Abstract
Collagen is the most abundant protein in animals and is a major component of the extracellular matrix in tissues such as skin and bone. A distinctive structural feature of all collagen types is a unique triple-helical structure formed by tandem repeats of the consensus sequence Xaa-Yaa-Gly, in which Xaa and Yaa frequently are proline and hydroxyproline, respectively. Hsp47/SERPINH1 is a procollagen-specific molecular chaperone that, unlike other chaperones, specifically recognizes the folded conformation of its client. Reduced functional levels of Hsp47 were reported in severe recessive forms of osteogenesis imperfecta, and homozygous knockout is lethal in mice. Here we present crystal structures of Hsp47 in its free form and in complex with homotrimeric synthetic collagen model peptides, each comprising one Hsp47-binding site represented by an arginine at the Yaa-position of a Xaa-Yaa-Gly triplet. Two of these three binding sites in the triple helix are occupied by Hsp47 molecules, which bind in a head-to-head fashion, thus making extensive contacts with the leading and trailing strands of the collagen triple helix. The important arginine residue within the Xaa-Arg-Gly triplet is recognized by a conserved aspartic acid. The structures explain the stabilization of the triple helix as well as the inhibition of collagen-bundle formation by Hsp47. In addition, we propose a pH-dependent substrate release mechanism based on a cluster of histidine residues.
Keywords: protein folding, protein–protein interactions, Serpins, protein stability
Collagen, the most abundant mammalian protein, is a major component of the extracellular matrix (ECM), which forms a specialized network that binds cells together, acts as reservoir for growth factors, and forms a framework through which differentiating cells move (1, 2). ECM defects can lead to abnormal development, e.g., Ehlers–Danlos syndrome or osteogenesis imperfecta (OI). So far, 28 different collagen types have been described. All are composed of three α-chains and comprise a unique tertiary structure in which three left-handed polyproline type II helices form a right-handed supercoil. This triple-helical structure is packed in such a way that, because of spatial constraints, a glycine residue is required in every third position, thus leading to repeating Xaa-Yaa-Gly triplets in the primary structure. The Xaa and Yaa positions frequently are occupied by proline and hydroxyproline, respectively. Collagen biosynthesis is a complex multistep process that includes intra- and extracellular events accompanied by numerous posttranslational modifications. One or more distinct procollagen α-chains are translocated into the endoplasmic reticulum (ER) where folding begins assisted by a number of chaperones or heat-shock proteins (HSPs) and accompanied by a multitude of posttranslational modifications. After initial trimerization the collagen triple helix folds in a zipper-like fashion from the C to the N terminus depending on previous proline hydroxylation. A large number of modifying enzymes such as proline-4-hydroxylase and chaperones such as Grp78/BiP and protein disulfide isomerase are involved in this process.
Hsp47, also called “SERPINH1,” is an ER-resident chaperone that is essential for the proper assembly of the triple-helical procollagen molecules, which eventually are transported across the Golgi apparatus to the extracellular space. This molecule, a 47-kDa, ER-resident collagen- or gelatin-binding protein, initially was identified independently in mice (as colligin) (3), in rats (as gp46) (4), and in chicken embryos, where it also was shown that its synthesis is increased strongly under heat-shock conditions (5). Unlike other heat shock proteins in general and other ER-resident proteins involved in collagen folding and maturation (e.g., protein disulfide isomerase or Grp78/BIP), Hsp47 does not possess other known client proteins. It is expressed in all collagen-synthesizing cells, and constitutive expression levels correlate strictly with the amounts of collagen being synthesized in the corresponding cell. Thus, elevated expression of Hsp47 is observed in a variety of fibrosis models (3, 5–7). Hsp47 binds to collagen of at least types I–V (8), recognizing an arginine at the Yaa position of an Xaa-Yaa-Gly triplet (9–14). Most interestingly, in striking contrast to other chaperones, Hsp47 preferentially recognizes the folded triple-helical conformation of its client (10–13, 15, 16).
Release of collagen from Hsp47 takes place in the cis–Golgi or ER–Golgi intermediate compartment and probably is accomplished by the lower pH in the Golgi compared with the ER (6.1 vs. 7.2), accompanied by a reversible pH-triggered conformational rearrangement of Hsp47 (14, 17). Free Hsp47 is transported rapidly back from Golgi to the ER by the KDEL receptors that recognize the RDEL motif at the C terminus of the chaperone.
To understand the molecular basis of the Hsp47–collagen interaction, we determined the crystal structures of Hsp47 alone and in complex with three different collagen model peptides (CMPs), Ac-PPGPPGPRGPPGPPG-NH2 (named “15-R8”), Ac-PPGPPGPTGPRGPPGPPG-NH2 (“18-T8R11”), and Ac-PPGPPGPPGPRGPPGPPG-NH2 (“18-R11”), each containing the important arginine at the Yaa position of a certain Xaa-Yaa-Gly triplet.
