Abstract
Oxidative stress regulates telomere homeostasis and cellular aging by unclear mechanisms. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is a key mediator of many oxidative stress responses, involving GAPDH nuclear translocation and induction of cell death. We report here that GAPDH interacts with the telomerase RNA component (TERC), inhibits telomerase activity, and induces telomere shortening and breast cancer cell senescence. The Rossmann fold containing NAD+ binding region on GAPDH is responsible for the interaction with TERC, whereas a lysine residue in the GAPDH catalytic domain is required for inhibiting telomerase activity and disrupting telomere maintenance. Furthermore, the GAPDH substrate glyceraldehyde-3-phosphate (G3P) and the nitric oxide donor S-nitrosoglutathione (GSNO) both negatively regulate GAPDH inhibition of telomerase activity. Thus, we demonstrate that GAPDH is regulated to target the telomerase complex, resulting in an arrest of telomere maintenance and cancer cell proliferation.
Keywords: telomerase reverse transcriptase, NAD+, gene expression, mutation, post-translational modification
Cell aging occurs at different rates in most mammalian cell types. Rapidly dividing cells have short lifespans of only 20–40 population doublings, followed by cell senescence (aging) and apoptosis. Although production of daughter cells costs energy, it remains largely elusive how energy metabolism regulates normal cell aging and lifespan. Considerable evidence suggests that metabolic waste causes premature senescence and apoptosis, shortening cellular lifespan. Accumulation of reactive oxygen species (ROS) damages cellular enzymes, organelle membranes, and chromosomal DNA including telomeres (the ends of chromosomes) (1, 2). Whereas enzymes that reduce the production of metabolic hazards may operate to extend cellular lifespan, recent studies demonstrate that the energy producing enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) that limits ROS production undergoes structural modifications and travel into the nucleus (3–7) to interact with telomeres directly and to potentially alleviate stress-induced cell aging (8, 9).
Human telomeres are composed of arrays of a few thousand tandem DNA repeats of TTAGGG and various binding proteins. Telomeres undergo progressive shortening as somatic cells divide, due to the inability of cells to replicate the extreme ends of chromosomes. When telomeres are critically short, cells exit the cell cycle and undergo cell senescence (10–13). As a measure to maintain telomeres for continuous renewal of germ lines, development of embryonic stem cells, clonal expansion of lymphocytes, and immortalization of neoplastic cells, telomerase is activated to synthesize and maintain telomeres by reverse transcription (10–13).
Telomerase is a large ribonucleoprotein complex containing the catalytic subunit telomerase reverse transcriptase (TERT) and an RNA template (reviewed in refs. 13 and 14). Human telomerase reverse transcriptase (hTERT) comprises 1,132 amino acid residues and human telomerase RNA (hTERC) is composed of 451 nucleotides (15–18). Reconstitution of telomerase activity in telomerase-negative cells can be achieved by expression of hTERT, demonstrating that hTERT is the rate-limiting component of the enzyme complex (19, 20).
GAPDH has long been recognized as an important enzyme in catalyzing ATP production through the anaerobic glycolysis of a monosaccharide (generally glucose) to pyruvate in the cytosol (21, 22). Intriguingly as a gene encoding a single 38-kDa protein without alternate splicing, GAPDH displays multiple functions that are independent of its role in energy generation (reviewed in refs. 5, 22–24). A large body of evidence indicates that GAPDH responds to various stress insults by translocation to the nucleus (3–7, 25). In the nucleus, GAPDH is the key component of a gene transcriptosome for cell division (6). Binding to nucleic acids, GAPDH is implicated in mediating RNA nuclear export (22), protecting mRNA from degradation through interaction with AU-rich elements (AREs) (26) and the repair of DNA (27). Moreover, studies have shown that GAPDH binds to telomeres directly (8, 9). These findings suggest that while catalyzing energy production, GAPDH also regulates chromosome stability and genome integrity by multiple mechanisms, including the maintainance of telomere homeostasis to facilitate longevity of cell proliferation (24). However, the mechanisms by which GAPDH is involved in telomere homeostasis are not known, and whether or not the regulation of telomere length by GADPH is dependent on telomerase requires further investigation. The present study provides clear evidence demonstrating that GAPDH binds TERC, that this binding is controlled by glyceraldehyde-3-phosphate (G3P) and S-nitrosoglutathione (GSNO), and that GAPDH regulates telomerase activity and cancer cell proliferation.
