movie S2), suggesting that ubiquilin-1 is able to delay the secretase processing of APP. To confirm that the transition from punctate to diffuse fluorescence was indeed indicative of APP–GFP processing, expression of APP–GFP and AICD–GFP was monitored by Western blotting. As shown in Fig. 2C Left, CHX treatment resulted in a loss of mature and immature APP–GFP, and a corresponding increase in AICD–GFP in cells that were not overexpressing ubiquilin-1. In contrast, ubiquilin-1 overexpression partially suppressed APP–GFP degradation and, more importantly, decreased the formation of AICD–GFP after CHX treatment (Fig. 2C, Right). Thus, the transition from punctate to diffuse fluorescence as determined in Fig. 2B is indicative of the secretase processing of APP–GFP. The transition from punctate to diffuse fluorescence can be quantified as the punctate/diffuse index (19). As shown in Fig. 2D, APP–GFP has a reduction in the punctate/diffuse index over time after CHX addition, indicating APP processing and liberation of GFP–AICD, as expected. In contrast, overexpression of ubiquilin-1 almost completely abrogates APP–GFP processing, whereas ubiquilin-1 knockdown accelerated APP–GFP processing (Fig. 2D). Essentially identical results were obtained with primary cortical neurons (Fig. 2E). These results are consistent with our previous studies showing that ubiquilin-1 expression inhibits the production of Aβ (18).
Fig. 2.
Ubiquilin-1 inhibits APP–GFP trafficking and secretase processing. (A) Schematic representation of the membrane topology of the APP–GFP construct used in this study. Secretase processing is predicted to release AICD–GFP from the membrane into the cytosol (resulting in diffuse fluorescence). (B) Fluorescent images showing APP–GFP distribution in NGF-differentiated PC12 cells before and after 40 min of CHX treatment in cells not expressing ubiquilin-1 (Upper) and cells expressing ubiquilin-1 (Lower). See also Movies S1 and S2. (C) Expression of mature APP–GFP, immature APP–GFP, and AICD–GFP in PC12 cells after CHX treatment. Optical-density AICD–GFP expression levels and the ratio of AICD–GFP to full-length APP expression are given below the blot. Note increasing ratio in APP–GFP-expressing cells, which is eliminated in cells overexpressing ubiquilin-1. (D) Quantification of the change in fluorescence from punctate to diffuse over time (punctate/diffuse index) in single PC12 cells. A reduction in this index indicates more homogeneous (i.e., diffuse) distribution of fluorescence. Solid line indicates no change. The ubiquilin-1 RNAi oligonucleotides have been described and validated elsewhere (18). (E) Punctate/diffuse index of APP–GFP in primary rat cortical neurons as in C. Similar results were seen in three independent experiments.
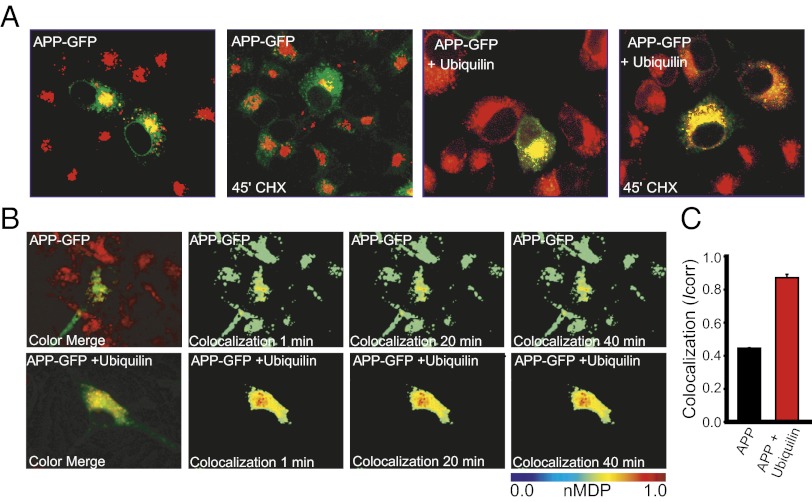
Because our results suggest that ubiquilin-1 may be regulating the exit of APP from the Golgi apparatus, we wished to analyze APP localization within Golgi compartments. We generated a stable cell line expressing the Golgi-resident enzyme N-acetylglucosaminyltransferase 1 (GlcNAc-T1) (20) fused to the mCherry red fluorescent protein, and transiently expressed APP–GFP in this cell line (Fig. 3A). Cells were treated with CHX or vehicle for 45 min, and subcellular localization was analyzed by confocal microscopy. Treatment with CHX resulted in a considerable reduction