Results
Hsp47 Binds to Two Binding Sites Without Conformational Change.
We determined the crystal structures of Hsp47 alone and in complex with different CMPs as well as the structure of one CMP alone. The Hsp47 structures are refined at resolutions between 2.3 and 2.9 Å, yielding satisfactory statistics with values of R/Rfree of about 0.22/0.24 (Table S1). The Hsp47:CMP complexes crystallize in three different space groups. The three crystal forms contain multiple copies of Hsp47:CMP complexes in the asymmetric unit that are all virtually identical, thus making crystal-packing artifacts unlikely. Except when noted otherwise, the following discussion focuses on the description of the triclinic crystal form with the shorter 15-R8 CMP, because of its higher resolution.
Hsp47 exhibits the typical serpin fold (18, 19), consisting of three β-sheets (A, B, and C) and nine α-helices (Fig. S1). There is no significant change in the Hsp47 conformation upon collagen binding, as can be inferred by comparing the unbound (apo) crystal form with all three complex crystal structures (Fig. S1): The rmsd is only about 0.5 Å for 1,150 main-chain atoms. The intact reactive center loop (RCL, residues 361–379) is flexible, and the segment 368–375 is not visible in the electron density map. There is no partial insertion of the RCL into sheet A. Although it is located close to the collagen-binding site (see below), there are no visible interactions between this stretch and the bound CMPs.
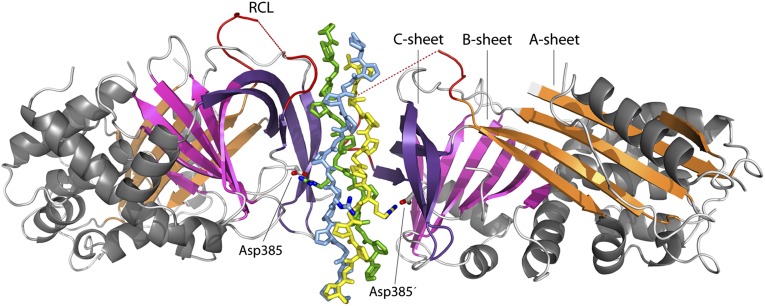
Two chaperones bind to one triple helix. The complex is formed by two Hsp47 molecules that attach in a head-to-head fashion via β-sheet C to two strands of one collagen trimer, i.e., the stoichiometry of (Hsp47)2:(CMP)3 (Fig. 1). The CMPs approximate the canonical 75-helical structure that usually is displayed by sequences rich in Pro-Pro/Hyp-Gly triplets (20) and are well defined in the electron density maps, except for the extreme termini, which are not in contact with the chaperone (Fig. S2). Comparison of the crystal structure of the 18-T8R11 CMP alone, crystallized from 3 M ammonium sulfate, and in the Hsp47 complex Hsp47:18-T8R11 does not reveal significant changes in the CMP conformation: An overlay of the main-chain atoms of the CMP structure alone and the four crystallographically independent copies in the Hsp47:18-T8R11 complex results in an rmsd of about 0.63 ± 0.25 Å. The triple helix exhibits the usual one-residue register shift (“stagger”) between the individual strands, thus defining a trailing, middle, and leading strand (21, 22). Here we adopt the original definition and that used by Bella (20), such that positions n in the leading strand, n+1 in the middle strand, and n+2 in the trailing strand are located at the same height along the axis of the triple helix.
Fig. 1.
Overall structure of the Hsp47:CMP complex. Hsp47 is shown in cartoon representation, and the CMP chains are depicted as sticks. α-Helices are drawn in gray. β-Sheet A is shown in orange, sheet B in magenta, and sheet C in violet. The segment corresponding to the RCL of inhibitory serpins is drawn in red. Asp385 is shown as sticks. The leading strand of the collagen triple helix is drawn in yellow, the middle strand in blue, and the trailing strand in green.
Hsp47 Docks via a Salt Bridge to Collagen and Recognizes all Three Strands.
In the leading and trailing strands, the important arginine at the position termed “Yaa0” in the Pro-Arg-Gly triplet (position 8 in the 15-R8 CMP and position 11 in the 18-R11 and 18-T8R11 CMPs) forms a salt bridge with the strictly conserved aspartate residue Asp385 of Hsp47 (Fig. 2). If either the leading or trailing strand or both bind to Hsp47, the corresponding position on the middle strand cannot be occupied because of steric restrictions. From inspection of our structure superimposed with Protein Data Bank entry 2DRT (23), 4(R)-hydroxyproline residues at the Yaa positions of the triple helix are compatible with binding to Hsp47, with the notable exception of the Yaa0 position of the trailing strand, where a clash occurs between the Oγ hydroxyl group and the aromatic ring of Tyr383. This finding is in agreement with previously published biochemical experiments (24).