Results and Discussion
Cancer Cell Senescence Induced by GAPDH.
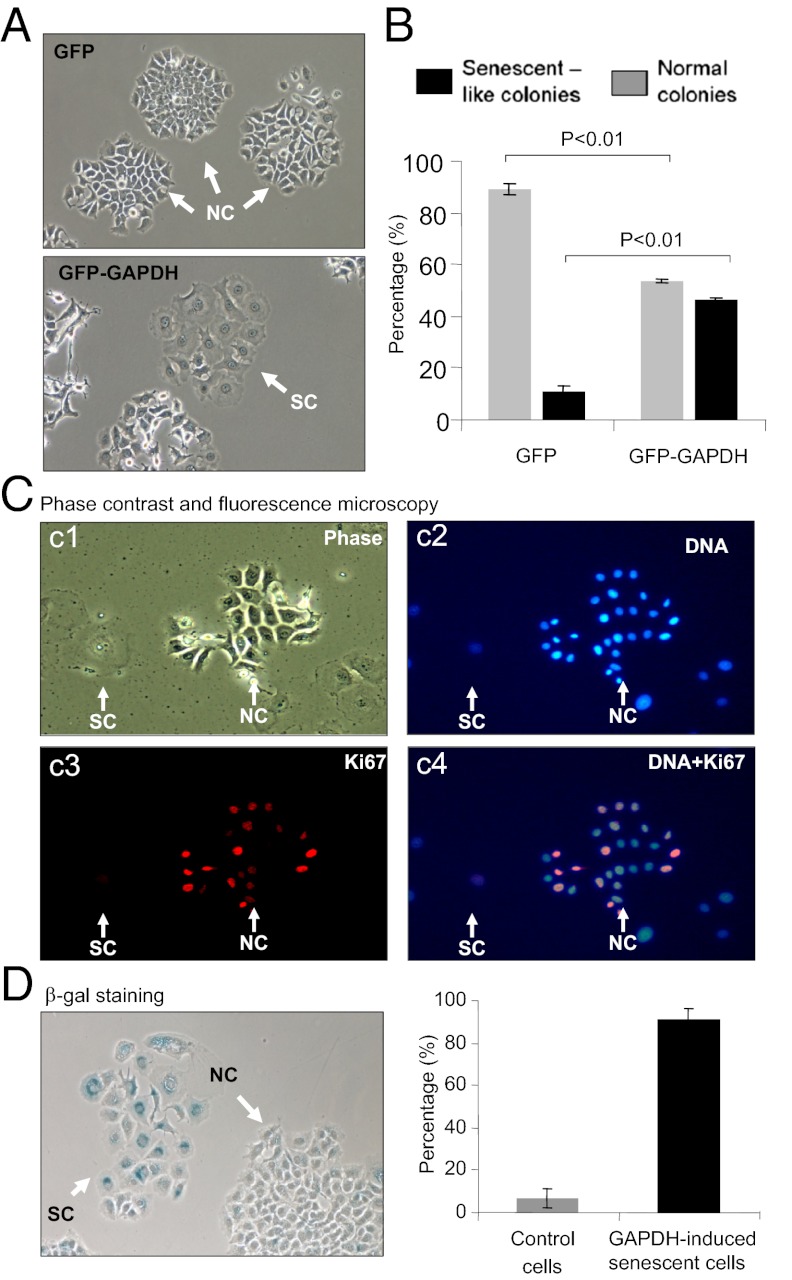
To investigate a potential role of GAPDH in telomere maintenance in breast cancer cells, we expressed recombinant GAPDH in MCF7 cells. Whereas we noted a reduced cell density in the MCF7 cell cultures stably expressing GFP–GAPDH consistent with previous findings of GAPDH-induced cancer cell death (3, 4, 7, 25, 28, 29), we found that there were significant changes in the morphology of MCF7 cells expressing GFP–GAPDH. Cells expressing exogenous GAPDH exhibited an enlarged and flattened morphology characteristic of typical stress-induced premature senescence cells (Fig. 1). To characterize the incidence of this cell-senescence–like phenotype, cells expressing either GFP–GAPDH fusion or GFP alone were seeded at 104 cells per 35-mm dish and allowed to proliferate and form colonies for 2 wk (Fig. 1A). In cells expressing GFP–GAPDH, there were ∼46% of cell-senescence–like colonies demonstrating an enlarged flattened cell morphology, compared with 11% in GFP-alone controls (P < 0.05) (Fig. 1B). Colonies without senescence-like morphology were fewer in stable cell cultures of GFP–GAPDH, compared with GFP-alone control cells (54 versus 89%, respectively, P < 0.05) (Fig. 1B). Staining of senescence-like cells for the proliferation marker Ki67 showed a clear absence of Ki67 consistent with cellular senescence (Fig. 1C). Further staining of cells expressing elevated GAPDH for senescence-associated β-galactosidase (SA-β-Gal) activity confirmed the cellular senescence phenotype (Fig. 1D, Left). Approximately 91% of cells with enlarged flattened morphology showed SA-β-Gal positivity vs. less than 10% in the controls (Fig. 1D, Right). These data uniquely demonstrate that increased gene expression of GAPDH triggers breast cancer cell senescence.
Fig. 1.
Gene expression of GFP–GAPDH induces MCF7 cell senescence. (A) Phase-contrast micrographs showing senescence-like cells. Stably transfected MCF7 cells expressing GFP or GFP–GAPDH were compared by microscopy. Cells with flattened morphology are indicated as senescent colonies (SC), compared with control normal colonies (NC). (B) Histogram summarizing data from four individual cultures of stably transfected cells expressing GFP or GFP–GAPDH. To score NCs and SCs more than 60 colonies were counted per well. Data shown are mean ± SEM. (C) Immunofluorescence staining for Ki67. c1, phase contrast micrograph showing the SC and NC cells. c2, DNA staining for the cell nuclei. c3, Ki67 immunoreactivity. c4, overlay of nuclear DNA and Ki67 staining. Ki-67 was stained red with anti–Ki-67 MAb. Nuclei were stained blue with DAPI. (D) SA-β-gal staining of cellular senescence in stably transfected cells. Histogram showing proportion of SA-β-gal+ cells (blue) within NCs and SCs (n > 100 cells/colony type, mean ± SEM).
GAPDH Inhibition of Telomerase Activity.
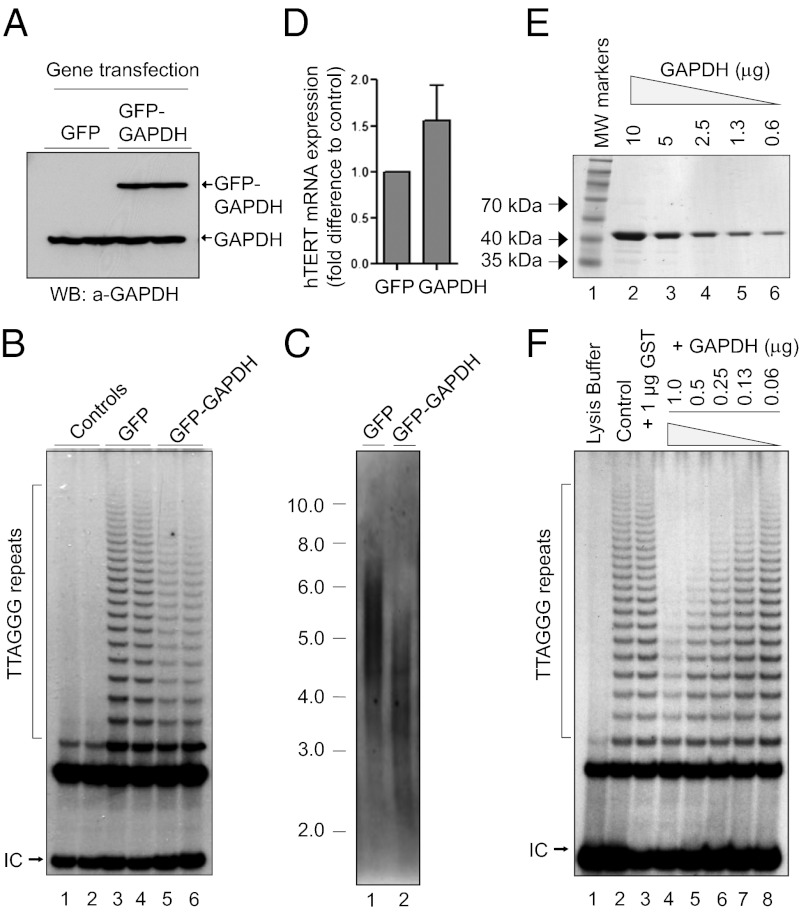
To further demonstrate breast cancer cell senescence, we measured telomerase activity and telomere length, as the reduction of both are among the key molecular events underpinning the permanent cell cycle arrest of cancer cell senescence (30, 31). In cultured cells expressing GFP–GAPDH, there was a significant inhibition of telomerase activity and shortening of telomere length, compared with that in cell cultures expressing GFP alone (Fig. 2 B and C). To determine whether elevated GAPDH gene expression might potentially alter the gene expression of hTERT, the catalytic subunit and rate-limiting component of telomerase (32, 33), we measured hTERT gene expression by real-time PCR and found that there were no significant changes in the hTERT gene expression levels (Fig. 2D). Because GAPDH has been shown to bind telomeric DNA directly (8, 9), we next determined whether purified human GAPDH has any effect on telomerase activity using breast cancer cell telomerase extracts. Interestingly, purified erythrocyte GAPDH (E-GAPDH) inhibited telomerase activity in a concentration-dependent manner in vitro (Fig. 2 E and F). The findings that GAPDH inhibits telomerase activity in vitro and in vivo suggest the possibility that a posttranslational mechanism is responsible for the inhibition of telomerase through a complex interaction between GAPDH and telomerase.