of colocalization of the two proteins, indicating maturation and exit of APP from the Golgi (Fig. 3A, Left and Center Left). In cells coexpressing APP–GFP and ubiquilin-1, colocalization persists throughout the time course (Fig. 3A, Right and Center Right). To analyze real time-colocalization of APP–GFP with mCherry–GlcNAc-T1, we performed live-cell imaging of single cells treated with CHX and quantified the colocalization by calculating the correlation index (Icorr) (21). In cells expressing APP–GFP and GlcNAc-T1, there was modest colocalization of the two proteins, which was dramatically increased by ubiquilin-1 overexpression (Fig. 3B). Quantification of Icorr indicated that ubiquilin-1 overexpression increased colocalization of APP–GFP and GlcNAc-T1 from ∼40% to almost 90% after 40 min of CHX treatment. Thus, ubiquilin-1 can function to retain APP in the Golgi apparatus.
Fig. 3.
Ubiquilin-1 colocalizes with and retains APP–GFP in the Golgi apparatus. (A) Confocal fluorescent images of APP–GFP localization in GlcNAc-T1–mCherry-expressing PC12 cells treated with CHX for 45 min with or without ubiquilin-1 overexpression, as indicated. (B) APP–GFP and GlcNAc-T1–mCherry colocalization without (Upper) or with (Lower) ubiquilin-1 overexpression as determined by the normalized mean deviation product (nMDP) (21) in living cells treated with CHX at time 0. (Upper Left and Lower Left) Raw, wide-field fluorescent image at time 0; other panels are nMDP values. nMDP panels are pseudocolored such that low values are cool colors, and high values are warmer colors (see scale bar). (C) Quantification of colocalization (Icorr) after 40 min CHX treatment. Data are presented as the mean ± SEM of 10 separate cells.
Ubiquilin-1 Inhibits Degradation of APP by Nonlysosomal Pathways.
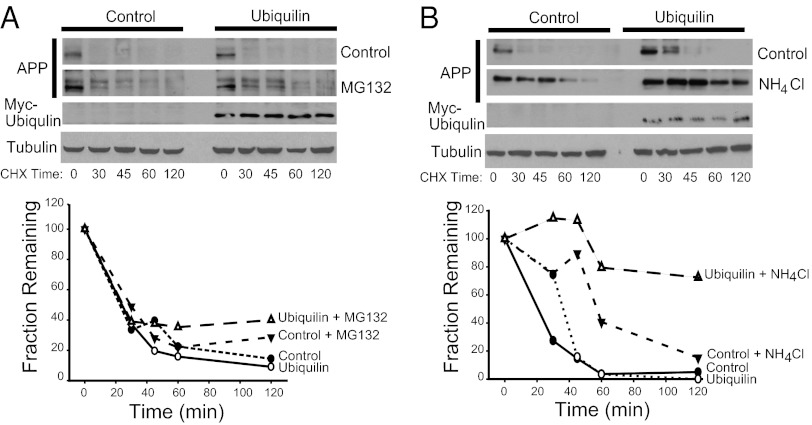
Our results indicate that ubiquilin-1 sequesters APP in the Golgi (Figs. 1 and 3) and delays secretase-dependent processing (Fig. 2). Ubiquilin-1 and other members of the ubiquitin-like (UBL)/ubiquitin-associated (UBA) family of proteins have been shown to regulate the degradation of various substrates (22–24). To further investigate whether ubiquilin-1 regulates the proteolysis of APP, its ability to modulate the half-life of endogenous APP was examined in the presence of the proteasome inhibitor MG132 or the lysosomal inhibitor NH4Cl. As shown in Fig. 4A, APP was rapidly degraded in the absence of MG132, and this degradation was partially attenuated by proteasome inhibition. Ubiquilin-1 overexpression had little effect on APP degradation in the presence of MG132. When the cells were treated with the lysosomal inhibitor ammonium chloride (NH4Cl), the degradation rate of APP was significantly delayed (Fig. 4B). These results are consistent with previous studies showing that a large proportion of APP is degraded in acidic compartments, including lysosomes (14, 15). Overexpression of ubiquilin-1 almost completely inhibited APP degradation in the presence of the lysosome inhibitor (Fig. 4B). This result suggests that nonlysosomal degradation of APP (as determined by half-life in the presence of NH4Cl) is significantly inhibited by ubiquilin-1 overexpression. Presumably the nonlysosomal degradation inhibited by ubiquilin-1 overexpression is secretase processing (as in shown in Fig. 2) and/or combined secretase and proteasomal degradation.