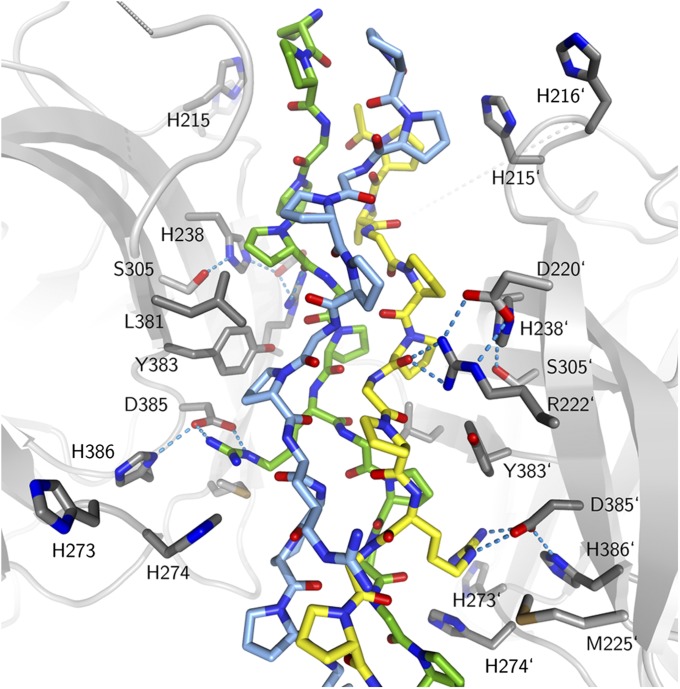
Fig. 2.
Close-up of the interaction surface. The CMP is color-coded as in Fig. 1. Key residues of Hsp47 with the CMP are numbered; prime numbers refer to the second Hsp47 molecule. Nitrogen atoms are shown in blue, oxygen atoms in red, and sulfur atoms in brown. Hydrogen bonds are indicated as cyan-colored dotted lines. The dotted grey segments indicate the disordered region of the RCL.
Each of the two Hsp47 molecules simultaneously forms numerous contacts to either the leading or trailing strand and to the middle strand of the collagen triple helix. Each Hsp47 also makes some contacts with the third strand. A total of 1,000 ± 150 Å2 of solvent-accessible surface is buried for each Hsp47 monomer/collagen triple helix upon complex formation. These contacts can be made only if collagen is in a proper folded, triple-helical conformation, thus explaining the preferred binding of the chaperone to the folded state of its client (10–13, 15).
In addition to the salt bridge between Asp385 and the arginine at Yaa0, the same collagen strand hydrogen bonds to the side-chain of Arg222 via its main-chain carbonyl oxygen atoms of Pro5 and Gly6, the former located at the so-called “Yaa−3” position (i.e., three amino acids before the important arginine residue). The side chain at this position influences the binding affinity, with threonine being the most favored naturally occurring amino acid (24). We can explain this effect from our structure of the Hsp47:18-T8R11 complex, in which the side chain of the threonine makes water-mediated contacts with the Ser305 side chain and the main chain of Ala303.
Furthermore, there are a number of hydrophobic interactions, especially involving Leu381 and Tyr383. We confirmed our crystallographic analysis by performing three different experiments using site-directed mutagenesis and pull-down experiments, solid-phase ELISA (Fig. 3), and surface plasmon resonance (SPR) spectroscopy (Fig. S3).
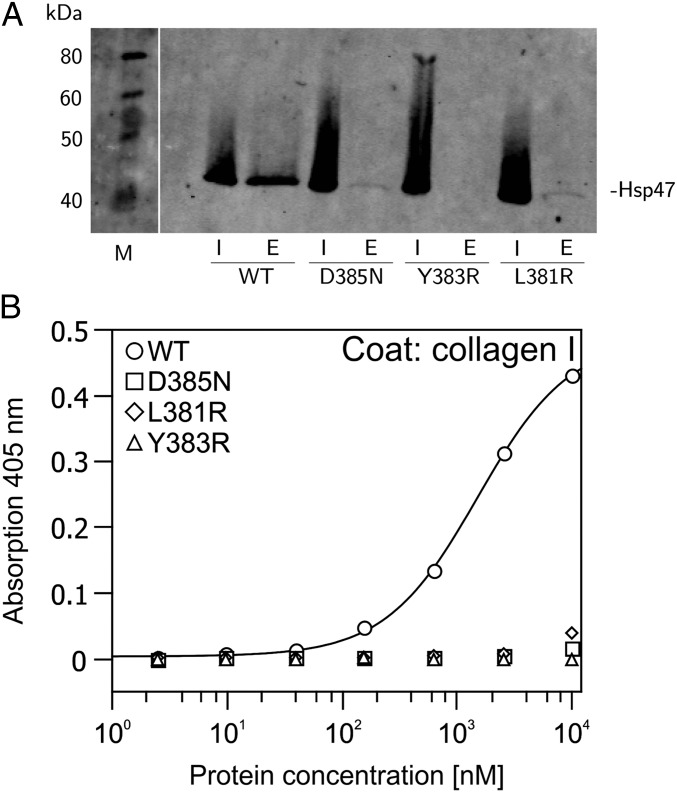
Fig. 3.