Fig. 2.
GAPDH induces telomerase inhibition and telomere shortening. (A) Levels of GFP–GAPDH and GFP gene expressions determined by immunoblotting. (B) Telomerase activity measured in the MCF7 cells expressing GFP only or GFP–GAPDH. Telomerase products and internal controls (ICs) are indicated. (C) Telomere restriction fragment (TRF) length measured from MCF7 cells expressing GFP or GFP–GAPDH. Scale shows DNA size ladder (kb). (D) Expression levels of hTERT in the MCF7 cells expressing GFP only or GFP–GAPDH. (E) Purified GST–GAPDH at different concentrations stained by Coomassie Blue. (F) Concentration-dependent inhibition of telomerase activity by GST–GAPDH in vitro. Telomerase products and ICs are indicated. Results are representatives of at least three experiments.
Role of the Rossmann Fold in GAPDH Binding to hTERC.
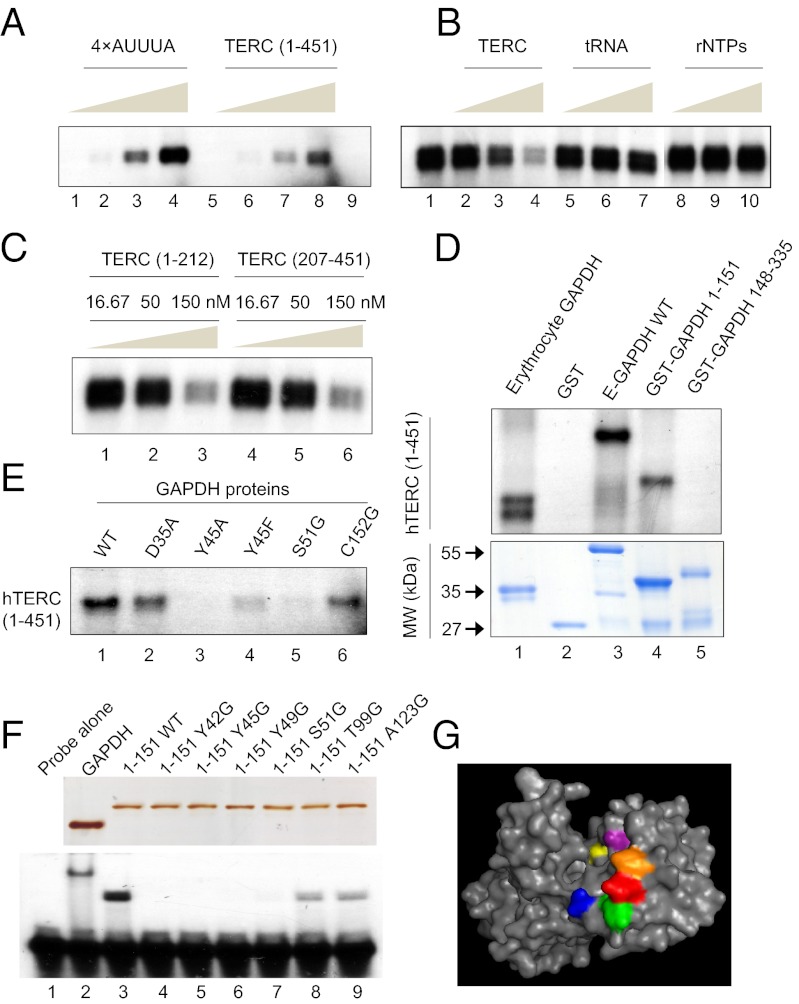
Because GAPDH inhibits telomerase directly in vitro and in vivo (Fig. 2), and influences the stability, location, and function of a variety of RNA species through direct interactions (reviewed in ref. 24), we examined whether an interaction between GAPDH and the telomerase RNA moiety hTERC exists. As a positive control for GAPDH RNA binding, an AU-rich RNA molecule (4× AUUUA) was used, as has been previously demonstrated to interact with GAPDH (26). As shown in Fig. 3 A and B, purified human GAPDH bound full-length hTERC (1–451) in a concentration-dependent manner. The hTERC binding appeared to be specific as unlabeled TERC, but not tRNA or rNTPs, competitively inhibited the binding of radioisotope-labeled 4× AUUUA in a dose-dependent manner (Fig. 3B). Both 5′ and 3′ halves of hTERC bound GAPDH in a concentration-dependent manner (Fig. 3C), suggesting that GAPDH may bind hTERC on multiple sites. To determine the structural specificity of GAPDH binding, we constructed recombinant GST–GAPDH fragments and found that the binding site of hTERC was within the N-terminal region of recombinant GST–GAPDH 1–151 but not in the C-terminal region of GST–GAPDH 148–335 (Fig. 3D).
Fig. 3.
GAPDH binds to telomerase RNA via its N-terminal region. (A) RNA binding assay demonstrating purified human E-GAPDH interaction with increasing amounts (0.1, 1, 5, and 10 × 104 cpm) of a radiolabeled 4× AUUUA RNA sequence (lanes 1–4) and full-length hTERC (lanes 5–8). Lane 9 is GAPDH without probe. (B) RNA binding competition assay performed with 0.2 μg GAPDH, 5 × 104 cpm 4× AUUUA probe (lane 1) and increasing concentrations (16.67, 50, and 150 nM) of cold competitor as indicated before UV cross-linking (lanes 2–10). Note that the rNTPs were conducted in a separate experiment. (C) Effect of cold hTERC (1–212) and hTERC (207–451) on GAPDH binding to radiolabeled 4× AUUUA RNA sequence. RNA binding competition assay performed with 0.2 μg GAPDH, 5 × 104 cpm 4× AUUUA probe and increasing concentrations (16.67, 50, and 150 nM) of cold competitor as indicated before UV cross-linking. (D) Binding of different GAPDH fragments to radioisotope-labeled hTERC. Upper, RNA binding assay of 1.0 μg indicated protein incubated with 5 × 104 cpm full-length hTERC probe. Lower, Coomassie blue staining of ∼1.0 μg indicated purified proteins. (E) Binding of wild-type and mutant GAPDH to radioisotope-labeled hTERC. RNA binding assay of 1.0 μg indicated protein incubated with 5 × 104 cpm full-length hTERC probe. (F, Upper,) Silver staining of SDS/PAGE gel containing ∼1.0 μg of E-GAPDH or indicated GST–GAPDH-1–151 variants. Lower, gel-shift assay of 3.33 nM C36 probe incubated with 1.0 μg indicated protein. (G) Structural rendering of GAPDH monomer (Protein Data Bank ID code 1ZNQ) highlighting targeted residues: Y42, orange; Y45, red; Y49, green; S51, blue; T99, purple; A123, yellow.