Fig. 4.
Ubiquilin-1 inhibits nonlysosomal degradation of APP. (A) Time course of APP degradation after addition of cycloheximide in the presence or absence of the proteasomal inhibitor MG132 (Upper). (Lower) Quantification of APP band intensities in an experiment representative of three separate determinations. (B) Time course of APP degradation after cycloheximide addition in the absence or presence of lysosomal inhibitor NH4Cl (Upper). (Lower) Quantification of band intensities in an experiment representative of three separate determinations. Tubulin is used as a loading control.
Ubiquilin-1 Regulates APP Trafficking and Degradation via Ubiquitination.
We next tested whether ubiquilin-1 regulates APP trafficking and degradation by stimulating ubiquitination. Polyubiquitin chains conjugated to lysine 29 (K29) or lysine (K48) of ubiquitin target a substrate protein for proteasomal degradation. However, lysine 63 (K63)-linked polyubiquitination and monoubiquitination are implicated in many nondegradative processes, such as protein trafficking, endocytosis, and DNA repair (25, 26). PC12 cells were transfected with HA-ubiquitin with or without ubiquilin-1 and MG132, and APP ubiquitination was investigated. As shown in Fig. 5A, ubiquilin-1 stimulated K63-linked polyubiquitination of APP as determined by linkage-specific ubiquitin antibodies. As an alternative approach to confirm linkage specificity, we used HA-ubiquitin constructs with all lysines mutated to arginine except for either K48 or K63. As shown in Fig. 5B, ubiquilin-1 overexpression potently stimulated K63-linked polyubiquitination of APP, and to a lesser extent, K48-linked polyubiquitination.
Fig. 5.
Ubiquilin-1 regulates APP trafficking and degradation through K63-linked polyubiquitination of lysine residue 688. (A) Ubiquitination of endogenous immunoprecipitated APP as determined by linkage-specific antibodies in the presence or absence of MG132 in cells coexpressing HA-ubiquitin (HA-Ubq) plus ubiquilin-1 or a control vector as indicated at the top. (B) Ubiquitination of endogenous APP in the presence of MG132 in cells coexpressing wild-type or mutant HA-ubiquitin constructs in which all lysine residues are mutated to arginine except K63 or K48. (C) Ubiquitination of overexpressed wild-type APP or a mutant in which lysine 688 is mutated to arginine (K688R). All lanes were preincubated with MG132. (D) Subcellular distribution of wild-type overexpressed APP in the presence or absence of ubiquilin-1 expression and the effects of CHX treatment as indicated. Expression of the ER-resident inositol 1,4,5-trisphosphate receptor type 1 (IP3R-1) is used as a loading control. (E) Subcellular distribution of K688R APP in the presence or absence of ubiquilin-1 expression and the effects of CHX treatment. (F) Confocal fluorescence images showing the subcellular distribution of wild-type and K688R APP–GFP before and 45 min after CHX treatment.