Binding of HSP47 mutants to collagen I. (A) Pull-down experiment on gelatin agarose. Hsp47 wild type and mutants were bound in equal amounts to gelatin agarose at pH 8. After extensive washing, bound proteins were eluted by shifting the pH to 5. E, eluate; I, total input. (B) ELISA binding assay. Collagen I (rat tail) was coated on 96-well plates and incubated with indicated concentrations of His-tagged HSP47 constructs. Binding was detected by a murine antibody directed against the His tag and visualized via an HRP-coupled secondary antibody. Every point was measured in triplicate.
Mutation of Asp385 to asparagine drastically weakens the interaction, as shown by pull-down experiments and ELISAs (Fig. 3), thus emphasizing the ionic nature of this interaction in agreement with previous studies on the optimal recognition sequence of collagen (9, 13, 24, 25). To verify further the importance of the identified interaction site, two additional mutations were introduced in the vicinity of Asp385, replacing Leu381 and Tyr383 by arginine. Although the wild type binds to collagen with a dissociation constant (Kd) of about 1 μM, as determined by ELISA and in agreement with IC50 values derived from competitive SPR data (8), all three mutants show drastically reduced binding (Fig. 3 and Fig. S3).
OI Missense Mutations Occur in the Hydrophobic Core Distant from the Binding Site.
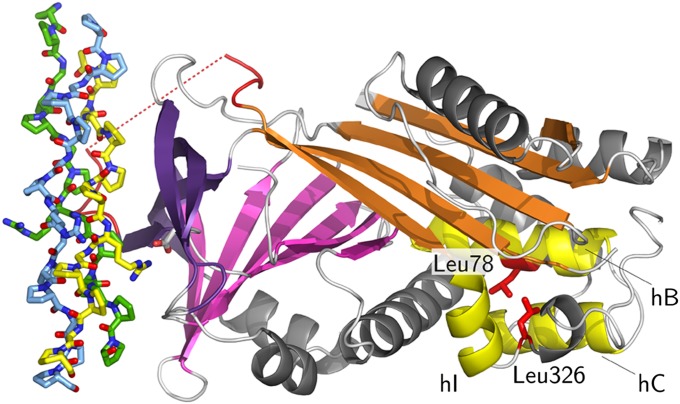
We also mapped the known missense mutation leading to recessive OI, i.e., L78P in humans and L326P in dogs (26, 27), on our experimental structures. As can be seen in Fig. 4, the mutations are located on the distant site of the collagen interaction surface. Leu78 is located in the middle of helix B, and Leu326 is located in a turn where it exhibits backbone dihedral angles of Φ ∼ −95° and Ψ ∼ +68°. The main-chain amide N-H groups of both residues donate hydrogen bonds to main-chain carbonyl oxygen atoms of preceding residues. Thus, replacement by proline, with its restricted Φ angle of about −60° and the absent polar hydrogen, most likely will lead to a severely altered structure. Indeed, Hsp47 levels were reduced drastically in an OI patient with the L78P mutation. We also found that attempts to express the L326P mutant in Escherichia coli resulted exclusively in insoluble protein, unlike the results with the wild-type protein.
Fig. 4.
Location of OI missense mutations. The mutations that lead to severe forms of recessive OI in Dachshunds (L326P) and a human patient (L78P) are located close together on the distant end of the collagen-binding site. The color coding is similar to Fig. 1, but mutated side chains are shown in red, and helices B, C, and I are drawn in yellow.
Discussion
Hsp47 Stabilizes the Collagen Triple Helix and Prevents Lateral Aggregation.
Our findings strongly support the notion that Hsp47 stabilizes the folded state of the collagen triple helix. Indeed, the CMPs used for crystallization are not triple-helical at room temperature at low or physiological ionic strength conditions, unlike their conformations observed in the crystal structures of the Hsp47:CMP structures (Fig. S4). Hsp47’s preferred binding to the folded state of collagen sheds light on a possible role in collagen folding and maturation. Both isolated collagen and procollagen are thermodynamically unstable at body temperature (28, 29). Collagen secreted by cells that either completely lack Hsp47 (30–32) or possess only reduced levels, as in OI patients (26), is protease sensitive in the triple-helical region, unlike collagen produced by normal cells. We propose that binding of Hsp47 prevents local unfolding (micro denaturation) of newly formed triple-helical regions at body temperature and misalignment or improper triple-helix formation during refolding. The preferred binding of Hsp47 to the folded state inevitably leads to a stabilization of the latter as compared with the unfolded state and probably would clamp the triple helix in the proper register.