The Rossmann fold in the N-terminal region of GAPDH mediates the binding of the dinucleotide NAD+ (34, 35). To determine the role of the Rossmann fold in GAPDH binding to hTERC, we carried out single amino acid substitutions in GAPDH by site-directed mutagenesis. As shown in Fig. 3E, a single amino acid mutation at D35, Y45, or S51 on GST–GAPDH dramatically reduced the binding of GST–GAPDH to hTERC 1–451, suggesting that the integrity of the Rossmann fold on GAPDH is required in mediating hTERC binding. To determine whether the Rossmann fold potentially binds telomeric DNA, we tested the binding capacity of radioisotope-labeled telomeric DNA to various GST–GAPDH fragments using an electrophoretic mobility shift assay (EMSA). Consistently, telomeric DNA oligonucleotides bound GST–GAPDH wild type and GST–GAPDH 1–151, but with a reduced level to GST–GAPDH 1–125. The oligonucleotides did not bind to GST–GAPDH 1–113, 1–71, 1–55, 1–45, or the C-terminal fragment 148–335 (Fig. S1). To further demonstrate a Rossmann fold-mediated binding of GAPDH to telomeric DNA, we used single amino acid mutations in the Rossmann fold of GST–GAPDH 1–151 and examined the interaction of these proteins with telomeric DNA oligonucleotides. As shown in Fig. 3F, whereas GAPDH full length and GAPDH 1–151 bound the telomeric oligonucleotides significantly, mutations at Y42, Y45, Y49, or S51 abolished this binding and mutations at the T99 or A123 reduced the binding. Thus, we have uniquely identified that the GAPDH Rossmann fold contains residues critically required for GAPDH to bind hTERC (Fig. 3G). Given the highly conserved structure of GAPDH (36) and telomeric RNA (17, 18) across different vertebrate species, it is possible that GAPDH–hTERC interaction is evolutionarily conserved. Our data also demonstrate that the GAPDH Rossmann fold can accommodate either hTERC or telomeric DNA.
To investigate a potential involvement of hTERT in hTERC interaction with GAPDH, we performed a pull-down assay using recombinant GST–hTERT fragments. GST–hTERT 423–658 that contains the hTERC binding site, and GST–hTERT 423–538 as controls, were incubated with breast cancer cell lysates, and after extensive washing, proteins bound to the GST–hTERT proteins were eluted with increasing concentrations of salt and concentrated for analysis by SDS/PAGE. A major protein was recovered at the molecular size of ∼38 kDa from the GST–hTERT 423–658 eluents but not that of GST 423–538 (Fig. S2A). Mass spectrometry experiments showed that 12 peptides derived from the purified 38-kDa protein covered 51–59% of GAPDH (Table S1), identifying the 38-kDa binding protein to be GAPDH. It is thus possible that GAPDH forms a complex with telomerase in vivo by a direct interaction with hTERC and hTERT. Consistent with GAPDH interaction with telomerase, the inhibition of telomerase activity induced by GFP–GAPDH in MCF7 cells was overcome by overexpression of hTERT but not by overexpression of dyskerin (a third component to hTERT and hTERC) (Fig. S2B).
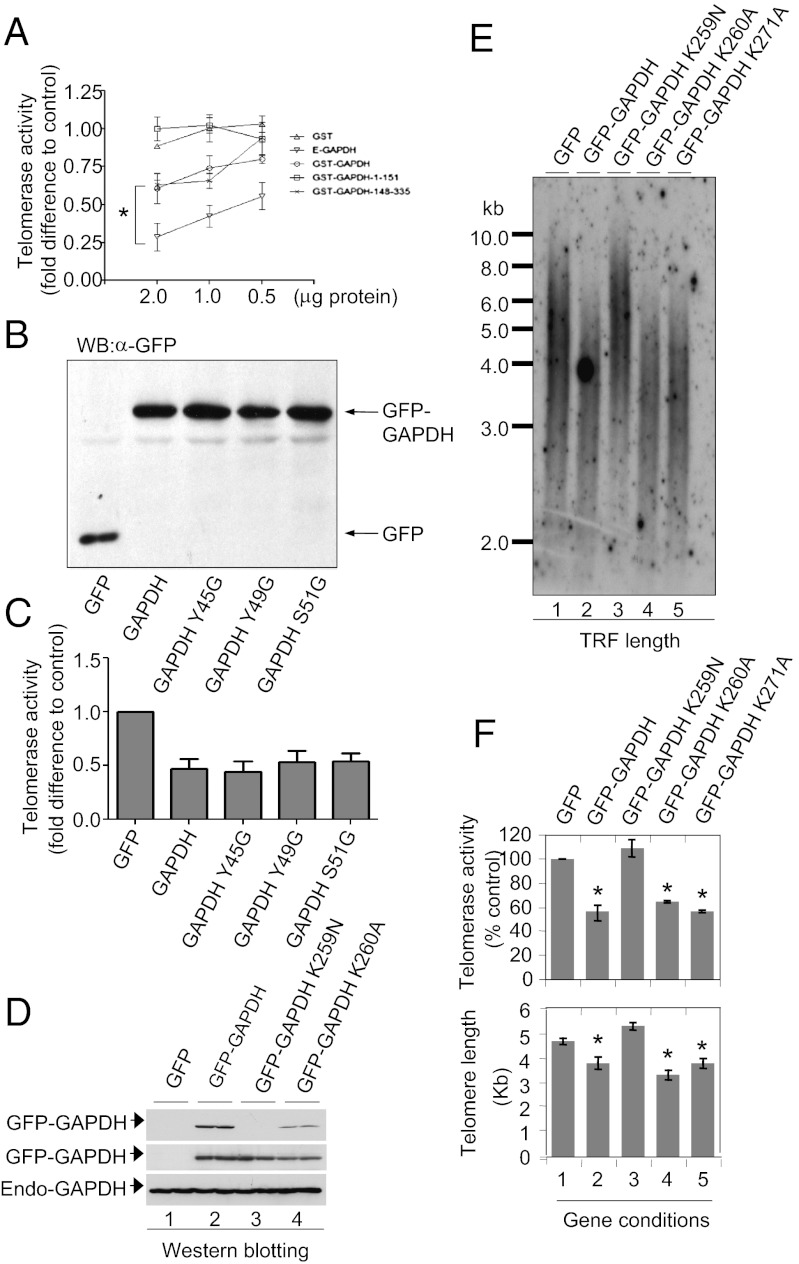
To determine the role of GAPDH binding to hTERC in GAPDH inhibition of telomerase activity, we tested for effects of different GST–GAPDH full-length and truncated forms on telomerase activity in vitro. The recombinant GST–GAPDH, GST–GAPDH 1–151, and GST–GAPDH 148–335 (Fig. 3D, Lower) showed varied levels of inhibition on telomerase activity (Fig. 4A). To our surprise, GST–GAPDH 1–151 bound hTERC (Fig. 3D) but had no telomerase inhibitory activity (Fig. 4A, square symbols), whereas GST–GAPDH 148–335 had no hTERC binding activity (Fig. 3D) but retained telomerase inhibitory activity comparable to wild-type GAPDH (Fig. 4A). To investigate the effects of the Rossmann fold mutants of GAPDH on telomerase activity, we expressed GFP–GAPDH wild type, Y45G, Y49G, and S51G in MCF7 cells (Fig. 4B). All of the three Rossmann fold mutants showed telomerase inhibitory activity with ∼50% inhibition similar to that of the wild-type GAPDH (Fig. 4C and representative raw data in Fig. S3). Examination of telomere length confirmed that GFP–GAPDH induced telomere shortening by ∼40%, but mutations of the Rossmann fold that inhibit hTERC binding did not prevent GAPDH-induced telomere shortening in breast cancer cells (Fig. S4). These data confirm the in vitro data that although the Rossmann fold is required for GAPDH binding to hTERC, it is not directly responsible for telomerase inhibition.