Ubiquilin-1 is a cytosolic protein and binds to the AICD (18). The AICD has five lysine residues that could potentially be ubiquitinated. We mutated all five lysine residues to arginine individually to determine which residue is targeted for ubiquitination by ubiquilin-1. One mutant, K688R, had significantly reduced ubiquilin-1–stimulated ubiquitination of APP (Fig. 5C). We reasoned that if ubiquilin-1 regulates APP trafficking by stimulating ubiquitination of K688, then ubiquilin-1 overexpression would have no effect on the subcellular localization of the K688R APP mutant. Thus, we examined the ability of ubiquilin-1 to affect the subcellular localization of overexpressed wild-type and K688R APP using subcellular fractionation. As shown in Fig. 5D, ubiquilin-1 increases the amount of overexpressed wild-type APP in both 10,000 and 100,000- × g fractions, which is consistent with the effects of ubiquilin-1 overexpression on endogenous APP shown in Fig. 1E. In contrast, ubiquilin-1 overexpression had no effect on the subcellular distribution of the K688R mutant (Fig. 5E). Strikingly, there was a complete absence of K688R mutant in the 100,000 × g pellet fraction, suggesting a dramatically altered subcellular localization. This was examined (as above) using APP–GFP and APP–GFP K688R mutant in GlcNAc-T1–mCherry cells treated with CHX. As shown in Fig. 5F, the K688R mutant had dramatically reduced Golgi colocalization both before and after CHX treatment, suggesting that ubiquitination of this residue by ubiquilin-1 is a critical regulator of APP retention in the Golgi apparatus.
Discussion
In the present study, we show that ubiquilin-1 delays the maturation of APP by sequestering it in the Golgi apparatus and preventing its transport to the cell surface for subsequent processing. Using a combination of biochemical and cell biological methodologies, our data suggests that ubiquilin-1 inhibits APP maturation and secretase processing by ubiquitinating lysine residue 688. These results indicate that ubiquilin-1 is a potent regulator of APP trafficking and secretase processing, and supports a model in which age-associated decreases in ubiquilin-1 levels as shown previously by our group (18) contribute to increased Aβ production in late-onset AD. Our results are in agreement with previous studies showing that reduced ubiquilin-1 protein levels correlate with accelerated maturation of APP and increased degradation rates (17), and provide a mechanistic basis by which ubiquilin-1 regulates APP trafficking. Because ubiquilin-1 has no obvious homology to known ubiquitin ligases, it is likely that ubiquilin-1 recruits an E3 ligase to APP while in the Golgi apparatus. Future studies will be necessary to identify the putative ligase(s) responsible for ubiquilin-1–stimulated APP degradation.
We have previously shown that ubiquilin-1 functions as a molecular chaperone for APP by binding to the AICD and preventing its aggregation (18). We also demonstrated, regardless of ubiquilin-1 genotype, that ubiquilin-1 levels were significantly reduced in late-onset AD patient brains (18, 27). Combined with the results presented here, our results suggest that ubiquilin-1 is a key regulator of APP biosynthesis, trafficking, and degradation, and likely functions as a key quality-control molecule to limit amyloidogenic processing of APP and associated neuronal dysfunction.
Materials and Methods
Expression Constructs.
To generate an APP–GFP fusion construct, APP695 was amplified using PCR, incorporating a 5′ HindIII restriction site and a 3′ SacII restriction site, and was cloned in frame to the HindIII-SacII sites of the pmaxFP Green-N mammalian expression vector (Amaxa). This vector has been described elsewhere, and the particular GFP variant has been shown to have no effect on proteasomal degradation (28). PCR primers were 5′-TTC AAG CTT CCA TGC TGC CCG GTT TGG CAC TG-3′ and 5′-TCC CCG CGG GTT CTG CAT CTG CTC AAA GAA CTT GTA G-3′. The GlcNAc-T1–mCherry fusion construct was kindly provided by Joachim Seemann (University of Texas Southwestern, Dallas, TX). This construct encodes the cytoplasmic and transmembrane domains of N-acetylglucosaminyltransferase 1, a Golgi-resident enzyme, fused to the mCherry red fluorescent protein, which replaces the C-terminal luminal catalytic domain of the enzyme. Wild-type and mutant HA-ubiquitin constructs were obtained from Addgene. Other expression constructs have been described previously (18). Ubiquilin-1 RNAi oligonucleotides used in Fig. 2D have been described and validated elsewhere (18).