Another hallmark of Hsp47-deficient cells is the formation of collagen aggregates in the ER (30, 31), although this type of aggregation was not observed in a particular OI patient (26). In part, this agglomeration can be explained by analogy with other proteins that are misfolded and associate via exposed hydrophobic patches. However, even correctly folded procollagen that has passed the ER quality control has an intrinsic propensity for forming lateral aggregates. In normal cells procollagen bundles are formed in the Golgi, where Hsp47 dissociates from its client (33, 34). Thus, Hsp47 also might prevent lateral aggregation of correctly folded procollagen in the ER. A similar effect is used in fibrillation assays, where Hsp47 inhibits collagen aggregation (14). Both effects are easily explainable by our structure, because the collagen triple helix is decorated with one or two chaperones per binding site. The long axes of these chaperones protrude outside each binding position, thereby inhibiting premature lateral multimerization of procollagen.
Chain Register and Number of Binding Sites.
The quaternary structure observed in our Hsp47:CMP complexes has implications for the number of chaperones bound to a given site and, in the case of heterotrimeric collagen molecules [e.g., collagen I with composition (α1)2(α2)1], depends on the register of the three strands. If the register leading:middle:trailing strand is α1:α2:α1, there will be two chaperones on a given binding site on the α1 chain, and, regardless of the local sequence of the α2 chain, no Hsp47 will be bound there. On the other hand, if the register is α1:α1:α2, as has been suggested recently (35), one or two Hsp47 molecules will be attached, depending on whether the local sequence of the α2 provides a Hsp47-binding site.
Does Hsp47 Serve as a Monitor of Quality?
The number of Hsp47-binding sites on any natural collagen has not yet been determined, but multiple sites have been identified in collagen chains α1(I), α2(I), and α1(II) by CNBr mapping (16). Interestingly, the highest-affinity sites were found in the N-terminal region of procollagen. Because triple-helix formation occurs in a zipper-like fashion proceeding from the C to the N terminus (36), binding of Hsp47 to folded segments at the N terminus could signal a successfully terminated folding event and mark this complex for passage to the Golgi. This proposal is in agreement with the finding that Hsp47−/− cells secrete collagen much more slowly and are deficient in N-propeptide processing (30).
Model for pH-Triggered Client Release.
The presumably pH-triggered release of collagen from Hsp47 in the Golgi prompted us to investigate the presence of groups with suitable pKa values located in or near the interface. An obvious candidate for pH-triggered release is Asp385, but the pKa value of an aspartic acid usually is about 4 and probably is even lower in the complex structure because of the salt bridge to the arginine at the Yaa0 position of collagen. Thus, we looked for other titratable groups within or near the binding interface with pKa values in the range of 6–7. Because the exact locations of binding sites on procollagen are not known, we considered only the Hsp47 partner. Six histidine residues are located in the vicinity of the collagen–Hsp47 interface: His215, His216, His238, His273, His274, and His386. From the pH of our crystallization media (pH = 8 for the complexes, 7 for the apo form) and because CMPs are bound, we deduce that our crystal structures faithfully represent the high-pH conformation that is competent for collagen binding. When pH is lowered, at least some of the histidines mentioned above should become protonated, and this protonation could cause client release. His273 is not strictly conserved and replaced by Asn or Tyr in chicken and zebrafish, respectively. His386, which most likely occurs in the protonated form even above pH 7, forms a salt bridge with the neighboring Asp385, positioning it for interaction with the essential arginine of the Pro-Arg-Gly triplet discussed above. His238 is the central part of a structure element that resembles the serine protease catalytic triad, involving Asp220 and Ser305. These residues form hydrogen bonds to the Nε1 and Nδ2 atoms of the imidazole ring of His238, respectively. Thus, His238 most likely occurs in the nonprotonated form at neutral pH. Asp220 positions Arg222 via a salt bridge for the interaction mentioned above with the carbonyl oxygen of the residue at the Yaa−3 position in the collagen peptide and therefore is important for high-affinity binding. Thus, protonation of His238 probably would lead to local rearrangements that affect binding affinity.
It is easily conceivable that protonation of all or some histidines, especially His238, and the resulting build-up of positive charges disrupt the binding interface crucially.
Materials and Methods
Expression and Purification.
A construct encoding amino acids 36–418 of the canine Hsp47 (canine SERPINH1 mRNA, NCBI accession NM_001165888), was cloned into the pET22-(b) vector (Novagen) with a C-terminal hexahistidine tag. Expression was induced in BL21(DE3) cells grown to an OD600 of 0.6–0.7 by adding 0.5 mM isopropyl-β-d-thio-galactoside and shaking for 5 h at 37 °C. Cells were resuspended in lysis buffer (300 mM NaCl, 10 mM imidazole, 20 mM Tris, pH 8) and lysed by sonication. Cleared lysate was purified by Ni-NTA affinity chromatography (Ni-NTA superflow; Qiagen). The eluate was reduced with 4 mM DTT, and 1.5 M ammonium sulfate was added to precipitate contaminants. The soluble fraction was concentrated and purified further by gel filtration (Superdex S75; GE Healthcare) in buffer A [20 mM Hepes (pH 7.5), 300 mM NaCl, 4 mM DTT]. The purified protein was concentrated to ∼15 mg/mL and stored at −80 °C for further use.