Fig. 4.
Structure–function relationship of GAPDH in telomerase inhibition and telomere shortening. (A) Effects of GAPDH fragments on telomerase activity in vitro. Telomerase activity was quantified in telomerase extracts preincubated with 0.5, 1.0, or 2.0 μg of indicated purified protein before TRAP assay. (B) Expression of GFP–GAPDH variants or GFP alone in MCF7 cells. (C) Graphical representation of telomerase activity in MCF7 cells expressing GFP–GAPDH variants with mutations that affect GAPDH binding to hTERC. (D) Western blot showing expression of various GAPDH proteins detected with two GAPDH antibodies. MAb, monoclonal antibody (6C5) targeted to GAPDH C-terminal epitope 250–269; PAb, polyclonal antibody for detection of GFP–GAPDH K259N. Endogenous GAPDH (“Endo-GAPDH”) is also shown as a loading control. (E) Representative autoradiograph showing the result of a TRF length analysis of MCF7 cells expressing GFP, GFP–GAPDH, and GFP–GAPDH mutants. (F) Histogram showing the mean telomerase activity (Upper) and TRF length (Lower) in cells stably expressing GFP, GFP–GAPDH, and mutant GFP–GAPDH. Data are shown as mean ± SEM from three replicate telomere length assays (*P < 0.05 versus GFP-alone control).
Role of the C-Terminal Region of GAPDH in Telomerase Inhibition.
With the combined evidence that the binding between the Rossmann fold and hTERC is not directly coupled to telomerase inhibition and that the C-terminal fragment of GAPDH retains telomerase inhibitory activity (Fig. 4 A–C), we next focused on an α-helical loop structure on the GAPDH C-terminal region that has been found to be important in a protein–protein interaction (37). Mutation of the GAPDH lysine residues 259 and 260 showed that GAPDH lysine 259 plays a critical role in GAPDH inhibition of telomerase activity (Fig. 4 D–F), consistent with the finding that this residue is important for the interaction of GAPDH with CRM1 (37). Expression of GAPDH K259N resulted in no change in telomere length or telomerase activity (Fig. 4 E and F), compared with those induced by GAPDH wild type or K260A. The reversal of GAPDH inhibition of telomerase activity and shortening of telomeres in breast cancer cells by mutating a single amino acid residue on the GAPDH C-terminal region further confirms the specific structure–function relationship of GAPDH in regulating telomere homeostasis, and suggests a complex molecular interaction between telomerase and GAPDH.
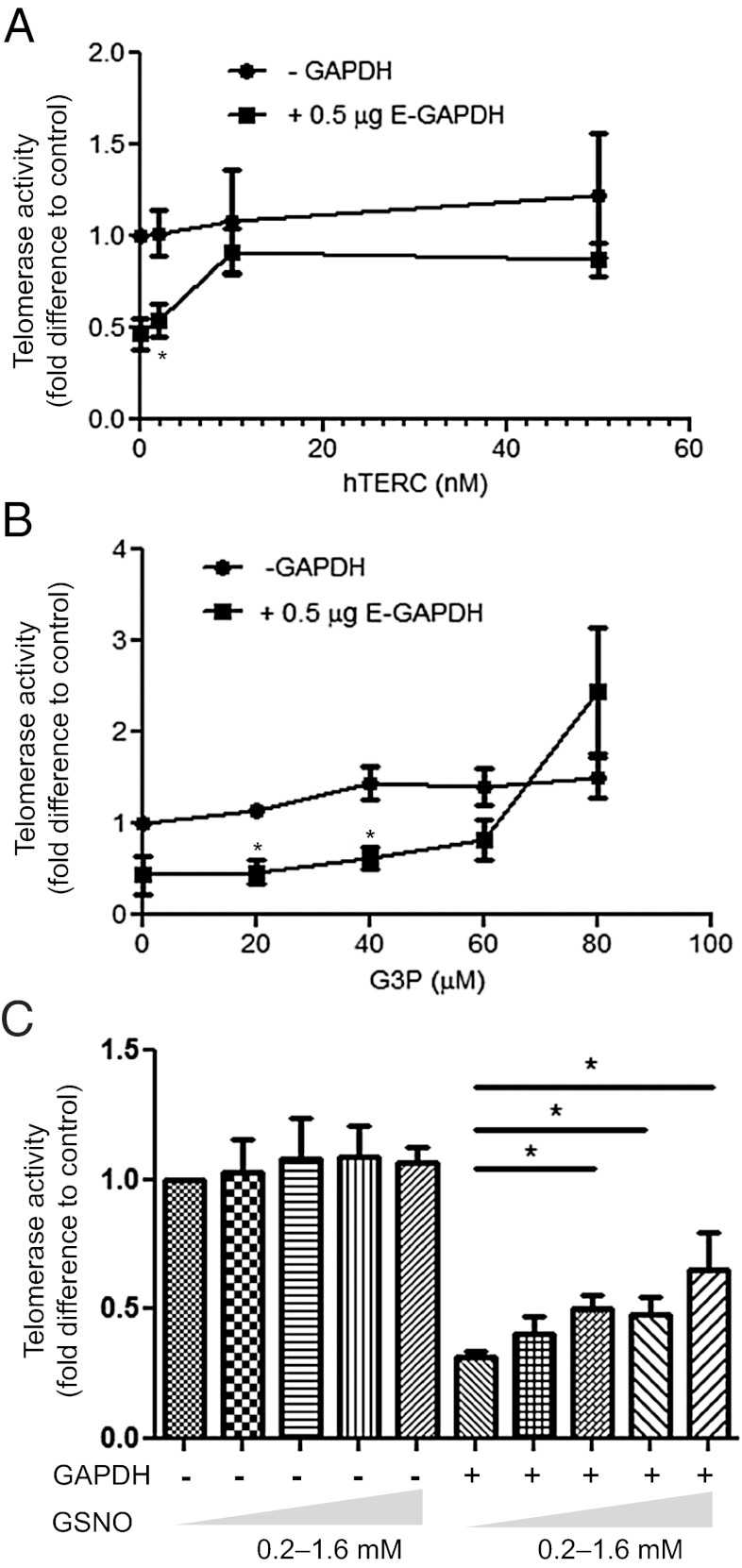
Reversible Regulation of GAPDH Binding to hTERC.