APP Degradation.
Cells were transfected with either myc-tagged ubiquilin-1 or empty vector (pcDNA3.1). At 1 d posttransfection, cells were treated with CHX. CHX was diluted in cell culture medium at a final concentration of 50 μg/mL before its addition. At the specified time points, one plate from each group was snap-frozen in liquid nitrogen, and plates were stored at −80 °C until processing. After completion of the time course, cells lysates were prepared and run on a 4–20% gradient gel. Control and treated samples were handled under identical conditions, and blots were exposed simultaneously on the same film. To examine the effects of ubiquilin-1 on APP degradation in the presence of lysosomal/proteasomal inhibitors, cells were treated for 15 min with the proteasome inhibitor MG132 (EMD Biosciences) at a final concentration of 10 μM or the lysosomal inhibitor ammonium chloride (Sigma) at a final concentration of 25 mM. Both inhibitor treatment groups were compared with control cells treated with vehicle.
Subcellular Fractionation.
PC12 cells were transfected with myc-tagged ubiquilin-1 or a control empty vector. The next day, cells were treated for 2 h with either CHX or vehicle (DMSO). Cells were then collected and snap-frozen in liquid nitrogen. Differential centrifugation and subsequent analysis was done as described (29).
Cell-Surface Biotinylation.
Cells were plated on 10-cm plates and transfected with myc-tagged ubiquilin-1 or a control empty vector. The next day, the media was removed, and cells were washed 3× with 10 mL sterile ice-cold PBS. A cell-impermeant biotinylation reagent (Sulfo-NHS-SS-Biotin; Pierce) was dissolved in ice-cold PBS/MgCl2/CaCl2 at a final concentration of 1 mM, and 6 mL of the biotin solution was added to each plate. Plates were gently agitated on an orbital shaker for 3 h at 4 °C to ensure even coverage with the labeling solution. Following incubation, the labeling solution was removed, and cells were incubated in 8.5 mL quenching solution [PBS (pH 8.0), 10 mM Tris, and 100 mM glycine] for 10 min at 4 °C. Cells were then lysed in 250 μL lysis buffer containing protease inhibitors. Lysates were cleared at 10,000 × g pellet for 20 min at 4 °C, and protein content of the supernatant was determined by the Bradford method. A total of 500 μg protein (∼1 mg/mL in lysis buffer) was added to 25 μL bed-volume NeutrAvidin agarose resin (Pierce), and mixtures were incubated overnight at 4 °C with rotation. The resin was washed 10× with the wash buffer containing protease inhibitors and boiled in 25 μL 2× SDS sample buffer, and eluted proteins were analyzed by SDS/PAGE and Western blotting.
Ubiquitination Assays.
For ubiquitination experiments, cells were transfected with HA-ubiquitin or mutants constructs as indicated in Fig. 5B. The next day, cells were treated with the proteasome inhibitor MG132 for 3 h to induce accumulation of ubiquitinated proteins before harvesting in lysis buffer containing protease inhibitors and the deubiquitinating enzyme inhibitor N-ethylmaleimide (Sigma) at 10 mM final concentration. Lysates (500 μg) were precleared by 30-min incubation at 4 °C with protein A from the bacteria Staphylococcus aureus (Sigma) before incubation with an APP antibody for 2 h at 4 °C. Immune complexes were recovered using 50 μL of protein A-Sepharose beads (Pierce). The beads were then washed with lysis buffer, boiled with SDS sample buffer, and analyzed by SDS/PAGE.
Live-Cell Imaging.