Crystallization and Structure Solution.
For cocrystallization, wild-type Hsp47 and either the 18-residues-long collagen peptide 18-R11 or 18-T8R11 or the shorter 15-residues-long peptide 15-R8 (Ac-PPGPPGPRGPPGPPG-NH2) (Biomatik) were mixed in a 1:1 molar ratio with respect to trimeric collagen at a concentration of about 0.2 mM. Crystals belonging to space group P1 (cocrystallization with 15-R8), P4122 (cocrystallization with 18-R11), and P2 (cocrystallization with 18-T8R11) were grown under the same crystallization conditions containing 8% (vol/vol) Tacsimate (Hampton Research) and 20% (wt/vol) PEG 3′350, pH 8, at 20 °C by vapor diffusion. Crystals of Hsp47 in the apo state were grown in 0.1 M Hepes (pH 7.5), 5% (vol/vol) Tacsimate (pH 7), 18% (wt/vol) PEG 3′350 in space group C2 at similar protein concentrations. Data from optimized crystals were collected at beamline X06DA, Swiss Light Source (Paul Scherer Institute, Villigen, Switzerland). For phasing, selenomethionine-labeled Hsp47 was produced by the pathway inhibition method (37), and data from cocrystals with 18-R11 in space group P4122 were collected at the Se K-edge peak. All data were processed using the program XDS (38). The selenium positions of this single-wavelength anomalous dispersion experiment were determined with SHELXD (39), and the initial model was built by SHELXE and phenix.autobuild (40). The P1, P2, and C2 structures were solved by molecular replacement using PHASER (41) with one Hsp47 monomer from the P4122 crystal form as search model. All structures were refined using iterative cycles of phenix.refine (42), COOT (43), and Buster (44).
Site-Directed Mutagenesis and Characterization of Mutants.
The mutations D385N, Y383R, and L381R were introduced according to an optimized mutagenesis protocol (45) using overlapping primers. The mutants were expressed and purified following the protocol used for the wild-type Hsp47. For the pull-down experiment, 100 μg Hsp47 in a volume of 200 μL (input) was added to 25 μL of gelatin-agarose (Sigma-Aldrich) in loading buffer [5 mM Tris (pH 8), 150 mM NaCl, 1 mM DTT] and incubated for 10 min at room temperature. The agarose beads were washed twice with 1 mL of washing buffer [5 mM Tris (pH 8), 400 mM NaCl, 1 mM DTT] and eluted in 80 μL of elution buffer [100 mM MES (pH 5.5), 150 mM NaCl, 1 mM DTT]. Samples were analyzed by SDS/PAGE and Western blot. Equal volumes from the input and elution fractions were loaded on the gel. For the Western blot, mouse anti-his5 (Qiagen) was used as primary antibody, and IRDye 800CW goat anti-mouse (LI-COR) was used as the secondary antibody. The green fluorescent signal of the secondary antibody was detected at 800 nm on an Odyssey infrared imaging system. The prestained molecular weight marker (Fermentas) was visualized at a wavelength of 700 nm.
For ELISA-style binding assays, rat tail collagen I (BD Bioscience) was diluted in TBS and coated at 10 μg/mL (500 ng per well) overnight at room temperature onto 96-well plates (MaxiSorp; Nunc). After washing with TBS/0.05% (vol/vol) Tween-20, plates were blocked for 1 h at room temperature with 5% (wt/vol) milk powder in TBS. His-tagged HSP47 proteins were added at concentrations ranging from 2.4 to 10,000 nM. After washing with TBS/0.05% (vol/vol) Tween-20, bound ligands were detected with a murine antibody directed against the His tag (Penta-His; Qiagen) and visualized with an alkaline phosphatase-coupled secondary antibody (GE Healthcare) and tetramethylbenzidine as substrate. The reaction was stopped by adding 10% (vol/vol) H2SO4, and absorption was measured at 450 nm. The data were analyzed using QTIPlot and fitted using a four-parameter logistic model (46). All measurements were done at least three times, and representative curves are shown.
Supplementary Material
Acknowledgments
We thank Sandro Waltersperger, Meitian Wang, and Vincent Olieric from beamline X06DA at Swiss Light Source, Paul Scherer Institute, for support. This work was supported by Grant 31003A-120174 from the Swiss National Science Foundation, by the Universities of Bern and Cologne, and by the German Research Council. Seed funding support was received from the Sonderforschungsbereich 829 “Molecular Mechanisms Regulating Skin Homeostasis.” The European Union Seventh Framework Programme European Light Source Activities (ELiSA) Grant Agreement 227616 provided travel support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4AU2, 4AU3, 4AU4, 4AWR, and 4AXY).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208072109/-/DCSupplemental.