To investigate the functional significance of the interplay between hTERC and GAPDH in telomerase inhibition, we determined whether an excess amount of hTERC is capable of preventing GAPDH from interacting with the telomerase complex. Through the provision of in vitro transcribed hTERC on GAPDH inhibition of telomerase, we found that excess hTERC completely reversed the inhibitory effect of GAPDH on telomerase activity in a dose-dependent manner (Fig. 5A). This finding suggests that addition of exogenous hTERC blocks the GAPDH interaction with endogenous hTERC in the telomerase complex. Thus, hTERC binding may play a central role in positioning GAPDH with telomerase, enabling the GAPDH C-terminal domain to inhibit telomerase activity. To further examine the mechanisms regulating GAPDH interactions with telomerase machinery, we investigated the effect of the GAPDH enzymatic substrate G3P on the ability of GAPDH to inhibit telomerase activity using in vitro telomerase activity TRAP assays. Addition of G3P reversed the inhibition of telomerase activity in a dose-dependent manner (Fig. 5B), consistent with our findings that the C-terminal catalytic region of GAPDH mediates its telomerase inhibitory function (Fig. 4). Thus, these data demonstrate that G3P binding to GAPDH blocks GAPDH inhibition of telomerase activity.
Fig. 5.
Reversal of GAPDH inhibition of telomerase activity by hTERC, G3P, and GSNO. (A) Telomerase activity determined from telomerase extracts following incubation with and without 0.5 μg GAPDH in the presence of increasing concentrations of hTERC (0–50 nM), followed by determination of telomerase activity. (B) Telomerase activity determined from telomerase extracts following incubation with and without 0.5 μg GAPDH in the presence of increasing concentrations of G3P (0–80 μM). (C) Telomerase activity determined from telomerase extracts following incubation with and without 0.5 μg GAPDH pretreated with increasing concentrations of GSNO (0–1600 μM) for 10 min. All data are presented as mean ± SEM of at least three independent experiments. Asterisk represents a significant difference with P value less than 0.05.
Under stress conditions, cells generate nitric oxide (NO), which leads to the S-nitrosylation of GAPDH cysteine residues, abolishing catalytic function and inducing the proapoptotic role of GAPDH (3, 38, 39). To test the hypothesis that the NO donor GSNO might compromise GAPDH inhibition of telomerase activity, GAPDH was pretreated with increasing concentrations of GSNO and then incubated with telomerase extracts in in vitro TRAP assays. Surprisingly, whereas control GAPDH inhibited telomerase activity by ∼68% of untreated controls, GAPDH that was treated with 1.6 mM GSNO inhibited telomerase activity by ∼35% of controls (Fig. 5C). These data indicate that GSNO modification impairs the ability of GAPDH to inhibit telomerase. Thus, whereas S-nitrosylation of GAPDH is required for GAPDH to enter the nucleus (4, 38–40), GAPDH inhibition of telomerase activity may require removal of S-nitrosylation from GAPDH, possibly by the recently demonstrated mechanism of trans–S-nitrosylation of GAPDH (38). Our findings suggest that GAPDH targeting of the telomerase complex is regulated by a complex modification of GAPDH including GSNO-induced modification of GAPDH as an additional layer of specificity control.
NAD+ Blocks GAPDH Binding but Does Not Inhibit Telomerase Activity.
Consistent with a possibly shared binding site for NAD+ and telomere in the Rossmann fold, GAPDH association with the telomeric DNA was inhibited by NAD+ in a concentration-dependent manner, with an IC50 and maximal inhibition being 13.1 and 50 μM, respectively (Fig. S5 A and B). Incubation of telomerase extracts with GAPDH in the presence of increasing concentrations of NAD+ showed no significant effect of NAD+ on the basal and GAPDH-inhibited telomerase activity (Fig. S5C), demonstrating a distinguishable activity of GAPDH between telomeric DNA binding and telomerase inhibition. Because the concentration of NAD+ in cytoplasm or the nucleus of healthy cells is ≥70 μM, GAPDH binding in the nucleus may be inhibited under normal conditions as NAD+ binding blocks the telomeric DNA binding to the Rossmann fold. Interestingly, it has been demonstrated that cellular stress depletes NAD+ almost completely (41–43). Thus, the Rossmann fold on GAPDH may provide a cognate binding site for the telomeric DNA in a manner critically dependent on the elimination of NAD+ from the Rossmann fold under stress conditions. Cellular depletion of NAD+ induced by oxidative stress may thus serve as a regulatory switch to enable the GAPDH Rossmann fold to be accessed by the telomeric DNA, consistent with stress-induced GAPDH S-nitrosylation and nuclear translocation (3, 4).
Summary and Conclusion.
Taken together, our findings demonstrate a unique phenomenon that GAPDH induces cancer cell senescence with significant inhibition of telomerase activity and shortening of telomere length. The molecular mechanisms of GAPDH inhibiting telomerase activity involve GAPDH N-terminal Rossmann fold binding to hTERC and C-terminal catalytic region inhibiting telomerase. We demonstrate that the interaction between GAPDH and telomerase is negatively regulated by the GAPDH substrate NAD+ and G3P, suggesting that the interaction between GAPDH and its substrate operates as an important check to prevent GAPDH switching from the metabolic pathway to telomere signaling. Furthermore, we demonstrate that GSNO also inhibits GAPDH suppressing telomerase, suggesting that NO regulates GAPDH structure and function by S-nitrosylation and indirect regulation of NAD+ production and binding to GAPDH. In conclusion, our results indicate that GAPDH interacts with the telomerase complex by directly binding the telomerase RNA moiety and subsequently inhibiting telomerase activity through its C-terminal region, ultimately resulting in telomere shortening and cell aging. Furthermore, this inhibition of telomerase activity is reversible and regulated by NAD+, G3P, and NO production.
Materials and Methods
Recombinant Fusion Proteins.