PC12 cells were cultured on 25-mm coverslips overnight and then cotransfected with APP–GFP plus ubiquilin-1 or a control vector at 1:1 ratio. At 5 h posttransfection, the media was changed to nerve growth factor-containing media to induce cells differentiation for an additional 24 h. The coverslips were transferred to an imaging chamber containing imaging media (DMEM without l-glutamine and phenol red supplemented with 10 mM Hepes and 1% FBS). For all experiments, transfection efficiency, as determined by fluorescence microscopy, was greater than 50%. All fields were imaged randomly, and all GFP-positive cells in a given field were imaged and quantified. To examine trafficking and degradation, cells were treated with CHX to inhibit de novo protein synthesis. The punctate/diffuse index was determined as described (19), and colocalization was determined using the Colocalization Colormap plugin in ImageJ (21).
Confocal Imaging.
For confocal imaging, PC12 cells stably expressing GlcNAc-T1–mCherry were plated on coverslips and the next day transiently transfected with APP–GFP with ubiquilin-1 or a control vector. At 24 h posttransfection, the cells were treated with CHX or vehicle for specified time points and then washed with PBS and fixed with 4% paraformaldehyde (wt/vol) in PBS solution for 20 min at room temperature. APP–GFP colocalization with GlcNacT1 was visualized with an Olympus FluoView confocal microscope.
Supplementary Material
Acknowledgments
We thank Dr. Hui Zheng for the APP695 cDNA construct; Drs. Eric J. Brown and Mervyn J. Monteiro for ubiquilin-1 cDNA constructs; and Dr. Joachim Seemann for the GlcNAc-T1–mCherry construct. We also thank Dr. Megan L. Landsverk and Xinmin Wang for assistance with preliminary aspects of this project. This work was supported by National Institutes of Health Grants R21AG031948 (to J.M.B. and D.B.) and F30AG030878 (to E.S.S.), as well as funds provided by the Jean C. and William D. Willis Neuroscience Research Endowment (to E.S.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206786109/-/DCSupplemental.
References
- 1.Weidemann A, et al. Identification, biogenesis, and localization of precursors of Alzheimer's disease A4 amyloid protein. Cell. 1989;57:115–126. doi: 10.1016/0092-8674(89)90177-3. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki T, Araki Y, Yamamoto T, Nakaya T. Trafficking of Alzheimer’s disease-related membrane proteins and its participation in disease pathogenesis. J Biochem. 2006;139:949–955. doi: 10.1093/jb/mvj121. [DOI] [PubMed] [Google Scholar]
- 3.Andersen OM, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci USA. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selkoe DJ, et al. The role of APP processing and trafficking pathways in the formation of amyloid beta-protein. Ann N Y Acad Sci. 1996;777:57–64. doi: 10.1111/j.1749-6632.1996.tb34401.x. [DOI] [PubMed] [Google Scholar]
- 5.De Strooper B, Annaert W. Proteolytic processing and cell biological functions of the amyloid precursor protein. J Cell Sci. 2000;113:1857–1870. doi: 10.1242/jcs.113.11.1857. [DOI] [PubMed] [Google Scholar]
- 6.Koo EH, et al. Precursor of amyloid protein in Alzheimer disease undergoes fast anterograde axonal transport. Proc Natl Acad Sci USA. 1990;87:1561–1565. doi: 10.1073/pnas.87.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez RG, et al. Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J Biol Chem. 1999;274:18851–18856. doi: 10.1074/jbc.274.27.18851. [DOI] [PubMed] [Google Scholar]
- 8.Tomita S, Kirino Y, Suzuki T. Cleavage of Alzheimer’s amyloid precursor protein (APP) by secretases occurs after O-glycosylation of APP in the protein secretory pathway. Identification of intracellular compartments in which APP cleavage occurs without using toxic agents that interfere with protein metabolism. J Biol Chem. 1998;273:6277–6284. doi: 10.1074/jbc.273.11.6277. [DOI] [PubMed] [Google Scholar]