References
- 1.Gelse K, Pöschl E, Aigner T. Collagens—structure, function, and biosynthesis. Adv Drug Deliv Rev. 2003;55:1531–1546. doi: 10.1016/j.addr.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Gordon MK, Hahn RA. Collagens. Cell Tissue Res. 2010;339:247–257. doi: 10.1007/s00441-009-0844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurkinen M, Taylor A, Garrels JI, Hogan BL. Cell surface-associated proteins which bind native type IV collagen or gelatin. J Biol Chem. 1984;259:5915–5922. [PubMed] [Google Scholar]
- 4.Cates GA, Nandan D, Brickenden AM, Sanwal BD. Differentiation defective mutants of skeletal myoblasts altered in a gelatin-binding glycoprotein. Biochem Cell Biol. 1987;65:767–775. doi: 10.1139/o87-100. [DOI] [PubMed] [Google Scholar]
- 5.Nagata K, Saga S, Yamada KM. A major collagen-binding protein of chick embryo fibroblasts is a novel heat shock protein. J Cell Biol. 1986;103:223–229. doi: 10.1083/jcb.103.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taguchi T, Razzaque MS. The collagen-specific molecular chaperone HSP47: Is there a role in fibrosis? Trends Mol Med. 2007;13:45–53. doi: 10.1016/j.molmed.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Takechi H, et al. Molecular cloning of a mouse 47-kDa heat-shock protein (HSP47), a collagen-binding stress protein, and its expression during the differentiation of F9 teratocarcinoma cells. Eur J Biochem. 1992;206:323–329. doi: 10.1111/j.1432-1033.1992.tb16930.x. [DOI] [PubMed] [Google Scholar]
- 8.Natsume T, Koide T, Yokota S, Hirayoshi K, Nagata K. Interactions between collagen-binding stress protein HSP47 and collagen. Analysis of kinetic parameters by surface plasmon resonance biosensor. J Biol Chem. 1994;269:31224–31228. [PubMed] [Google Scholar]
- 9.Koide T, Takahara Y, Asada S, Nagata K. Xaa-Arg-Gly triplets in the collagen triple helix are dominant binding sites for the molecular chaperone HSP47. J Biol Chem. 2002;277:6178–6182. doi: 10.1074/jbc.M106497200. [DOI] [PubMed] [Google Scholar]
- 10.Koide T, et al. Specific recognition of the collagen triple helix by chaperone HSP47: Minimal structural requirement and spatial molecular orientation. J Biol Chem. 2006;281:3432–3438. doi: 10.1074/jbc.M509707200. [DOI] [PubMed] [Google Scholar]
- 11.Ono T, Miyazaki T, Ishida Y, Uehata M, Nagata K. Direct in vitro and in vivo evidence for interaction between Hsp47 protein and collagen triple helix. J Biol Chem. 2012;287:6810–6818. doi: 10.1074/jbc.M111.280248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tasab M, Batten MR, Bulleid NJ. Hsp47: A molecular chaperone that interacts with and stabilizes correctly-folded procollagen. EMBO J. 2000;19:2204–2211. doi: 10.1093/emboj/19.10.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tasab M, Jenkinson L, Bulleid NJ. Sequence-specific recognition of collagen triple helices by the collagen-specific molecular chaperone HSP47. J Biol Chem. 2002;277:35007–35012. doi: 10.1074/jbc.M202782200. [DOI] [PubMed] [Google Scholar]
- 14.Thomson CA, Ananthanarayanan VS. Structure-function studies on hsp47: pH-dependent inhibition of collagen fibril formation in vitro. Biochem J. 2000;349:877–883. doi: 10.1042/bj3490877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koide T, Aso A, Yorihuzi T, Nagata K. Conformational requirements of collagenous peptides for recognition by the chaperone protein HSP47. J Biol Chem. 2000;275:27957–27963. doi: 10.1074/jbc.M003026200. [DOI] [PubMed] [Google Scholar]
- 16.Thomson CA, Tenni R, Ananthanarayanan VS. Mapping Hsp47 binding site(s) using CNBr peptides derived from type I and type II collagen. Protein Sci. 2003;12:1792–1800. doi: 10.1110/ps.0236903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saga S, Nagata K, Chen WT, Yamada KM. pH-dependent function, purification, and intracellular location of a major collagen-binding glycoprotein. J Cell Biol. 1987;105:517–527. doi: 10.1083/jcb.105.1.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Löbermann H, Lottspeich F, Bode W, Huber R. Interaction of human alpha 1-proteinase inhibitor with chymotrypsinogen A and crystallization of a proteolytically modified alpha 1-proteinase inhibitor. Hoppe Seylers Z Physiol Chem. 1982;363:1377–1388. doi: 10.1515/bchm2.1982.363.2.1377. [DOI] [PubMed] [Google Scholar]