Plasmid constructs expressing GST-fusion proteins were constructed by PCR cloning of desired GAPDH fragments onto pGEX-4T1 plasmid. The oligonucleotides used to produce the hTERT cDNA fragments were GGGGAATTCGCCGGTGTCTCTGCC and GGGGTCGACCTCACGCAGACGGTG for GST–hTERT 423–538 and GGGGAATTCGCCGGTGTCTCTGCC and CGCAGAGTGGAGCTCCCA for GST–hTERT 423–658. Transformed BL21 DE3 Escherichia coli cells were grown overnight at 37 °C, induced with isopropyl-β-d-1-thiogalactopyranoside (IPTG) for 3–5 h and lysed in ice-cold GST lysis buffer. Glutathione sepharose 4B beads (GE Life Sciences) were incubated at 4 °C with lysates for 2–4 h followed by three to six washes in cold GST lysis buffer. Proteins were eluted by resuspending beads in 200–500 μL elution buffer (EB) with end-over-end rotation for 15 min at room temperature. Elution was repeated three times and pooled eluates loaded into Amicon buffer exchange columns (Millipore) to concentrate proteins. A total of 3 mL EB without glutathione was then added and centrifuged again. Solutions were stored at −80 °C until use. Purified GST–hTERT proteins attached to the beads were mixed with breast cancer PMC42 cell lysates (30–40 mg). GST–hTERT-binding proteins were eluted from GST–hTERT proteins in an open column with increasing concentrations of salt followed by a glycine-HCl (0.1 M, pH 2.8) wash. Eluted binding proteins were concentrated by lyophilization and resolved by SDS/PAGE and silver staining. Proteins were digested in the gel for mass spectrometry. The gene expression plasmid pGex4T–GAPDH was a gift from Yan Luo (Institute of Molecular and Cell Biology, Singapore) (6).
Mammalian Cell Culture, Senescence Analysis, and Gene Transfection.
The human breast cancer epithelial cell line MCF7 was cultured in DMEM (Invitrogen) supplemented with 4 mM l-glutamine, 3.7 mg/mL sodium bicarbonate, and 10% heat-inactivated FBS (vol/vol) (Gibco BRL) at 37 °C in 5% CO2 humidified incubator. β-Galactosidase staining was performed for cell senescence by incubating cells with 2 mL of staining solution (1 mg/mL X-gal, 40 mM citric acid/Na phosphate buffer, pH 6.0, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 150 mM MgCl2). The stained plates were wrapped with parafilm to protect against pH changes and incubated overnight at 37 °C. The cells were rinsed and stored in PBS and analyzed by microscopy (Nikon). Cells were transfected with various amounts of plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocols. Stable MCF7 cell lines were generated following transfection with specified pcDNA3–GFP–GAPDH plasmids (a kind gift from William E. Evans, St. Jude Children’s Research Hospital, Memphis, TN) (37). and subsequent selection with 1 mg/mL G418 (Sigma-Aldrich). Single amino acid mutations of GAPDH were conducted by site-directed mutagenesis of the pcDNA3–GFP–GAPDH gene expression constructs.
Telomerase Activity and Telomere Length Analyses.
Telomere repeat amplification protocol assays and telomere length analysis were performed as previously described (44) with minor modifications (45, 46).
Immunofluorescence Microscopy and Western Blotting Analysis.
Cells were seeded in chamber slides (Lab-Tek II; Nalgene Nunc International) at 60–80% confluence and transfected with gene expression plasmids. Analysis was conducted at indicated times, fixed with 4% paraformaldehyde in PBS for 15 min, permeabilized in 0.1% Triton X-100/PBS, blocked in 1% BSA. Cells were incubated with primary antibodies at 4 °C overnight and detected by Cy3-conjugated antimouse IgG secondary antibodies. Antibodies used were: mouse anti-GAPDH monoclonal antibody (clone 6C5; Ambion), rabbit GAPDH polyclonal antibody (sc-2035; Santa Cruz Biotechnology); β-actin and β-tubulin antibodies were from Chemicon. After washing with PBS–Tween, the membrane was incubated with a 1:20,000 dilution of peroxidase-conjugated goat antimouse IgG or porcine antirabbit IgG (DAKO) in PBS.
In Vitro Transcription and RNA Binding Assays.
Radiolabeled and unlabeled competitor RNA were produced using the MAXIScript T7 in vitro transcription kit (Ambion) according to manufacturer instructions. Before use in synthesis reactions, template DNA was excised from plasmids using restriction endonucleases and purified. For RNA binding assays 0.1 × 104–10 × 104 cpm RNA was incubated with 0.2–1.0 μg protein in the presence or absence of unlabeled competitor RNA for 20 min at 30 °C. Samples were exposed to 254 nm UV light for 10 min for UV cross-linking, followed by addition of 1 μL RNase mixture (Invitrogen) and incubation for 15 min at 37 °C. Samples were then subjected to SDS/PAGE on a 12% acrylamide TBE gel, dried between cellophane, and exposed to X-ray film overnight.
RNA Isolation and Gene Expression Analyses.
Total RNA was isolated using the High Pure RNA Isolation kit (Roche Diagnostics). cDNA was reverse transcribed from 2 μg of RNA, using the ThermoScript RT-PCR kit (Invitrogen). Real-time PCR was conducted with SYBR Green Mastermix (Invitrogen) on an ABI Prism 7900 HT system using primers: hTERT, CCATTTCATCAGCAAGTTTGGA and GCGACATCCCTGCGTTCTT and β-actin, TCCCTGGAGAAGAGCTACGA and AGGAAGGAAGGCTGGAAGAG.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (2012CB911201), the National Health and Medical Research Council of Australia, and the Cancer Council of Victoria. A.R.P. was the recipient of a Monash University postgraduate scholarship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206672109/-/DCSupplemental.