- 9.Small SA, Gandy S. Sorting through the cell biology of Alzheimer’s disease: Intracellular pathways to pathogenesis. Neuron. 2006;52:15–31. doi: 10.1016/j.neuron.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerr ML, Small DH. Cytoplasmic domain of the beta-amyloid protein precursor of Alzheimer’s disease: Function, regulation of proteolysis, and implications for drug development. J Neurosci Res. 2005;80:151–159. doi: 10.1002/jnr.20408. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt V, et al. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem. 2007;282:32956–32964. doi: 10.1074/jbc.M705073200. [DOI] [PubMed] [Google Scholar]
- 12.Spoelgen R, et al. Interaction of the cytosolic domains of sorLA/LR11 with the amyloid precursor protein (APP) and beta-secretase beta-site APP-cleaving enzyme. J Neurosci. 2006;26:418–428. doi: 10.1523/JNEUROSCI.3882-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Traub LM. Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim Biophys Acta. 2005;1744:415–437. doi: 10.1016/j.bbamcr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Caporaso GL, Gandy SE, Buxbaum JD, Greengard P. Chloroquine inhibits intracellular degradation but not secretion of Alzheimer beta/A4 amyloid precursor protein. Proc Natl Acad Sci USA. 1992;89:2252–2256. doi: 10.1073/pnas.89.6.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knops J, Lieberburg I, Sinha S. Evidence for a nonsecretory, acidic degradation pathway for amyloid precursor protein in 293 cells. Identification of a novel, 22-kDa, beta-peptide-containing intermediate. J Biol Chem. 1992;267:16022–16024. [PubMed] [Google Scholar]
- 16.Nunan J, et al. Proteasome-mediated degradation of the C-terminus of the Alzheimer’s disease beta-amyloid protein precursor: Effect of C-terminal truncation on production of beta-amyloid protein. J Neurosci Res. 2003;74:378–385. doi: 10.1002/jnr.10646. [DOI] [PubMed] [Google Scholar]
- 17.Hiltunen M, et al. Ubiquilin 1 modulates amyloid precursor protein trafficking and Abeta secretion. J Biol Chem. 2006;281:32240–32253. doi: 10.1074/jbc.M603106200. [DOI] [PubMed] [Google Scholar]
- 18.Stieren ES, et al. Ubiquilin-1 is a molecular chaperone for the amyloid precursor protein. J Biol Chem. 2011;286:35689–35698. doi: 10.1074/jbc.M111.243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR. The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat Cell Biol. 2000;2:156–162. doi: 10.1038/35004029. [DOI] [PubMed] [Google Scholar]
- 20.Kumar R, Yang J, Larsen RD, Stanley P. Cloning and expression of N-acetylglucosaminyltransferase I, the medial Golgi transferase that initiates complex N-linked carbohydrate formation. Proc Natl Acad Sci USA. 1990;87:9948–9952. doi: 10.1073/pnas.87.24.9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaskolski F, Mulle C, Manzoni OJ. An automated method to quantify and visualize colocalized fluorescent signals. J Neurosci Methods. 2005;146:42–49. doi: 10.1016/j.jneumeth.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao L, et al. Interaction with a ubiquitin-like protein enhances the ubiquitination and degradation of hepatitis C virus RNA-dependent RNA polymerase. J Virol. 2003;77:4149–4159. doi: 10.1128/JVI.77.7.4149-4159.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: Disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 25.Lim KL, Dawson VL, Dawson TM. Parkin-mediated lysine 63-linked polyubiquitination: A link to protein inclusions formation in Parkinson’s and other conformational diseases? Neurobiol Aging. 2006;27:524–529. doi: 10.1016/j.neurobiolaging.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 26.Chiu RK, et al. Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet. 2006;2:e116. doi: 10.1371/journal.pgen.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boehning D, Barral JM. Protein quality control in Alzheimer's disease: The contentious role of ubiquilin-1. Future Neurology. 2012;7:5–8. [Google Scholar]
- 28.Baens M, et al. The dark side of EGFP: Defective polyubiquitination. PLoS ONE. 2006;1:e54. doi: 10.1371/journal.pone.0000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boehning D, et al. Cytochrome c binds to inositol (1,4,5) trisphosphate receptors, amplifying calcium-dependent apoptosis. Nat Cell Biol. 2003;5:1051–1061. doi: 10.1038/ncb1063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.