- 19.Whisstock JC, et al. Serpins flex their muscle: II. Structural insights into target peptidase recognition, polymerization, and transport functions. J Biol Chem. 2010;285:24307–24312. doi: 10.1074/jbc.R110.141408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bella J. A new method for describing the helical conformation of collagen: Dependence of the triple helical twist on amino acid sequence. J Struct Biol. 2010;170:377–391. doi: 10.1016/j.jsb.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. Structural basis of collagen recognition by integrin alpha2beta1. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- 22.Hohenester E, Sasaki T, Giudici C, Farndale RW, Bächinger HP. Structural basis of sequence-specific collagen recognition by SPARC. Proc Natl Acad Sci USA. 2008;105:18273–18277. doi: 10.1073/pnas.0808452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuyama K, et al. Unique side chain conformation of a Leu residue in a triple-helical structure. Biopolymers. 2007;86:212–221. doi: 10.1002/bip.20724. [DOI] [PubMed] [Google Scholar]
- 24.Nishikawa Y, et al. A structure-activity relationship study elucidating the mechanism of sequence-specific collagen recognition by the chaperone HSP47. Bioorg Med Chem. 2010;18:3767–3775. doi: 10.1016/j.bmc.2010.04.054. [DOI] [PubMed] [Google Scholar]
- 25.Koide T, et al. Specific recognition of the collagen triple helix by chaperone HSP47. II. The HSP47-binding structural motif in collagens and related proteins. J Biol Chem. 2006;281:11177–11185. doi: 10.1074/jbc.M601369200. [DOI] [PubMed] [Google Scholar]
- 26.Christiansen HE, et al. Homozygosity for a missense mutation in SERPINH1, which encodes the collagen chaperone protein HSP47, results in severe recessive osteogenesis imperfecta. Am J Hum Genet. 2010;86:389–398. doi: 10.1016/j.ajhg.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drögemüller C, et al. A missense mutation in the SERPINH1 gene in Dachshunds with osteogenesis imperfecta. PLoS Genet. 2009;5:e1000579. doi: 10.1371/journal.pgen.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leikina E, Mertts MV, Kuznetsova N, Leikin S. Type I collagen is thermally unstable at body temperature. Proc Natl Acad Sci USA. 2002;99:1314–1318. doi: 10.1073/pnas.032307099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makareeva E, Leikin S. Procollagen triple helix assembly: An unconventional chaperone-assisted folding paradigm. PLoS ONE. 2007;2:e1029. doi: 10.1371/journal.pone.0001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishida Y, et al. Type I collagen in Hsp47-null cells is aggregated in endoplasmic reticulum and deficient in N-propeptide processing and fibrillogenesis. Mol Biol Cell. 2006;17:2346–2355. doi: 10.1091/mbc.E05-11-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuoka Y, et al. Insufficient folding of type IV collagen and formation of abnormal basement membrane-like structure in embryoid bodies derived from Hsp47-null embryonic stem cells. Mol Biol Cell. 2004;15:4467–4475. doi: 10.1091/mbc.E04-01-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagai N, et al. Embryonic lethality of molecular chaperone hsp47 knockout mice is associated with defects in collagen biosynthesis. J Cell Biol. 2000;150:1499–1506. doi: 10.1083/jcb.150.6.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonfanti L, et al. Procollagen traverses the Golgi stack without leaving the lumen of cisternae: Evidence for cisternal maturation. Cell. 1998;95:993–1003. doi: 10.1016/s0092-8674(00)81723-7. [DOI] [PubMed] [Google Scholar]
- 34.Trelstad RL, Hayashi K. Tendon collagen fibrillogenesis: intracellular subassemblies and cell surface changes associated with fibril growth. Dev Biol. 1979;71:228–242. doi: 10.1016/0012-1606(79)90166-0. [DOI] [PubMed] [Google Scholar]
- 35.Brondijk TH, Bihan D, Farndale RW, Huizinga EG. Implications for collagen I chain registry from the structure of the collagen von Willebrand factor A3 domain complex. Proc Natl Acad Sci USA. 2012;109:5253–5258. doi: 10.1073/pnas.1112388109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bächinger HP, Bruckner P, Timpl R, Prockop DJ, Engel J. Folding mechanism of the triple helix in type-III collagen and type-III pN-collagen. Role of disulfide bridges and peptide bond isomerization. Eur J Biochem. 1980;106:619–632. doi: 10.1111/j.1432-1033.1980.tb04610.x. [DOI] [PubMed] [Google Scholar]
- 37.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 40.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 45.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Findlay JW, Dillard RF. Appropriate calibration curve fitting in ligand binding assays. AAPS J. 2007;9:E260–E267. doi: 10.1208/aapsj0902029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.