References
- 1.Browner WS, et al. The genetics of human longevity. Am J Med. 2004;117:851–860. doi: 10.1016/j.amjmed.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 2.Sung HJ, et al. Mitochondrial respiration protects against oxygen-associated DNA damage. Nat Commun. 2010;1:5. doi: 10.1038/ncomms1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sen N, et al. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hara MR, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 5.Chuang DM, Hough C, Senatorov VV. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2005;45:269–290. doi: 10.1146/annurev.pharmtox.45.120403.095902. [DOI] [PubMed] [Google Scholar]
- 6.Zheng L, Roeder RG, Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
- 7.Sawa A, Khan AA, Hester LD, Snyder SH. Glyceraldehyde-3-phosphate dehydrogenase: Nuclear translocation participates in neuronal and nonneuronal cell death. Proc Natl Acad Sci USA. 1997;94:11669–11674. doi: 10.1073/pnas.94.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundararaj KP, et al. Rapid shortening of telomere length in response to ceramide involves the inhibition of telomere binding activity of nuclear glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 2004;279:6152–6162. doi: 10.1074/jbc.M310549200. [DOI] [PubMed] [Google Scholar]
- 9.Demarse NA, et al. Direct binding of glyceraldehyde 3-phosphate dehydrogenase to telomeric DNA protects telomeres against chemotherapy-induced rapid degradation. J Mol Biol. 2009;394:789–803. doi: 10.1016/j.jmb.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 11.Shay JW. Toward identifying a cellular determinant of telomerase repression. J Natl Cancer Inst. 1999;91:4–6. doi: 10.1093/jnci/91.1.4. [DOI] [PubMed] [Google Scholar]
- 12.Reddel RR, Bryan TM. Alternative lengthening of telomeres: Dangerous road less travelled. Lancet. 2003;361:1840–1841. doi: 10.1016/S0140-6736(03)13538-6. [DOI] [PubMed] [Google Scholar]
- 13.Pinto AR, Li H, Nicholls C, Liu JP. Telomere protein complexes and interactions with telomerase in telomere maintenance. Front Biosci. 2011;16:187–207. doi: 10.2741/3683. [DOI] [PubMed] [Google Scholar]
- 14.Liu JP, et al. Regulation of telomerase activity by apparently opposing elements. Ageing Res Rev. 2010;9:245–256. doi: 10.1016/j.arr.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura TM, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 16.Meyerson M, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 17.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 18.Chen JL, Greider CW. An emerging consensus for telomerase RNA structure. Proc Natl Acad Sci USA. 2004;101:14683–14684. doi: 10.1073/pnas.0406204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 20.Sarin KY, et al. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruns GA, Gerald PS. Human glyceraldehyde-3-phosphate dehydrogenase in man-rodent somatic cell hybrids. Science. 1976;192:54–56. doi: 10.1126/science.176725. [DOI] [PubMed] [Google Scholar]
- 22.Sirover MA. New insights into an old protein: The functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta. 1999;1432:159–184. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 23.Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of GAPDH: Views from different subcellular compartments. Cell Signal. 2011;23:317–323. doi: 10.1016/j.cellsig.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholls C, Li H, Liu JP. GAPDH: A common enzyme with uncommon functions. Clin Exp Pharmacol Physiol. 2011 doi: 10.1111/j.1440-1681.2011.05599.x. [DOI] [PubMed] [Google Scholar]
- 25.Huang Q, et al. Akt2 kinase suppresses glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-mediated apoptosis in ovarian cancer cells via phosphorylating GAPDH at threonine 237 and decreasing its nuclear translocation. J Biol Chem. 2011;286:42211–42220. doi: 10.1074/jbc.M111.296905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonafé N, et al. Glyceraldehyde-3-phosphate dehydrogenase binds to the AU-Rich 3′ untranslated region of colony-stimulating factor-1 (CSF-1) messenger RNA in human ovarian cancer cells: Possible role in CSF-1 posttranscriptional regulation and tumor phenotype. Cancer Res. 2005;65:3762–3771. doi: 10.1158/0008-5472.CAN-04-3954. [DOI] [PubMed] [Google Scholar]
- 27.Krynetski EY, Krynetskaia NF, Bianchi ME, Evans WE. A nuclear protein complex containing high mobility group proteins B1 and B2, heat shock cognate protein 70, ERp60, and glyceraldehyde-3-phosphate dehydrogenase is involved in the cytotoxic response to DNA modified by incorporation of anticancer nucleoside analogues. Cancer Res. 2003;63:100–106. [PubMed] [Google Scholar]
- 28.Dastoor Z, Dreyer JL. Potential role of nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase in apoptosis and oxidative stress. J Cell Sci. 2001;114:1643–1653. doi: 10.1242/jcs.114.9.1643. [DOI] [PubMed] [Google Scholar]
- 29.Hara MR, et al. Neuroprotection by pharmacologic blockade of the GAPDH death cascade. Proc Natl Acad Sci USA. 2006;103:3887–3889. doi: 10.1073/pnas.0511321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kipling D, et al. Telomere-dependent senescence. Nat Biotechnol. 1999;17:313–314. doi: 10.1038/7827. [DOI] [PubMed] [Google Scholar]
- 31.Abdallah P, et al. A two-step model for senescence triggered by a single critically short telomere. Nat Cell Biol. 2009;11:988–993. doi: 10.1038/ncb1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harley CB. Telomerase and cancer therapeutics. Nat Rev Cancer. 2008;8:167–179. doi: 10.1038/nrc2275. [DOI] [PubMed] [Google Scholar]
- 33.Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: The senescence connection. Nat Rev Cancer. 2008;8:450–458. doi: 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagy E, Rigby WF. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold) J Biol Chem. 1995;270:2755–2763. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- 35.Liu ZJ, et al. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat Struct Biol. 1997;4:317–326. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- 36.Fermani S, et al. Crystal structure of the non-regulatory A(4 )isoform of spinach chloroplast glyceraldehyde-3-phosphate dehydrogenase complexed with NADP. J Mol Biol. 2001;314:527–542. doi: 10.1006/jmbi.2001.5172. [DOI] [PubMed] [Google Scholar]
- 37.Brown VM, et al. A novel CRM1-mediated nuclear export signal governs nuclear accumulation of glyceraldehyde-3-phosphate dehydrogenase following genotoxic stress. J Biol Chem. 2004;279:5984–5992. doi: 10.1074/jbc.M307071200. [DOI] [PubMed] [Google Scholar]
- 38.Kornberg MD, et al. GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol. 2010;12:1094–1100. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakravarti R, Aulak KS, Fox PL, Stuehr DJ. GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc Natl Acad Sci USA. 2010;107:18004–18009. doi: 10.1073/pnas.1008133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goodwin PM, Lewis PJ, Davies MI, Skidmore CJ, Shall S. The effect of gamma radiation and neocarzinostatin on NAD and ATP levels in mouse leukaemia cells. Biochim Biophys Acta. 1978;543:576–582. doi: 10.1016/0304-4165(78)90312-4. [DOI] [PubMed] [Google Scholar]
- 42.Shieh WM, et al. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 43.Nakamura J, et al. Quantitation of intracellular NAD(P)H can monitor an imbalance of DNA single strand break repair in base excision repair deficient cells in real time. Nucleic Acids Res. 2003;31:e104. doi: 10.1093/nar/gng105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim NW, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 45.Sumer H, et al. Chromosomal and telomeric reprogramming following ES-somatic cell fusion. Chromosoma. 2010;119:167–176. doi: 10.1007/s00412-009-0245-1. [DOI] [PubMed] [Google Scholar]
- 46.Cassar L, et al. Bone morphogenetic protein-7 inhibits telomerase activity, telomere maintenance, and cervical tumor growth. Cancer Res. 2008;68:9157–9166. doi: 10.1158/0008-5472.CAN-08-1323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.